I am deeply honored to receive the Allan Award from the American Society of Human Genetics and to join the ranks of the 38 illustrious awardees who have preceded me. The last time Arno Motulsky and I were both together on this podium, I was introducing him on his receipt of the Society’s Education Award. That was a great pleasure for me and quite appropriate, because he was the teacher and I was the student. Now, things are the other way around, and it is an even greater pleasure for the student to be introduced by his teacher.
I have been present at nearly all of the Allan Award addresses, the first one being given by Oliver Smithies as an after-dinner talk at the Society banquet in 1964. Oliver’s presentation was memorable for me because he gave us a little flute concert after concluding his talk. For a fleeting second I thought that I might emulate him and play my 'cello for you—but you will be happy to hear that I thought better of it.
There have been many other memorable talks over the years, and one that is still particularly vivid in my mind was given by Jerome Lejeune in 1969, ten years after the discovery of trisomy 21 (Lejeune 1970). The 1969 meeting, which was in San Francisco, was notable for more that Lejeune’s talk, since it had the added attraction of a San Francisco special—a gentle earthquake! More about all of this later.
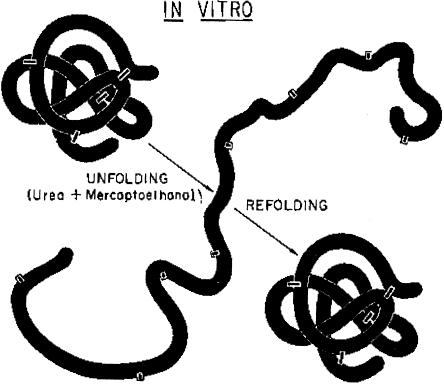
My first real exposure to human genetics came in conversations with Kurt Benirschke while I was in medical school. Kurt has been the legendary geneticist of the San Diego Zoo. It was from him that I learned about the sex life of nine-banded armadillos, which he allegedly kept in his basement, and heard about Tjio and Levan’s work on the human karyotype and about the early discoveries of the chromosomal basis of Down syndrome and other disorders. However, my turn toward genetics as a career came about in a somewhat unusual way. When I appeared for the first time in Christian Anfinsen’s laboratory in the National Heart Institute, I asked him what he wanted me to do. All that he said to me was, “Refold trypsin,” and he then promptly went on a trip to Israel. What he meant was that I was supposed to reduce the six disulfide bonds of trypsin with mercaptoethanol, denature it completely in urea, and then see whether I could get it to reacquire enzymatic activity and, by inference, its native structure (fig. 1). All of this was by way of adding further support to Anfinsen’s theory, for which he subsequently got the Nobel Prize, that the three-dimensional structure of a protein is determined by the linear sequence of amino acids in the polypeptide chain (Anfinsen 1973). While now part of the dogma of molecular biology, this was a very controversial issue at the time. Anfinsen’s challenge to me and the work that ensued from it were my first introduction to genetics in action, since what was really being studied was the genetic control of protein structure.
Figure 1.
The experimental denaturation (UNFOLDING) and renaturation (REFOLDING) of a disulfide bond–containing protein, to demonstrate that the three-dimensional structure of the protein is determined by its primary amino acid sequence. Reprinted, by permission, from Epstein et al. (1963). (© 1963 by Cold Spring Harbor Laboratory Press.)
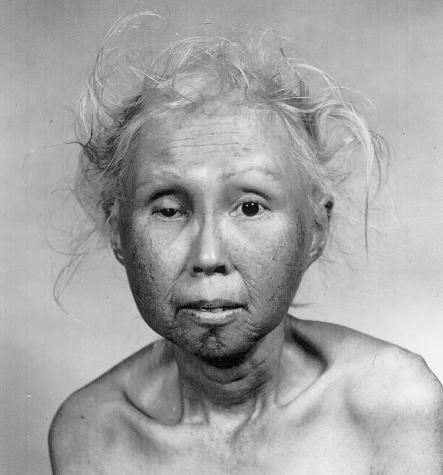
During the middle of my tenure at the NIH [the National Institutes of Health], I spent a year in Seattle as a fellow with Arno Motulsky. This was to be my only formal training in human and medical genetics. It was during this time that we began our work together—along with George Martin—on Werner syndrome. This work was stimulated by the woman in figure 2, who was 48 years old when the picture was taken. As hard as we tried to think about what the genetic defect might be (Epstein et al. 1966), it took another thirty years before it was identified, by the Seattle group, as a mutation in a helicase gene (Yu et al. 1996).
Figure 2.
Forty-eight-year-old woman with Werner syndrome, an autosomal recessive disorder caused by a helicase mutation that produces a caricature of aging. Reprinted, by permission, from Epstein et al. (1966). (© 1966 by Lippincott Williams & Wilkins.)
Arno’s mentorship has been instrumental in much that I have been able to accomplish since then. Both Chris Anfinsen and Arno Motulsky had a similar style of supervising their trainees—they led by example and by establishing expectations and standards of excellence. It was up to the trainees to figure out how to get things done.
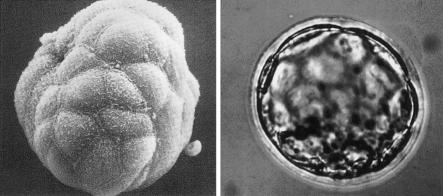
My time in Seattle convinced me that I did not want to be a protein chemist—even a genetic protein chemist—so, eventually, my wife, Lois, our two and a half children, and I headed west to San Francisco where I started down two parallel tracks. The first track involved a considerable gamble on my part. Without any prior experience or training, I switched to the mouse as an experimental system. To be more precise, I chose to work on the preimplantation mouse embryo (fig. 3). While still at the NIH, I had been very much taken by the work of many of the mouse developmental geneticists of the time—Salome Waelsch, Beatrice Mintz, Tibby Russell, and Dorothea Bennett were among the leaders. Of these, Beatrice Mintz was particularly influential in that she showed how both genetics and embryo manipulation could be used in combination to study various aspects of development. I was quite impressed by her ability to generate chimeric mice by the aggregation of early embryos, a technique that I was to use later in my own work (Epstein et al. 1982b) (fig. 4).
Figure 3.
Scanning and transmission micrographs of a preimplantation mouse embryo at the blastocysts stage. Reprinted, by permission, from Calarco and Epstein (1973).
Figure 4.
A trisomy 17 ↔ diploid chimera generated by preimplantation embryo aggregation, by the method developed by Beatrice Mintz (1972). Reprinted, by permission, from Epstein et al. (1982b). (© 1982 by the National Academy of Sciences, U.S.A.)
