Abstract
The CLN6 gene that causes variant late-infantile neuronal ceroid lipofuscinosis (vLINCL), a recessively inherited neurodegenerative disease that features blindness, seizures, and cognitive decline, maps to 15q21-23. We have used multiallele markers spanning this ∼4-Mb candidate interval to reveal a core haplotype, shared in Costa Rican families with vLINCL but not in a Venezuelan kindred, that highlighted a region likely to contain the CLN6 defect. Systematic comparison of genes from the minimal region uncovered a novel candidate, FLJ20561, that exhibited DNA sequence changes specific to the different disease chromosomes: a G→T transversion in exon 3, introducing a stop codon on the Costa Rican haplotype, and a codon deletion in exon 5, eliminating a conserved tyrosine residue on the Venezuelan chromosome. Furthermore, sequencing of the murine homologue in the nclf mouse, which manifests recessive NCL-like disease, disclosed a third lesion—an extra base pair in exon 4, producing a frameshift truncation on the nclf chromosome. Thus, the novel ∼36-kD CLN6-gene product augments an intriguing set of unrelated membrane-spanning proteins, whose deficiency causes NCL in mouse and man.
Introduction
The neuronal ceroid lipofuscinoses (NCLs) are the most common neurodegenerative disorders of childhood (Rider and Rider 1999). This group of diseases is characterized by recessively inherited symptoms, including progressive blindness, seizures, and dementia (Kohlschutter et al. 1993), and by hallmark proteolipid pigment deposits in lysosomes of neurons and other cell types (Wisniewski et al. 1988; Prasad et al. 1996). Eight NCL subtypes, distinguished by age at clinical onset and storage-body appearance (granular, fingerprint, and curvilinear), reflect known genetic heterogeneity (Dyken 1989; Gardiner 2000).
The severe infantile (INCL; CLN1 [MIM 256730]) and late infantile (LINCL; CLN2 [MIM 204500]) forms are caused by deficiency of lysosomal palmitoyl-protein thioesterase (Vesa et al. 1995) and of pepstatin-insensitive peptidase (Sleat et al. 1997), respectively. Juvenile-onset, or Batten disease (The International Batten Disease Consortium 1995), which is the most common NCL worldwide (JNCL; CLN3 [MIM 204200]) (Rider and Rider 1988), Finnish progressive epilepsy with mental retardation (EPMR; CLN8) (Haltia et al. 1998), and Finnish variant late-infantile NCL (vLINCL; CLN5 [MIM 256731]) (Savukoski et al. 1994) are due to defects in novel membrane-spanning proteins (The International Batten Disease Consortium 1995; Savukoski et al. 1998; Ranta et al. 1999). Turkish variant LINCL (Wheeler et al. 1998) (vLINCL; CLN7 [MIM 600143]) (Wheeler et al. 1999) may be allelic to CLN8, although no disease-causing mutations have been identified (Mitchell et al. 2001), and CLN6, linked to 15q21-23 markers, causes variant late-infantile disease (vLINCL; CLN6 [MIM 601780]) in families of Indian ancestry (Sharp et al. 1997) and in descendants of Spanish settlers in Costa Rica (Haines et al. 1998).
Costa Rican vLINCL, clinically straddling LINCL and JNCL, features small mixed storage bodies (curvilinear and fingerprint profiles), intermediate onset of symptoms at age 5–7 years, and a course that leads to death in the affected individual's mid 20s (Boustany 1997; Pena et al. 2001). The causative CLN6 defect maps to an ∼6-cM segment of chromosome 15, between D15S1020 and D15S1000 (Auger et al. 1999; Sharp et al. 1999), although allele sharing in two affected siblings, not common to other CLN6 kinships, has suggested an ∼4-cM subinterval (D15S988–D15S983) (Sharp et al. 1999).