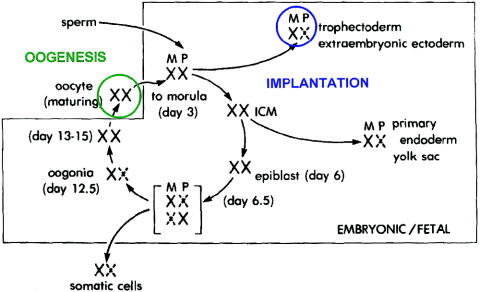
To get into the preimplantation mouse system, I followed the lead of Ralph Brinster, who had been able to measure enzyme activities in just one or a few preimplantation mouse embryos (Brinster 1965, 1966). Since I knew, from my earlier protein chemistry days, how to measure enzyme activities, this was a comfortable way to begin. The trick was to find really sensitive assays, and two that turned out to be particularly useful were for the X-linked enzymes: glucose-6-phosphate dehydrogenase and hypoxanthine-guanine phosphoribosyltransferase. Using these assays, we were able to show that embryonic X-inactivation starts at about the time of implantation, initially with the formation of the trophectoderm, and is reversed during oogenesis (fig. 5) (Epstein 1969, 1972, 1981; Epstein et al. 1978). The effect of the lack of inactivation during oogenesis is to make the eggs from XO females functionally aneuploid and haploinsufficient, which, I believe, accounts for their premature degeneration. This work on the X chromosome was my first foray into matters of gene dosage.
Figure 5.
The life cycle of the X chromosome, showing that both X chromosomes are active during oogenesis and early embryonic development, with X-inactivation first occurring at the time of implantation. Reprinted, by permission, from Epstein (1981). (© 1981 by Lippincott Williams & Wilkins.)
While all of this was going on, I was engaged in the second track of my academic life, as a clinical geneticist. In this capacity, I encountered the entire gamut of genetic disorders and birth defects and became involved in the establishment of a prenatal-diagnosis service and of several satellite genetic-counseling clinics. Of the many clinical disorders that I dealt with, the one that most captured my research attention was Down syndrome, and Edward Schneider, a postdoctoral fellow at the time, prepared matched pairs of trisomic and sibling fibroblasts for some early experiments (Schneider and Epstein 1972). With these cells in hand, we were in a position to begin to look at the effects of a change in gene dosage in Down syndrome when Chris Tan, Jay Tischfield, and Frank Ruddle mapped the first two genes to human chromosome 21 in 1973 (Tan et al. 1973). Although they had different names at the time, we now know these two genes to be SOD1, the gene for CuZn superoxide dismutase (SOD), and IFNAR1, the gene for the binding subunit of the interferon-alpha receptor. Because my wife, Lois, had by then become an internationally recognized expert on interferon and knew how to carry out the appropriate interferon-response assays, we chose to pursue the interferon-receptor gene first.
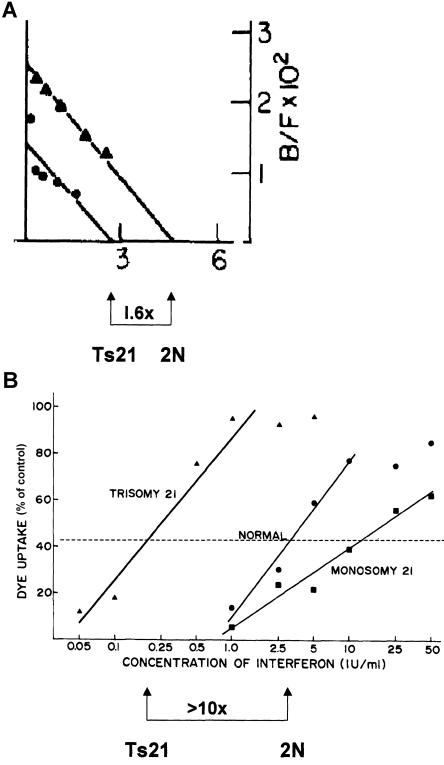
Our investigations revealed that trisomy 21 cells bind more interferon—just about 50% more, in fact (fig. 6) (Epstein et al. 1982a), as was expected—and, as a result, are more sensitive to it (Epstein and Epstein 1976; Epstein et al. 1980). In fact, the trisomic cells could be as much as 10 times more sensitive to interferon (Weil et al. 1980). This was taken to be a concrete demonstration of how a change in the dosage of a single gene of the degree occurring in trisomy 21 could have demonstrable functional consequences. (The actual situation may be somewhat more complicated because both the interferon-binding and signal-transducing subunits of the interferon-α receptor are encoded by contiguous loci on chromosome 21.) It was legitimate, therefore, to think in terms of the effects of the increased dosage of specific genes in producing the aneuploid phenotype.
Figure 6.
Interferon-α binding and antiviral sensitivity, in trisomy 21 cells. A, The binding of interferon-α to the interferon-α receptor in matched trisomy 21 and diploid fibroblasts. In the experiment shown, the trisomic cells bound 1.6 times as much interferon as do the diploid cells, which is very close to the theoretical value of 1.5 that is expected from the increase in copy number of the receptor gene from 2 to 3. Reprinted, by permission, from Epstein et al. (1982a). (© 1982 by Academic Press.) B, Increased sensitivity of trisomy 21 fibroblasts to the antiviral effect of interferon-α. In the experiment shown, sensitivity was increased by greater than 10-fold, but in several experiments the median increase was close to 6-fold. Reprinted, by permission, from Weil et al. (1980). (© 1980 by Mary Ann Liebert, Inc.)
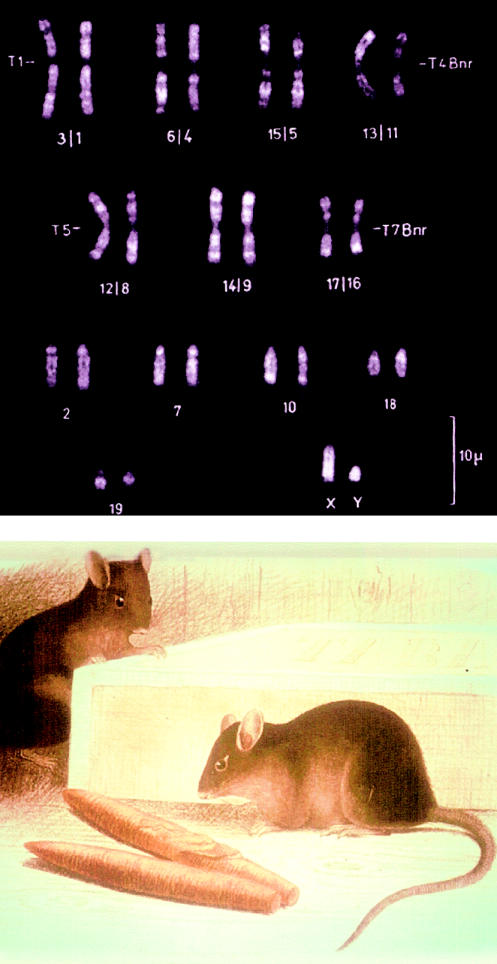
At this point, the two lines of research—mouse genetics and human trisomy 21—converged, with the bridge between the two being provided by the work of Alfred Gropp, a German pathologist interested in hunting and cytogenetics. These interests brought him to an isolated valley in the Italian Alps in which lives Mus poschiavinus, the so-called “tobacco mouse,” which has only 26 chromosomes, rather than the usual 40, with seven pairs of Robertsonian fusion chromosomes (fig. 7). Gropp transferred many of these Robertsonians to regular laboratory mice and then devised a method for generating each of the mouse trisomies at a relatively high frequency (Gropp et al. 1975). This made it possible to consider, for the first time, the development of a mouse model of human trisomy 21.