We have now employed a panel of multiallele polymorphic DNA markers, pursuing our location-cloning strategy. A core Costa Rican haplotype, distinct from a Venezuelan chromosome that suggests multiple origins for the disease, narrowed the human candidate region, localizing the CLN6 defect within a small gene-rich interval. Sequence comparisons revealed differences in FLJ20561, encoding a novel putative membrane-spanning protein. We identified distinct alterations—a truncating change and a codon deletion, on the Costa Rican and Venezuelan haplotypes, respectively, and a third defect, a frameshift-truncating change, in the nclf mouse featuring NCL-like disease with fingerprint deposits, retinal atrophy, and paralysis (Bronson et al. 1998). The presence of these alterations on disease chromosomes but not on normal chromosomes strongly suggests that FLJ20561 is the CLN6 gene and that its deficiency causes NCL in mouse and in man.
Material and Methods
vLINCL Samples
Studies were performed with prior approval of the institutional review boards (IRBs) at Massachusetts General Hospital, Harvard Medical School, and Duke University Medical Center. Samples were collected from 13 Costa Rican families with LINCL and from a single Venezuelan sibship, comprising both parents and their affected child. Lymphoblastoid cell lines were established for most family members with Epstein-Barr virus (Anderson and Gusella 1984). The clinical diagnosis of vLINCL was determined by a single physician (R.-M.N.B.) using standardized criteria (Haltia et al. 1998) and was confirmed by mixed rectilinear- and fingerprint-profile deposits observed by electron-microscopic examination of skin-biopsy samples. Eight Costa Rican families were used in previous linkage studies, demonstrating that the disease locus is allelic to CLN6 in 15q21-23 (Haines et al. 1998; Auger et al. 1999). All of these families were collected pursuant to Duke IRB-approved protocols.
Genotyping and Haplotype Analysis in Families with vLINCL
Genomic DNA was extracted from blood samples and lymphoblastoid cell lines by a standard protocol (The International Batten Disease Consortium 1995). D15S213, D15S153, D15S988, D15S1015, D15DS983, D15S1025, D15S1000, and D15S216 were genotyped by primer pairs and PCR-amplification conditions recommended by the Whitehead Institute/MIT Center for Genome Research. Five new dinucleotide markers—MADH3gt, MAP2K5ca, ITGA11ca, poly YE, and poly YG—were generated from human 15q23 genomic-DNA sequence (UCSC Human Genome Project Working Draft). The specific primer sets and PCR-amplification conditions are presented in table 1, together with the allele frequencies found on normal chromosomes, obtained by genotyping the unrelated parents of CEPH (Foundation Jean Daussett CEPH) and Venezuelan reference pedigrees. For all markers, PCR analyses were performed with 100 ng genomic DNA, 20 pmol each primer, 0.5 U Taq polymerase, and [32P]-dGTP, with optimized buffer, temperature, and cycling conditions. PCR products, electrophoresed on 6% denaturing polyacrylamide gels, were detected by autoradiography using XAR film (Kodak).
Table 1.
New CLN6-Region Polymorphisms (and Frequencies) on Normal Chromosomes
|
Primer(5′→3′) |
|||||||
| Marker | Forward | Reverse | TAa(°C) | Alleleb | Nc | Hzd | CEPH Standard (Alleles) |
| MADH3gt | GGCCCACCATGTTATTCAG | AAGTGCATGACTGCTGTATGTG | 52.5 | 1 (.143) |
12 | .69 | 2801 (4, 4), 2802 (5, 7) |
| 2 (.012) | 1 | ||||||
| 3 (.024) | 2 | ||||||
| 4 (.345) | 29 | ||||||
| 5 (.31) | 26 | ||||||
| 6 (.000) | 0 | ||||||
| 7 (.167) | 14 | ||||||
| MAP2K5ca | TACTTGGTGGGCTGAGGTG | ATGCGATGAACAGTGAGAATG | 52.6 | 1 (.016) | 1 | .58 | 2801 (2, 4), 2802 (4, 6) |
| 2 (.145) | 9 | ||||||
| 3 (.065) | 4 | ||||||
|
4 (.662) |
41 | ||||||
| 5 (.000) | 0 | ||||||
| 6 (.113) | 7 | ||||||
| ITGA11ca | GGAAGCATGTGGATGACAC | AAGGATGGGAGGCAGTGTG | 56.5 | 0 (.025) | 2 | .85 | 2801 (18, 18), 2802 (14, 14) |
| 1 (.288) | 23 | ||||||
| 10 (.013) | 1 | ||||||
| 11 (.013) | 1 | ||||||
| 12 (.013) | 1 | ||||||
| 14 (.10) |
8 | ||||||
| 15 (.038) | 3 | ||||||
| 16 (.075) | 6 | ||||||
| 17 (.113) | 9 | ||||||
| 18 (.088) | 7 | ||||||
| 19 (.025) | 2 | ||||||
| 20 (.075) | 6 | ||||||
| 21 (.050) | 4 | ||||||
| 22 (.025) | 2 | ||||||
| 23 (.025) | 2 | ||||||
| 24 (.025) | 2 | ||||||
| Poly YE | TCTATCAATGGACAAATGGGTG | TCCAGGCTCATCCATGTT | 50.8 | 1 (.125) | 7 | .57 | 2801 (2, 2), 2802 (2, 5) |
| 2 (.321) |
18 | ||||||
| 3 (.089) | 5 | ||||||
| 4 (.018) | 1 | ||||||
| 5 (.071) | 4 | ||||||
| 6 (.089) | 5 | ||||||
| 7 (.268) | 15 | ||||||
| 8 (.018) | 1 | ||||||
| Poly YG | TTTAGTGGGCTCAATTATGACC | TACGTTGCCCAGGATGAT | 51.5 | −1 (.015) | 1 | .71 | 2801 (9, 12), 2802 (12, 12) |
| 1 (.015) | 1 | ||||||
| 2 (.059) | 4 | ||||||
| 3 (.074) | 5 | ||||||
| 4 (.015) | 1 | ||||||
| 5 (.103) | 7 | ||||||
| 7 (.015) | 1 | ||||||
| 8 (.029) | 2 | ||||||
| 9 (.162) | 11 | ||||||
| 10 (.176) | 12 | ||||||
| 11 (.015) | 1 | ||||||
| 12 (.309) |
21 | ||||||
| 13 (.015) | 1 | ||||||
Annealing temperature.
Data in parentheses are allele frequencies. Alleles in boldface italic are on the Costa Rican disease haplotype; underlined alleles are on the Venezuelan haplotype.
No. of normal chromosomes.
Heterozygosity.
Marker haplotypes on normal and disease chromosomes were determined by tracking the inheritance of alleles in families with vLINCL. Table 1 shows alleles found on Costa Rican and Venezuelan disease haplotypes. Disease chromosomes in Costa Rican pedigrees share alleles at MADH3gt and ITGA11ca, found only rarely on normal chromosomes, which is consistent with suggested linkage disequilibrium (Haines et al. 1998; Auger et al. 1999). The Venezuelan disease chromosome exhibits marker alleles common to nondisease chromosomes.