Figure 7.
Fluorescent-banded karyotype (top) and somewhat fanciful picture of M. poschiavinus, the tobacco mouse (bottom). These pictures were obtained from the late Prof. Alfred Gropp, of Lübeck, Germany.
There were several reasons for wanting to have such a model. For both practical and ethical reasons, we were restricted in our ability to study humans with Down syndrome. Only a few types of cells could be looked at, and there was no real access to the CNS. By contrast, all cells and tissues—including, in particular, the brain—could be studied in the mouse, and we would be able to exercise tight genetic control to reduce the effects of a variable background. And, although we did not know it at the time, we would eventually have the ability to manipulate the mouse genome almost at will. Finally, and perhaps of greatest importance, an appropriate model would permit potential therapeutic approaches based on a detailed knowledge of relationship between genotype and phenotype to be rationally designed and tested.
We were ready to start on the development of a model, but there were two questions that had to be answered. First, given the number of chromosomal rearrangements that have occurred since the evolutionary divergence of humans and mice, would enough human chromosome 21 genes be syntenic in the mouse to make any single mouse trisomy a useful model? And, if so, which mouse chromosome most closely resembles human chromosome 21?
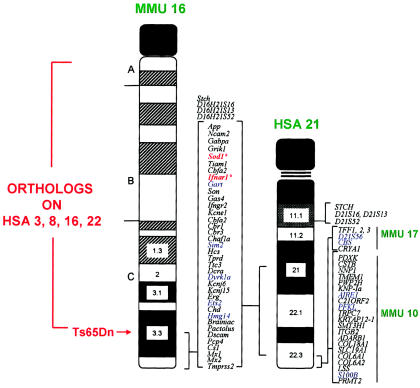
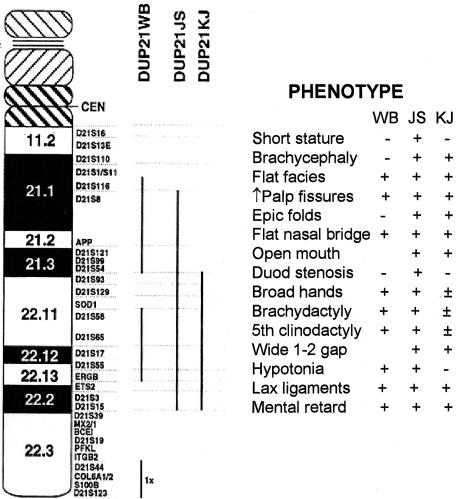
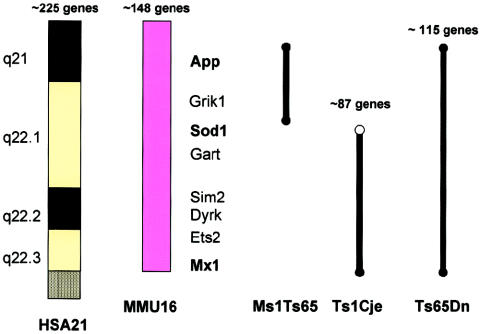
The answers to both questions required the mapping of known human chromosome 21 genes into the mouse, and, when David Cox (then a fellow in my laboratory), Lois Epstein, and I began this work, we still knew of only two genes—the same SOD and interferon-receptor genes I mentioned earlier. However, good fortune smiled on our efforts, and it turned out that these two genes are both on mouse chromosome 16 (Cox et al. 1980). With time, it has been shown that all of the genes on the long arm of human chromosome 21 down to the Mx locus in mid-21q22.3 are syntenic on mouse chromosome 16 (fig. 8). From examining the complete sequence of human chromosome 21, we now know that this corresponds to more than half of the ∼225 known and inferred genes on this chromosome (Roger Reeves, personal communication). The remaining human chromosome 21 genes are present on mouse chromosomes 10 and 17. However, from the work of Julie Korenberg, who had been analyzing the relationships between phenotype and genotype in the rare persons with segmental trisomy 21, we learned that many aspects of the Down syndrome phenotype—including dysmorphic changes in the craniofacies, hands, and feet; duodenal stenosis; hypotonia; lax ligaments; and mental retardation—can also occur with segmental trisomy—as in patients JS and KJ, in fig. 9—for just that region of human chromosome 21 that is in common with mouse chromosome 16.
Figure 8.
Syntenic region in common between human chromosome 21 (HSA 21) and mouse chromosome 16 (MMU 16) that forms the basis for the development of the trisomy 16 mouse models of Down syndrome. The other mouse chromosome regions orthologous to human chromosome 21 and other human chromosome regions orthologous to mouse chromosome 16 are shown, as is the location of the Ts65Dn breakpoint above App. The first two human chromosome 21 loci mapped to mouse chromosome 16 by Cox et al. (1980)—SOD1 and IFNAR—are marked with by asterisks (*). This figure is a modification of comparative maps kindly provided by Roger Reeves and published in Richtsmeier et al. (2000). Reprinted by permission. (© 2000 by Wiley-Liss, Inc., a subsidiary of John Wiley & Sons, Inc.)
Figure 9.
Components of Down syndrome phenotype present in persons with segmental trisomy 21. The regions of trisomy in JS and KJ, indicated by the vertical bars, are quite similar to that present in the Ts65Dn mouse. Modified and reprinted, by permission, from Korenberg et al. (1994). (© 1994 by the National Academy of Sciences, U.S.A.)
All of this suggested that mouse trisomy 16 would be a valid genetic model for human trisomy 21, and the phenotype appeared to confirm this—congenital heart disease with an atrioventricular canal and conotruncal anomalies, thymic hypoplasia, delayed maturation of the immune system, craniofacial abnormalities, and midgestational edema (fig. 10) (Epstein et al. 1985; Epstein 1986; Berger and Epstein 1989). If anything, the defects in the trisomic mouse were even more severe than in Down syndrome, and it was eventually realized that the trisomy 16 mouse was too good to be true—not because the human chromosome 21 genes were not there, but because too many other genes were! The principal problem is that mouse chromosome 16, in addition to having a large proportion of the human chromosome 21q genes, also has appreciable amounts of sequence homologous to regions of four other human chromosomes in its proximal 80% (fig. 8). Therefore, while still quite useful for studying various aspects of the effects of aneuploidy, complete mouse trisomy 16 was not the best model of human trisomy 21. What would be better would be a mouse with segmental trisomy 16—trisomic for just that region of mouse chromosome 16 that is homologous to human chromosome 21—and this is just what Muriel Davisson, at the Jackson Laboratory, was able to produce.
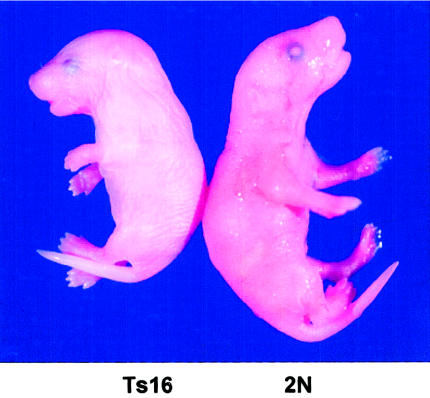
Figure 10.