Gene Identification and Mutation Analyses
The locations of DNA markers and genes within the CLN6 region were determined by our reported 15q22-15q23 YAC contig (Auger et al. 1999) and by search of the 15q23 genomic-DNA sequence, provided by the Human Genome Project, for annotated known genes, expressed-sequence tags (ESTs), cDNAs, and predicted gene segments (UCSC Human Genome Project Working Draft). Mutation scanning was performed by use of specific primer sets, to PCR-amplify exons and splice junctions. Single-strand conformational polymorphism (SSCP) analysis was performed according to the method of Orita et al. (1989), and PCR products, purified by a QIAGEN PCR-purification kit, were sequenced by a BigDyeTerminator Cycle Sequencing Kit (Applied Biosystems), on an ABI Prism 377 automated sequencer. The genotyping assay for the CLN6 G317T mutation was as follows: the 228-bp exon 3 product, amplified, with 5′-ACTGGGAGAAGGTGGTCAG-3′ and 5′-TGTGTCCCTGGAGCCAGTGAC-3′, for 30 cycles of 94°C for 40 s, 50°C for 40 s, and 72°C for 1 min, was restricted with XhoI and was electrophoresed on 3% agarose gels. For the CLN6 exon 5 codon–deletion assay, PCR products, amplified, by 5′-ACTGGGAGAAGGGTGGTCAG-3′ and 5′-TGTGTCCCTGAGCCAGTGAC-3′, for 30 cycles of 94°C for 40 s, 50°C for 40 s, and 72°C for 1 min, the 226-bp mutant and 229-bp normal PCR products, labeled by incorporation of [32P]-dGTP, were displayed on 6% polyacrylamide gels. The parents of CEPH (Foundation Jean Daussett CEPH) and Venezuela DNA genetic-reference pedigrees served as a source of unrelated non-disease chromosomes.
The coding region of the mouse FLJ20561 homologue (mCG9290 [Celera sequence 55053941–55061138]) was analyzed by DNA sequencing of exon-specific PCR products amplified from various wild-type mouse and nclf mouse genomic DNA (stock JR2648; The Jackson Laboratories). For the nclf-mutation assay, exon 4 PCR product was amplified, by 5′ CATACTAGTTTGGGTGACAGTAGACAAACAG-3′ and 5′-CCACCATGATCAGTAGCTAGGCTG-3′, for 30 cycles of 94°C for 40 s, 58°C for 40 s, and 72°C for 1 min. The 510-bp mutant and 509-bp wild-type products were purified, and their DNA sequences were determined.
Northern Blot and RT-PCR Analyses
Multiple-tissue northern blots were hybridized according to the manufacturer's protocol (Clontech). Probes were generated by PCR amplification of human exon 7 and mouse exons 2–7. PCR products were purified by the QIAQuick PCR purification kit (Qiagen) and were [32P]-labeled with the RadPrime DNA labeling system (Gibco BRL). Hybridization was performed with ExpressHyb Hybridization Solution (Clontech) at 65°C. RT-PCR analysis was performed by The One Step system (Invitrogen).
Results
Refining the CLN6 Region
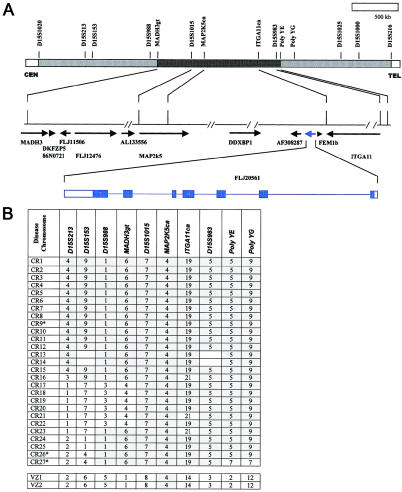
The CLN6 candidate region, defined by recombination events in families with vLINCL, spans ∼4 Mb of 15q22-23 DNA between D15S1020 and D15S1000 (Auger et al. 1999; Sharp et al. 1999). To refine this region, we followed the strategy depicted in figure 1A, initially using a panel of multiallele DNA markers to reveal the haplotypes of the disease chromosomes in our collection of 13 Costa Rican families with vLINCL and a single Venezuelan nuclear sibship with vLINCL. The data, summarized in figure 1B, indicate that all Costa Rican disease chromosomes exhibit a common haplotype, whereas both Venezuelan disease chromosomes share an apparently unrelated haplotype, suggesting that there are two independent mutational origins of vLINCL. Moreover, within the Costa Rican disease haplotype, a core region of shared marker alleles places CLN6 between two of our new polymorphic markers, MADH3gt and polyYE (see the “Material and Methods” section).
Figure 1.
Narrowing the CLN6 region on human chromosome 15. A, Schematic of locations of DNA markers that delimit the genomic 15q22-23 DNA that must contain the CLN6 gene (top), drawn on the basis of data from the Human Genome Project (UCSC Human Genome Project Working Draft). Reported recombination events in families with vLINCL place the gene between D15S1020 and D15S216 (gray-stippled sections). The shared core Costa Rican haplotype points to the segment between MADH3gt and poly YE (black-stippled section). The middle line depicts genomic 15q23 DNA sequence, with the location of gaps (paired slashes), ESTs, and predicted and known genes (arrows) (UCSC Human Genome Project Working Draft). The location of FLJ20561 (blue) is shown, and its predicted exon-intron structure is given, with the coding region shaded (bottom). B, Results of haplotype analysis in Costa Rican and Venezuelan families. Alleles for DNA markers that span the CLN6 region are shown for 27 independent Costa Rican (CR1–CR27) and 2 Venezuelan (VZ1 and VZ2) disease chromosomes. Three chromosomes, distinguished by apparent recombination events (indicated by asterisks [*]), are from two affected Costa Rican siblings. Shared alleles (gray-shaded boxes) indicate a common haplotype that, for chromosomes CR1–CR15 also includes D15S1020, D15S1025, D15S1000, and D15S216 (not shown). Lack of allele sharing (unshaded boxes) at MADHG3gt and poly YE delimits a core Costa Rican haplotype and reveals an unrelated Venezuelan vLINCL chromosome. The “21”' allele at ITGA11ca is likely to be derived from the “19” allele by slippage. The MAP2K5ca “4”' allele is also common to normal chromosomes (see the “Material and Methods” section).
Identification of a Candidate CLN6 Gene
The narrowed CLN6 genetic region, near the centromeric portion of our cloned YAC contig (Auger et al. 1999), is represented in 15q22-23 DNA sequence reported by the Human Genome Project. Searches using the Genome Browser server (UCSC Human Genome Project Working Draft) revealed that MADH3gt and poly YE flank an ∼1.6-Mb interval that contains at least 11 annotated genes encoding known or predicted proteins (fig. 1A).
To identify potential CLN6 mutations, we systematically scanned the coding segments of each candidate gene, for disease-specific DNA sequence changes, by comparing the PCR products amplified from patients and normal controls. Direct DNA sequencing and SSCP analyses did not reveal differences in any of the known genes—MADH3 (mothers against decapentaplegic homologue), MAP2K5 (mitogen-activated protein kinase kinase5), DDXBP1 (DEAD/H box binding protein), FEM1B (feminization factor), and ITGA11 (integrin, alpha 11)—or in three (GenBank accession numbers AF308287, AL13356, and FLJ12476) of five novel mRNAs in GenBank. By contrast, alterations were identified in a hypothetical protein, FLJ20561, that is located between DDXBP1 and FEM1B (fig. 1A). This novel full-length (2.3-kb) cDNA comprises seven exons that span ∼22.7 kb of 15q23 genomic DNA, with, in exon 1, an ATG start codon predicting a novel 311-amino-acid protein.
FLJ20561 Coding Sequence Mutations on vLINCL Chromosomes
Figure 2 presents the FLJ20561 DNA sequence changes that we identified on disease chromosomes. The Costa Rican haplotype carries a G→T nucleotide change at bp 317 in exon 3 (fig. 2A), evident both in PCR products from an affected child (homozygous) and, in a heterozygous state, in a transmitting parent. This transversion, creating a premature stop codon, was present on all 28 Costa Rican disease chromosomes but was not found in the Venezuelan family. Furthermore, as shown, a direct genotyping assay, involving XhoI restriction to distinguish the transversion, confirmed recessive coinheritance of the alteration and the disease in Costa Rican families. This change was not found on 332 normal chromosomes (from 150 unrelated individuals and 32 independent FLJ20561 sequences in databases), providing evidence that the transversion causes vLINCL in patients with the Costa Rican haplotype.