Mouse fetus with trisomy 16 (Ts16; left) and diploid littermate (2N; right). The small size, short snout, open eye lids, and thick neck (the residual of midgestational edema) caused by trisomy 16 are visible. Picture provided by David Holtzman (Washington University, St. Louis).
The new mouse, designated “Ts65Dn” (“Ts65” for short), was the result of a radiation-induced translocation between parts of chromosomes 16 and 17 that gave rise to a tertiary trisomy (Reeves et al. 1995; Akeson et al. 2001). This mouse is viable but tends to be small. Its immune system is in reasonably good shape, and there are no gross congenital malformations—in particular, no heart disease. As in human trisomy 21, the males are sterile, but it is possible to breed Ts65 mice, with some difficulty, from trisomic females, although not on an inbred background.
How good is Ts65 as a model for Down syndrome? Quite good, I would say, and, in at least two areas, the CNS and the craniofacial skeleton, the Ts65 mouse exhibits very interesting and highly relevant abnormalities. Among the several abnormalities of the nervous system that have been found, we have been examining, in a longtime collaboration with William Mobley (formerly of UCSF [University of California–San Francisco] and now at Stanford), the status of the basal forebrain cholinergic neurons in the medial septal nucleus. These neurons are of particular interest because of their vulnerability in Alzheimer disease, a late consequence of trisomy 21. Mobley and his colleagues have shown that, in the trisomic brains, there is a decrease, with time, in the relative number and size of functional cholinergic neurons (Holtzman et al. 1993, 1996; Cooper et al. 2001). Nerve growth factor (NGF) is a trophic factor made in the hippocampus that is transported by retrograde axonal transport to the cell bodies in the basal forebrain. When the rate of this retrograde transport was determined, it was found to be significantly decreased in the trisomic brain (Cooper et al. 2001). This finding suggested an experiment with obvious therapeutic implications. If the loss of cholinergic neurons is indeed the result of a deficiency of NGF, what effect would treatment with NGF have? This experiment was carried out by introducing NGF into the ventricles of the Ts65 mice through a cannula attached to an osmotic minipump, and the effect was quite gratifying (Cooper et al. 2001). The atrophy of the cholinergic neurons was partially reversed, as is indicated by the arrow in figure 11, and the number of neurons actually increased, indicating that, although they had disappeared functionally, they had not really disappeared anatomically. Dr. Mobley and his group are continuing these studies.
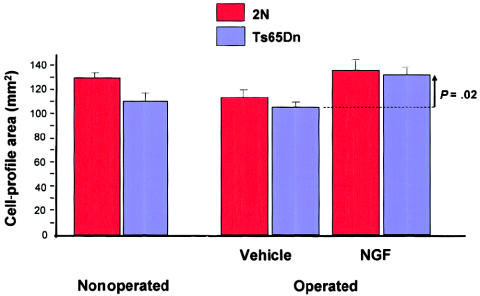
Figure 11.
Restoration of size of basal-forebrain cholinergic neurons by infusion of nerve growth factor in aged Ts65n mice. The decrease in cell-profile area in the nonoperated Ts65Dn mice is significant at P=.009, and the increase in cell-profile area in treated Ts65Dn animals (vertical arrow) is significant at P=.02. Modified and reprinted, by permission, from Cooper et al. (2001). (© 2001 by the National Academy of Sciences, U.S.A.)
In our studies of the interferon-response system that I described earlier, it was possible to demonstrate specific functional consequences of the increased dosage of just a single gene. Furthermore, an analysis of the specificity and reproducibility of the patterns of abnormalities in the various trisomies and monosomies, even in the face of individual variability, has convinced me that the aneuploid phenotypes that we see are truly the result of the particular genes that are genetically unbalanced when a duplication or deletion is present and are not the result of some kind of random noise or general loosening up of developmental homeostasis (Epstein 1986). Therefore, although a phenotype is undoubtedly produced by the interaction of the effects—some large, some small—of many of the unbalanced genes, it is valid to investigate the contributions of individual loci to the phenotype. This conclusion suggested a second approach to the modeling of Down syndrome, and that was by investigating the imbalance of genes individually by the generation of transgenic mice.
When Yoram Groner and his colleagues at the Weizmann Institute in Israel reported the cloning of the first human chromosome 21 gene—our old friend, SOD1—it became possible to do just that, and we collaborated to produce the first series of mice transgenic for CuZn SOD (Epstein et al. 1987). Groner used these mice to look for abnormalities that might resemble those found in persons with Down syndrome, and he found several—abnormalities of the myoneural junctions in the tongue, an impairment in the uptake of serotonin by platelets, and a decreased synthesis of prostaglandins D2 and E2 (Groner 1995). We, on the other hand, set out to test Pierre-Marie Sinet’s hypothesis that an increase in CuZn SOD activity might have deleterious effects on the nervous system because of an enhancement of the production of hydrogen peroxide from the superoxide that is generated within cells, the conversion of O2− to H2O2 being the function of the enzyme (Sinet 1982). This proved not to be the case, at least in the mouse brain, but these studies led us in a direction entirely different from Down syndrome. Having these transgenic animals in hand, we became quite interested in the role of the SODs themselves—I use the plural because there are actually three different dismutases in mammals (CuZn SOD, Mn SOD, and EC SOD)—in longevity and in protecting against various forms of oxidative stress, and what we found is that, contrary to what was observed in fruit flies and proclaimed in the headlines a few years back (Kotulak and Gorner 1992), an increase in CuZn SOD activity by three- to fivefold did not prolong life span in transgenic mice (fig. 12) (Huang et al. 2000). However, studies carried out in collaboration with many different groups have indicated that, with just one exception (Fullerton et al. 1998), increased CuZn SOD activity does protect, in a number of different experimental paradigms, against acute oxidative stress induced by a wide variety of physical and chemical agents (Huang et al. 1999).
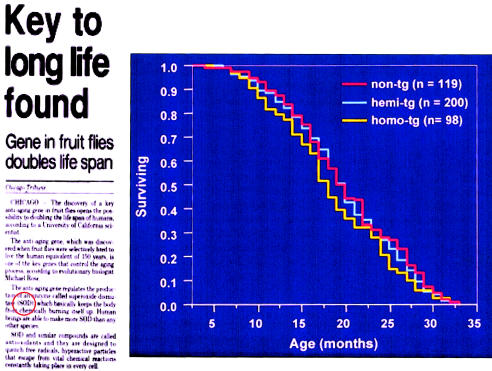
Figure 12.
Association between CuZn SOD activity and life span. Although an association between elevated CuZn SOD activity and prolongation of life in Drosophila has been observed, transgenic mice with threefold (hemi-Tg) and fivefold (homo-Tg) increased CuZn SOD activity do not have an increased life span or an increased mean survival. Reprinted, by permission, from the Chicago Tribune and Huang et al. (2000). (© 2000 by the Gerontological Society of America.)