Figure 2.
Candidate CLN6 mutations in human and mouse FLJ20561. A, G317T mutation, in exon 3, found on Costa Rican disease chromosomes. On the left are ABI Prism 377 sequence traces showing the reverse strand of exon 3 PCR products amplified from a normal control, from a Costa Rican patient (homozygote), and from a transmitting parent (heterozygote). The C→A change (underlined) results in the loss of an XhoI site, permitting a direct genotyping assay, the results of which are shown on the right. The ethidium bromide–stained agarose gel reveals two XhoI fragments (of 175 bp and 53 bp) for the normal allele and a single XhoI fragment (228 bp) for the mutant allele. The results demonstrate coinheritance of the mutant allele and clinically diagnosed disease (black square), inherited with the disease chromosome transmitted from each carrier parent (symbols containing dots), which is also detected in an unaffected carrier sibling. B, CTA deletion in exon 5 (producing the ΔY171 change in the protein), which is found on both Venezuelan vLINCL chromosomes. On the left are ABI Prism 377 sequence traces of the exon 5 products amplified from a normal control, from the affected Venezuelan child who is homozygous for the ΔY171 change, and from a heterozygous transmitting parent. The deletion produces a shorter (226 bp) PCR product, permitting a direct genotyping assay, the results of which are shown on the right. The autoradiograph shows radiolabeled exon 5 PCR products—226 bp for the mutant allele and 229 bp for the normal allele—found in the parents. C, Extra C-nucleotide insertion in exon 4 (producing the R103 frameshift change in the protein), which is found on the nclf chromosome. Shown are ABI Prism 377 sequence traces (for the reverse strand) of exon 4 PCR products amplified from wild-type mouse DNA and from nclf/nclf mouse DNA. Direct sequencing did not reveal this change in products amplified from several inbred strains of mice, from outbred mice, or from wild mice.
The Venezuelan vLINCL chromosome possesses an exon 5 CTA codon deletion at bp 613, predicted to eliminate tyrosine 171 (ΔY171). As shown in figure 2B, the affected child has inherited the change from both the mother and the father. This change was not detected on Costa Rican chromosomes, and a direct genotyping assay (fig. 2B) did not reveal the alteration on any of 324 normal chromosomes (from 148 unrelated people and 28 independent ESTs). Thus, consistent with a distinct haplotype, vLINCL in the Venezuelan kindred appears to be caused by a second mutation, confirming the existence of independent origins of this disease in man.
FLJ20561 Sequence Change on the Murine nclf Chromosome
nclf, a spontaneous mouse mutation causing a recessively inherited NCL-like disease with hallmark storage deposits, retinal atrophy, and paralysis, maps to a mouse chromosome 9 region that is syntenic with human 15q21-23, making it a likely CLN6 homologue (Bronson et al. 1998). Searches of GenBank and the Celera Mouse Gene database with the human FLJ20561 sequence disclosed matches to mouse chromosome 9 genomic sequence, disclosing identities to a predicted gene, mCG9290. The mouse FLJ20561 homologue, as illustrated in figure 3, encodes a predicted 308-amino-acid protein that is highly identical (90.3% overall amino acid identity [BLAST 2]) to its human counterpart. Furthermore, the chromosome 9 matches reveal both an exon-intron organization that mirrors the human gene and, for the mouse homologue, a location between the murine Ddxbp1 (mCG9281 [Celera sequence 55084931–55182946]) and Fem1b (mCG9291 [Celera sequence 55005793–55020906) homologues, which is flanked by Map2k5 (mCG9283 [Celera sequence 55363607–55576330]) and Itga11 (mCG10167–mCG10169 [Celera sequence 54887579–54992508]) and which is in a region of synteny with the human 15q23 CLN6 region (fig. 1A). This location is consistent with the genetic position of nclf in a 0.18-cM interval determined with 535 backcross and 2,803 intercross progeny: D9Mit164–0.37 cM–D9Mit234–0.05 cM–D9Mit208,B5,B3–0.06 cM–B6–0.16 cM–B2,B4,nclf–0.02 cM–B1–0.08 cM–D9Mit338,D9Mit105,B7–0.26 cM–D9Mit106 (B1–B7 microsatellite primer information is available from J.R.F.).