To further this line of investigation, we decided to knock out Sod1, the gene for CuZn SOD. It should have been easy—and the technical aspects of the homologous recombination were—but when it came to generating a Sod1 null mouse from our knockout heterozygotes, we just could not seem to do it. Something was very wrong, but, fortunately, Haru Sago, a visiting fellow from Japan, found out what it was. In the course of generating the knockout, my colleague Ting-Ting Huang had inadvertently produced a reciprocal translocation between the end of chromosome 12 and the distal part of chromosome 16 just proximal to the knocked-out Sod1 gene (Sago et al. 1998). This can be seen in the left panel of figure 13 in which the Gart gene, which is distal to Sod1, has moved—as the yellow arrow indicates—from end of chromosome 16 to the end of chromosome 12, whereas the more proximal App gene (fig. 13, middle panel), is still on chromosome 16, where it belongs. This was bad news for the SOD research—although Dr. Huang subsequently did produce a usable Sod1 knockout—but it was great news for the Down syndrome–model work. We now had available to us the means to produce a mouse with a second type of segmental trisomy 16, shown in the panel on the right. This animal, which has been designated as Ts1Cje, has three copies of the Sod1 gene, one of which has been knocked out, as well as of all of the genes distal. It is, therefore, functionally trisomic for the segment of mouse chromosome 16 distal to Sod1.
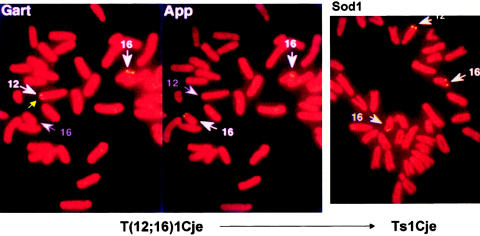
Figure 13.
Reciprocal translocation and the rise of Ts1Cje. The identification of the T1Cje translocation by FISH. The left and middle panels identify the locations of Gart and App, respectively. Whereas the two App signals are located on chromosome 16, one of the more distal Gart signals is located on chromosome 12, indicating the presence of a reciprocal 12;16 translocation with a breakpoint between the two markers. In the trisomic Ts65Dn progeny of the balanced translocation carriers (right panel), there are three Sod1 signals, two on chromosome 16 and one on chromosome 12. The gene represented by the Sod1 signal on chromosome 12, which is at or just distal to the breakpoint, has been knocked out by the same gene-targeting event that apparently caused the translocation. The figures, prepared as described by Sago et al. (1998), were provided by Dr. Haru Sago.
The reason that the serendipitous generation of this mouse was so exciting is that it put us in the position of at last being able to do what we really wanted to do, and that was to use a subtractive approach to correlate genes with effects. In this approach, we start with the phenotype exhibited by the Ts65Dn mouse. We then look at what happens to the phenotype when the degree of trisomy is reduced, in this case from about 115 genes in Ts65Dn to about 81 genes in Ts1Cje (fig. 14). I say “about” because Roger Reeves (personal communication) has assured me that the numbers of genes change daily, as annotation of the human and mouse genomes proceeds. I favor the subtractive approach because it begins with a defined phenotype and then looks at how the phenotype changes when a particular gene or group of genes is removed. This is in contrast to the transgenic approach, which looks at the appearance of an abnormal phenotype as extra genes are added. Because of the difficulties in controlling the location, copy number, and regulation of transgenes, the findings are often difficult to interpret.
Figure 14.
The three currently existing segmental trisomies for mouse chromosome 16. The numbers of genes in each trisomic segment represent approximations, subject to change, that were provided by Dr. Roger Reeves. Reprinted, by permission, from Epstein (2001). (© 2001 by the McGraw-Hill Companies.)
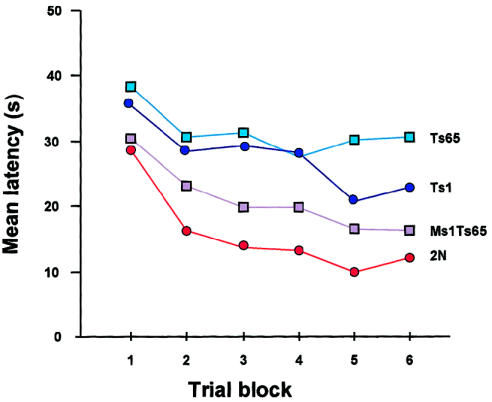
We have used the subtractive approach in two experimental systems so far. The first is with a test of spatial learning carried out in the Morris water maze, a small swimming pool in which a mouse seeks to locate a platform hidden under the water. Several groups have studied Ts65 mice by this procedure, and the unanimous conclusion has been that these animals are slow to learn where the platform is located (Reeves et al. 1995; Coussons-Read and Crnic 1996; Sago et al. 1998, 2000). This is regarded as a defect in the function of the hippocampus. Further, having finally learned where the platform is, the trisomic mice then have great difficulty in learning its new location when it is moved unbeknownst to them to a new place. Rather than improve their performance in repeated trials, as do diploid mice, the Ts65 mice tend to perseverate and take nearly as long to find the relocated platform in the last set of trials as they do in the first. This is considered to involve an impairment of function of the hippocampus and prefrontal cortex. Using a somewhat complicated breeding scheme, Elaine Carlson and Haru Sago produced Ts65; Ts1; Ms1Ts65, the segmental trisomy for the region of difference between the [aforementioned] two; and euploid animals on the same, albeit somewhat heterogeneous, genetic background (fig. 14) (Sago et al. 2000). The results were quite clear: the performance of Ts1Cje mice in the maze was almost as compromised as that of Ts65Dn, with the exception that the Ts1Cje animals did a bit better in learning the location of platform after it had been moved (fig. 15). By contrast, the Ms1Ts65 animals, while certainly not normal, performed much better than did either of the two other segmental trisomics. Therefore, it can be inferred that the impaired performance in the water maze is largely, but not entirely, the result of the imbalance of genes distal to Sod1.
Figure 15.
Latency (time to reach platform) in reverse hidden-platform test in the Morris water maze. The best performance is by 2N animals, and the poorest performance is by Ts65Dn. The performance by Ts1Cje is somewhat better than that by Ts65Dn, whereas that by Ts1Cje is better than that by the other two segmental trisomics but still not as good as that by the 2N mice. All mice were generated from the same Ts65Dn × T1Cje cross. Modified and reprinted, by permission, from Sago et al. (2000). (© 2000 by the International Pediatric Research Foundation, Inc.)
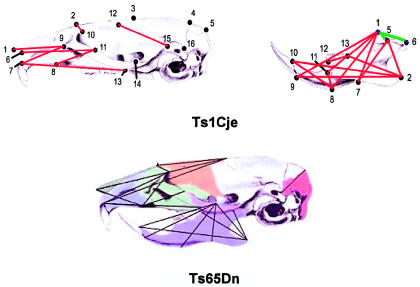
The second system in which Ts65 and Ts1 have been compared is in the development of the skull and craniofacial skeleton, a process that is altered in a quite consistent manner in trisomy 21 to produce the characteristic appearance of Down syndrome. These studies were done in collaboration with Joan Richtsmeier, at Penn State, and Roger Reeves, at Johns Hopkins, who had already set out to determine whether the morphogenic abnormalities in the Ts65 mouse modeled those in humans with Down syndrome. Although mouse and human skulls look quite different from one another, they do share the same overall bony architecture and have the same landmarks. This makes it possible to compare statistically the quantitative differences and similarities between trisomic and nontrisomic skulls in humans and mice. Thus, in the skull of a human with trisomy 21, there is an overall reduction in the size of the skull, a flattened occiput, brachycephaly, a small midface, reduced maxilla and mandible, and a reduced interorbital distance. What Richtsmeier et al. (2000) found is that Ts65 mice also have smaller skulls overall, with a disproportionately reduced midface and anterior neurocranium and a broader calvarium (fig. 16). Thus, as remarkable as it seems, the changes, from normal, in the shape of the skull that occur in the trisomic mouse directly parallel those seen in the skulls of persons with Down syndrome. The similarity of the effects resulting from imbalance affecting orthologous regions of the human and mouse genomes suggests that the genetic pathways determining the morphogenesis of the skull have been evolutionarily conserved even as these two species diverged. I take great comfort from this!
Figure 16.
Comparison between skulls of Ts1Cje (top) and Ts65Dn (bottom) mice, demonstrating that there is a similar shortening of distances between landmarks (numerals) relative to diploid controls in the two segmental trisomics. Modified and reprinted, by permission, from Reeves et al. (2001) (© 2001 by Elsevier Science) and Richtsmeier et al. (in press) (© 2001 by Wiley-Liss, Inc., a subsidiary of John Wiley & Sons, Inc.).
When Joan Richtsmeier went on to look at Ts1Cje mice, she found that for more than 80% of these measurements, the Ts1Cje’s differ (fig. 16, top, in red) or resemble (in blue) their euploid littermates in ways very similar to what had been found to define the differences and similarities between Ts65Dn mice and their littermates (fig. 16, bottom) (Richtsmeier et al., in press). Although there are differences between Ts65Dn and Ts1Cje, they are in the minority. Overall, what is altered in the one is altered in the other. It would appear, therefore, that just as with the impairment in learning in the Morris water maze, the craniofacial dysmorphology is largely, but not entirely, the result of imbalance of the genes distal to Sod1 on mouse chromosome 16.
Having narrowed down the region of major effects on behavior in the water maze and formation of the skull by about a quarter, where do we go from here? Our present strategy is to extend the subtractive approach by making a series of nested deletions in chromosome 16 using the cre-lox approach and then to assess their effects on defined components of the trisomic phenotype. This should ultimately permit us to determine, by what will undoubtedly be an iterative process, which genes are doing what.
Insofar as this is all directed to trying to understand the genesis of Down syndrome, perhaps the greatest problem that we will confront is that we cannot think about cognition in a mouse in the same manner that we think about cognition in humans, and we certainly cannot speak about speech and language. Nevertheless, the ways in which the brain works—the basic neural processes that govern its ability to function and the genes that control this—are very similar among the mammals. In fact, conservation of function even between quite disparate species is more the norm than the exception, and neuroscientists have been willing to study these processes in organisms as lowly as Aplysia, the sea slug. Therefore, I believe that there is every reason to expect that the perturbations of form and function found in the trisomic mice are relevant to trisomic humans. [Following delivery of the Allan Award address, Eric Kandel’s Nobel Prize essay was published {Kandel 2001}. In this essay, Kandel makes several comments that are supportive of my point of view. Of note is his assertion, characterized as a leap of faith, that elementary forms of learning are common to all animals with an evolved nervous system. This belief has been bolstered by his findings that different forms of learning present in a simple invertebrate are paralleled by corresponding forms of learning in higher vertebrates and humans and that there is conservation of the relevant biochemical mechanisms. Therefore, if it is possible to consider an invertebrate to be a legitimate model for learning and memory in humans, it certainly requires a much smaller leap of faith to place the mouse in this role.]
In a curious way, our problem at this time is more on the human than on the mouse side. Despite [a] the length of time that Down syndrome has been known as an entity and studied and [b] the elegance of the methods for studying the nervous system that have been developed, we actually know very little about what is abnormal about the brains and nervous systems of people with Down syndrome—about whether, for example, there are abnormalities of the retrograde transport of NGF or of long-term potentiation and depression or even of pain perception—all of which are abnormal in the mouse (Cao et al. 1999; Martinez-Cue et al. 1999; Siarey et al. 1999; Cooper et al. 2001). This is not really surprising, because our hands seem to be very much tied when it comes to studying what is happening within the black box of the human cranium. Perhaps one benefit of working with the mouse will be to point our attention to relevant problems that need to be studied in humans and force us to develop methods for doing so.
I now want to return to the Allan Award lecture given by Jerome Lejeune in 1969 (Lejeune 1970). Since I was the local organizer for that meeting, I was charged with taking Lejeune to dinner. What should have been an enjoyable experience for me turned out to be less than pleasurable. I had just started the prenatal-diagnosis program at UCSF and thought that it would be a boon to genetic counseling. Lejeune, believing that human life begins at conception and strenuously opposed to abortion, thought otherwise, and he forcefully articulated his position both at dinner and the next day in his Allan Award address that turned out to be the second earthquake of the meeting. With obvious reference to those who were promoting prenatal diagnosis, he described what he called “The National Institute of Death … a new facility for research and applied eugenics” (Lejeune 1970). As stipulated in Article II.E, the function of this institute was to “destroy, delete, or decry any human condition” deemed undesirable and any embryos “not fitting standard requirements.” He concluded his talk by asking, “Should we capitulate in the face of our own ignorance and propose to eliminate those we cannot help?” It was strong rhetoric—especially since he invoked the dreaded notion of eugenics—but I was not persuaded and went on about my business.
Now, more than 30 years later, I still do not believe that we are engaged in eugenics in the sense that Lejeune implied, although it is certainly true that we seek to help people have children who are indeed well born—which is the literal meaning of “eugenic.” However, by virtue of my research on Down syndrome, I have been very much involved with volunteer organizations—the National Down Syndrome Society, in particular—that have an interest in promoting the welfare of persons with Down syndrome, and this involvement has given me personal contact with literally hundreds of people with Down syndrome, of all ages, and with their families. Furthermore, I have also dealt, in the clinic, with patients and families confronting an enormous variety of disabilities and have interacted with organizations representing their interests. Even though I do not regard abortion as the most desirable way to prevent genetic diseases, none of these encounters has dissuaded me from the belief that prospective parents should have the right to exercise the options that prenatal diagnosis makes possible.
However, I must say that, with time, I have become more sensitive to many of the concerns of Lejeune and others about the applications of modern genetics and believe that we need to be more attuned to the messages that are implicit in our various programs for the prevention of genetic disorders. The disability-rights movement asserts that the willingness to terminate a pregnancy because of fetal abnormality constitutes a rejection of those who have such an abnormality and, in the extreme, a denial of their very right to exist (Parens and Asch 1999). I do not go along with this position, but I do think that we need to listen to what is being said about how people with disabilities, genetic and otherwise, feel they are being regarded. The fact that we have state-supported maternal serum-screening and prenatal-diagnosis programs that are justified on the basis of cost:benefit calculations—dollars per trisomic fetus detected or prevented (i.e., aborted)—says a great deal about how we, as geneticists and a society, view Down syndrome and the other conditions for which we screen (Cunningham and Tomkinison 1999). As much as we talk about neutrality and nondirectiveness in genetic counseling, the message that these programs convey is that it is really not all right to give birth to a child with serious abnormalities.