Figure 3.
Human and mouse proteins encoded by FLJ20561 cDNA homologues. The top line in each pair shows the predicted 311-amino-acid open reading frame of the FLJ20561 cDNA. Below this line are differences unique to the 308-amino-acid mouse protein, predicted on the basis of the murine cDNA sequence. Sequences were aligned by BLAST 2 Sequences (also see the NCBI BLAST web site). Dashes (–) represent gaps in the match. The positions of six predicted introns (numbered 1–6), delineating seven exons, were determined by alignment of cDNA and genomic-DNA sequences (UCSC Human Genome Project Working Draft and Celera database). The stop mutation found on Costa Rican–haplotype disease chromosomes is indicated by an “at” symbol (@), the ΔY171 deletion mutation on Venezuelan disease chromosomes is indicated by an arrow, and the position of the frameshift, produced by the extra C nucleotide, on the nclf chromosome is indicated by an asterisk (*). The latter change leads to a stop codon after a run of 61 novel amino acids (PNAAAVYSLCQHHHFHHGSQHPPGGRLSQPSPALQWVPAPPVRQ REPHYQEPQAGDSDRLL&) that are not shown.
Sequencing of murine FLJ20561 exons, amplified from nclf/nclf genomic DNA, revealed that the nclf chromosome carries, in exon 4, an extra cytidine (C) that extends a normal run of six Cs to seven nucleotides (fig. 2C), predicting a frameshift that results in a premature stop codon. This change was not found by direct sequence analysis of PCR products amplified from either inbred mouse strains (C57BL/6, BALB/c, and C3H), outbred CD1 mice, or wild-mouse stocks. Nor was it found in mouse (or human) FLJ20561 ESTs or cDNAs identified in the databases. Thus, a single-base insertion in the FL20561 gene is the likely cause of recessively inherited NCL-like disease in the nclf mouse.
CLN6-Gene Products
Northern blot analyses, shown in figure 4A, reveal expression of FLJ20561 mRNA in mouse and human, both in adult and embryonic brain and in peripheral tissues. A major ∼2.4-kb band matches the size of the full-length FLJ20561 cDNA, and the wide expression pattern is consistent with ESTs and cDNAs in human and mouse databases. Notably, some of these ESTs and cDNAs lack exons 4 or 5, suggesting some alternative splicing. Furthermore, as shown for the mouse, full-length cDNA probes also reveal an ∼5.5-kb band that may represent either a much larger FLJ20561 isoform or, alternatively, a product from a related gene. The FLJ20561 ATG codon in exon 1 is located within an optimal Kozak consensus sequence, suggesting that Met1, shown in figure 3, is the start of translation. The genomic-DNA sequence upstream of exon 1 lacks a canonical TATA box but features a GC-rich genomic-DNA sequence that may be a promoter region (MatInspector program).
Figure 4.