Why have I have juxtaposed these troublesome thoughts about prenatal diagnosis and disability rights to a description of my research into the mechanisms by which extra chromosomes cause their deleterious effects? My reason is quite simple: It is to remind us that the operative word in “human genetics” is “human.” Human genetics is about human beings—about humanity and humaneness.
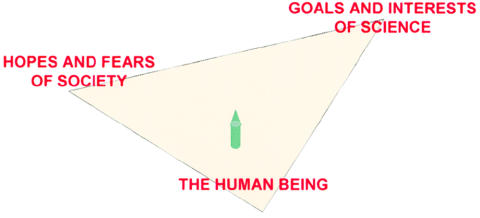
In my Presidential Address in 1996, I described the challenge facing human geneticists as being to find the proper balance between the hopes and fears of society and the goals and interests of our science in discovering new knowledge, improving health, and curing disease (fig. 17) (Epstein 1997). The current debate over cloning and stem cells is a perfect case in point. Now I want to broaden the challenge to add a third element to the balance—the individual, the human being—whose personal interests, whether as an affected patient, a parent of an abnormal fetus, or a clinically normal person, may or may not be consonant with the interests of government, business, the genetic establishment, or society at large (fig. 18). With the major push toward the development and patenting and marketing of genetic tests and products, which many—certainly those who write in the business pages of the newspapers—seem to think the postgenomic era is all about, the individual already stands the risk of becoming merely grist for the mill—the consumer to be targeted. With the ever-increasing power of genomics, transgenesis, and manipulative reproductive technologies, the stakes are becoming even greater. There is no question that the potential for doing good is increasing, but so is the potential to do harm. In the end, and perhaps with greater urgency in these perilous times in which we now find ourselves, it is the human being that must be our ultimate point of reference.
Figure 17.
The challenge facing human geneticists, as articulated by Epstein (1997).
Figure 18.
The challenge facing human geneticists, with consideration of the human being entered into the balance.
Acknowledgments
I must acknowledge the efforts and dedication of the very large number of colleagues, fellows, technicians, counselors, students, collaborators, and patients and their families, with whom I have worked over the past forty years on the projects that I have mentioned, as well as many others. The fact that only a few could be mentioned by name in the foregoing text does not in the least diminish the importance of their contributions to my clinical, research, and organizational activities, and I am truly indebted to all of them. And, even though already mentioned in the narrative, my wife and collaborator in both science and life, Lois Barth Epstein, M.D., deserves special recognition for all that she has done. The research reported in this paper was supported by grants from many sources—particularly, the National Institute of Child Health and Human Development and the National Institute on Aging. Current grants to myself and collaborators include AG16998, AG16999, 1F33DE/HD05706, HD24605, HD31498, HD38384, and NS38869.
Footnotes
Previously presented at the annual meeting of The American Society of Human Genetics, in San Diego, on October 15, 2001.
References
- Akeson EC, Lambert JP, Narayanswami S, Gardiner K, Bechtel LJ, Davisson MT (2001) Ts65Dn: localization of the translocation breakpoint and trisomic gene content in a mouse model for Down syndrome. Cytogenet Cell Genet 93:270–276 [DOI] [PubMed] [Google Scholar]
- Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230 [DOI] [PubMed] [Google Scholar]
- Berger CN, Epstein CJ (1989) Delayed thymocyte maturation in the trisomy 16 mouse fetus. J Immunol 143:389–396 [PubMed] [Google Scholar]
- Brinster RL (1965) Lactate dehydrogenase activity in the preimplanted mouse embryo. Biochim Biophys Acta 110:439–441 [DOI] [PubMed] [Google Scholar]
- ——— (1966) Glucose-6-phosphate dehydrogenase activity in the preimplanted mouse embryo. Biochem J 101:161–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco PG, Epstein CJ (1973) Cell surface changes during preimplantation development in the mouse. Dev Biol 32:208–213 [DOI] [PubMed] [Google Scholar]
- Cao Y, Carlson EJ, Epstein CJ, Basbaum AI (1999) Acute thermal and mechanical nociceptive responses in Ts65Dn mice, a mouse model for Down syndrome. Paper presented at the Ninth World Congress on Pain, Vienna, August 22–27 [Google Scholar]
- Cooper J, Salehi A, Delcroix J-D, Howe CL, Belichenko PV, Chua-Couzens J, Kilbridge JK, Carlson EJ, Epstein CJ, Mobley WC (2001) Failed retrograde transport of NGF in a mouse model of Down’s syndrome: reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc Natl Acad Sci USA 98:10439–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussons-Read ME, Crnic LS (1996) Behavioral assessment of the Ts65Dn mouse, a model for Down syndrome: altered behavior in the elevated plus maze and the open field. Behav Genet 26:7–13 [DOI] [PubMed] [Google Scholar]
- Cox DR, Epstein LB, Epstein CJ (1980) Genes coding for sensitivity to interferon (IfRec) and soluble superoxide dismutase (SOD-1) are linked in mouse and man and map to mouse chromosome 16. Proc Natl Acad Sci USA 77:2168–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham GC, Tompkinison DG (1999) Cost and effectiveness of the California triple marker prenatal screening program. Genet Med 1:199–206 [DOI] [PubMed] [Google Scholar]
- Epstein CJ (1969) Mammalian oocytes: X-chromosome activity. Science 163:1078–1079 [DOI] [PubMed] [Google Scholar]
- ——— (1972) Expression of the mammalian X-chromosome before and after fertilization. Science 175:1467–1468 [DOI] [PubMed] [Google Scholar]
- ——— (1981) Inactivation of the X-chromosome. In: Ritzen M, Hall K, Aperia A, Larsson A, Zetterburg A, Zetterstrom R (eds) Biology of normal human growth. Raven, New York, pp 79–90 [Google Scholar]
- ——— (1986) The consequences of chromosome imbalance: principles, mechanisms, and models. Cambridge University Press, New York [Google Scholar]
- ——— (1997) 1996 ASHG Presidential Address: toward the 21st century. Am J Hum Genet 60:1–9 [PMC free article] [PubMed] [Google Scholar]
- ——— (2001) Down syndrome (trisomy 21). In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw Hill, New York, pp 1123–1256 [Google Scholar]
- Epstein CJ, Avraham KB, Lovett M, Smith S, Elroy-Stein O, Rotman G, Bry C, Groner Y (1987) Transgenic mice with increased CuZn-superoxide dismutase activity: an animal model of dosage effects in Down syndrome. Proc Natl Acad Sci USA 84:8044–8048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Goldberger RF, Anfinsen CB (1963) The genetic control of tertiary protein structure: studies with model systems. Cold Spring Harbor Symp Quant Biol 28:439–449 [Google Scholar]
- Epstein CJ, Hofmeister BG, Yee D, Smith SA, Philip R, Cox DR, Epstein LB (1985) Stem cell deficiencies and thymic abnormalities in fetal mouse trisomy 16. J Exp Med 162:695–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG (1966) Werner's syndrome: a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine 45:177–221 [DOI] [PubMed] [Google Scholar]
- Epstein CJ, McManus NH, Epstein LB, Branca AA, D'Alessandro SB, Baglioni C (1982a) Direct evidence that the gene product of the human chromosome 21 locus, IFRC, is the interferon-α receptor. Biochem Biophys Res Commun 107:1060–1066 [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Smith S, Travis B, Tucker G (1978) Both X-chromosomes function prior to visible X-chromosome inactivation in female mouse embryos. Nature 274:500–503 [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Smith SA, Zamora T, Sawicki JA, Magnuson TR, Cox DR (1982b) Production of viable adult trisomy 17 ↔ diploid mouse chimeras. Proc Natl Acad Sci USA 79:4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LB, Epstein CJ (1976) Localization of the gene AVG for the antiviral expression of immune and classical interferon to the distal portion of the long arm of chromosome 21. J Infect Dis Suppl 133:A56–A62 [DOI] [PubMed] [Google Scholar]
- Epstein LB, Lee SHS, Epstein CJ (1980) Enhanced sensitivity of trisomy 21 monocytes to the maturation-inhibiting effects of interferon. Cell Immunol 50:191–194 [DOI] [PubMed] [Google Scholar]
- Fullerton HJ, Ditelberg JS, Chen SF, Sarco DP, Chan PH, Epstein CJ, Ferriero DM (1998) Copper/zinc superoxide dismutase transgenic brain accumulates hydrogen peroxide after perinatal hypoxia ischemia. Ann Neurol 44:357–364 [DOI] [PubMed] [Google Scholar]
- Groner Y (1995) Transgenic models for chromosome 21 gene dosage effects. Prog Clin Biol Res 393:193–212 [PubMed] [Google Scholar]
- Gropp A, Kolbus U, Giers D (1975) Systematic approach to the study of trisomy in the mouse. II. Cytogenet Cell Genet 14:42–62 [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Li YW, Chen K, Gage FH, Epstein CJ, Mobley WC (1993) Nerve growth factor reverses neuronal atrophy in a Down syndrome model of age-related neurodegeneration. Neurology 43:2668–2673 [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Santucci D, Kilbridge J, Chua-Couzens J, Fontana DJ, Daniels SE, Johnson RM, Chen K, Sung Y, Carlson E, Alleva E, Epstein CJ, Mobley WC (1996) Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc Natl Acad Sci USA 93:13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Carlson EJ, Gillespie AM, Shi Y, Epstein CJ (2000) Ubiquitous overexpression of CuZn superoxide dismutase does not extend life span in mice. J Gerontol A Biol Sci Med Sci 55:B5–B9 [DOI] [PubMed] [Google Scholar]
- Huang TT, Carlson EJ, Raineri I, Gillespie AM, Kozy H, Epstein CJ (1999) The use of transgenic and mutant mice to study oxygen free radical metabolism. Ann NY Acad Sci 893:95–112 [DOI] [PubMed] [Google Scholar]
- Kandel ER (2001) The molecular biology of memory storage: a dialogue between genes and synapses. Science 294:1030–1038 [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen X-N, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, Graham JM Jr, Hudgins L, McGillivray B, Miyazaki K, Ogasawara N, Park JP, Pagon R, Pueschel S, Sack G, Say B, Schuffenhauer S, Soukup S, Yamanaka T (1994) Down syndrome phenotypes: the consequences of chromosome imbalance. Proc Natl Acad Sci USA 91:4997–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotulak R, Gorner P (1992) Fruit flies provide hint on aging. Chicago Tribune, February 9 [Google Scholar]
- Lejeune J (1970) The William Allan Memorial Award lecture: on the nature of man. Am J Hum Genet 22:121–128 [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cue C, Baamonde C, Lumbreras MA, Vallina IF, Dierssen M, Florez J (1999) A murine model for Down syndrome shows reduced responsiveness to pain. Neuroreport 10:1119–1122 [DOI] [PubMed] [Google Scholar]
- Mintz B (1972) Allophenic mice of multi-embryo origin. In: Daniel JC Jr (ed) Methods in mammalian embryology. Freeman, San Francisco, pp 186–214 [Google Scholar]
- Parens E, Asch A (1999) The disability rights critique of prenatal genetic testing: reflections and recommendations. Hastings Cent Rep 29:S1–S22 [PubMed] [Google Scholar]
- Reeves RH, Baxter LL, Richtsmeier JT (2001) Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet 17:83–88 [DOI] [PubMed] [Google Scholar]
- Reeves RH, Irving N, Moran T, Wohn A, Kitt C, Sisodia S, Schmidt C, Bronson R, Davisson M (1995) A mouse model for Down syndrome exhibits learning and behavior deficits. Nat Genet 11:177–184 [DOI] [PubMed] [Google Scholar]
- Richtsmeier JT, Baxter LL, Reeves RH (2000) Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev Dyn 217:137–145 [DOI] [PubMed] [Google Scholar]
- Richtsmeier JT, Zumwalt A, Carlson EJ, Epstein CJ, Reeves RH. Craniofacial anomalies in segmentally trisomic mouse models for Down syndrome. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, Mobley WC, Epstein CJ, Huang T-T (1998) Ts1Cje, a new partial trisomy mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci USA 95:6256–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Rubin EM, Crnic LS, Huang T-T, Epstein CJ (2000) Genetic dissection of the region associated with behavioral abnormalities in mouse models of Down syndrome. Pediatr Res 48:606–613 [DOI] [PubMed] [Google Scholar]
- Schneider EL, Epstein CJ (1972) Replication rate and lifespan of cultured fibroblasts in Down's syndrome. Proc Soc Exp Biol Med 141:1092–1094 [DOI] [PubMed] [Google Scholar]
- Siarey RJ, Carlson EJ, Epstein CJ, Balbo A, Rapoport SI, Galdzicki Z (1999) Increased synaptic depression in the Ts65Dn mouse, a model for mental retardation in Down syndrome. Neuropharmacology 38:1917–1920 [DOI] [PubMed] [Google Scholar]
- Sinet P-M (1982) Metabolism of oxygen derivatives in Down's syndrome. Ann NY Acad Sci 396:83–94 [DOI] [PubMed] [Google Scholar]
- Tan YH, Tischfield J, Ruddle FH (1973) The linkage of genes for the human interferon-induced antiviral protein and indophenoloxidase-B traits to chromosome G-21. J Exp Med 137:317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil J, Epstein LB, Epstein CJ (1980) Synthesis of interferon-induced polypeptides in normal and chromosome 21-aneuploid human fibroblasts: relationship to relative sensitivities in antiviral assays. J Interferon Res 1:111–124 [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD (1996) Positional cloning of the Werner's syndrome gene. Science 272:258–262 [DOI] [PubMed] [Google Scholar]