Comparison of human and mouse CLN6-gene products. A, Results of human and mouse multiple-tissue northern blots (Clontech), demonstrating wide expression, in brain and peripheral tissues. The human-adult-tissue northern blot was probed with an ∼700-bp FLJ20561 exon 7 probe, revealing an ∼2.4-kb mRNA, matching the size of the full-length cDNA. The mouse-adult tissue and whole-embryo northern blots were hybridized with a ∼1.1-kb probe (exons 2–7), which detects the 2.4-kb mRNA and an unexpected ∼5.5-kb band. A full-length FLJ20561 probe also detects a 5.5-kb band in human tissues (data not shown). P = pancreas; K = kidney; skM = skeletal muscle; Lv = liver; Lg = lung; Pl = placenta; B = brain; H = heart; T = testis; Sp = spleen; E17 = embryonic day 17; E15 = embryonic day 15; E11 = embryonic day 11; E7 = embryonic day 7. B, Hydropathy-plot overlay of predicted human and mouse proteins. Hydropathy plots, drawn according to the Kyte-Doolittle x-1 method (window length 19), by use of the Weizmann Institute of Science Genome and Bioinformatics, Protein Hydrophilicity/Hydrophobicity Search and Comparison server, are nearly identical for the human and mouse proteins, with the exception of the amino terminus. Positive values indicate calculated hydrophobic values. Conserved serine/threonine phosphorylation sites are indicated by circles. The location of the “Costa Rican” exon 3 stop codon is indicated by an “at” symbol (@); the “nclf” mouse exon 4 frameshift lesion is indicated by an asterisk (*), and the “Venezuelan” exon 5 ΔY171 change is indicated by an arrow. Below the plot is an alignment of the amino-terminal regions of the human and mouse proteins. A conserved putative N-myristoylation site (“lollipop” symbol) predicted by PredictProtein may become exposed on cleavage (vertical bar) of a putative presequence predicted by the PSORT II and CBS TargetP programs.
FLJ20561 is novel, and, in addition to the mouse gene, EST-sequence identities indicate likely homologues in a variety of species, including cow (GenBank accession number BE483775), pig (GenBank accession number BE014837), chicken (GenBank accession number BI389620), and Xenopus laevis (GenBank accession number AW644932). The mouse and human versions share high amino acid identity across the entire protein, including, as illustrated by the hydropathy plot in figure 4B, approximately six predicted membrane-spanning domains that strongly suggest an integral membrane protein. The exception is the relatively poorly conserved amino-terminal segment, encoded by exon 1, which may represent a mitochondrial presequence predicted by PSORT II, MITOPROT II (Claros and Vincens 1996), and CBS TargetP (score 0.86) (Nielsen et al. 1997; Emanulelsson et al. 2000). These programs predict a cleavage site, after a conserved histidine, that may expose the internal glycine residue to N-myristoylation (PredictProtein). Conserved serine and threonine residues may be potential sites of phosphorylation by casein kinase II and PKC, although N-glycosylation sites are not predicted.
Impact of the Mutations
RT-PCR amplification of RNA isolated from lymphoblastoid cells of an affected Costa Rican patient reveals that the truncating mutation in exon 3 does not eliminate mRNA expression from the disease allele (data not shown). Thus, although a quantitative effect at the mRNA level is not ruled out, the exon 3 change, as well as the exon 4 and exon 5 mutations that we have identified, may, instead, critically alter the protein product. The Costa Rican CLN6 and nclf mRNAs are expected to encode severely truncated products—of 72 and 103 residues (plus 61 novel amino acids), respectively—which, if stable, are likely to be nonfunctional. Loss of a single tyrosine residue, one of an unusual triplet at a transmembrane junction, causes similar disease in the Venezuelan child, implying a site that is absolutely critical to the activity of this novel integral membrane protein.
Discussion
The discovery of two mutations on different vLINCL chromosomes, both in Costa Rican families and in a Venezuelan kinship, and of a third defect in the nclf mouse suggest that the CLN6 gene has been identified. The clinical and neuropathologic features of the delayed late-infantile-onset form of NCL, therefore, stem from disruption of a novel ∼36-kD membrane-spanning protein. Historically, mutations that cause vLINCL in man have occurred on at least two occasions and, given the lack of allele sharing in families of Asian and Indian ancestry that have vLINCL (Sharp et al. 1999), it is likely that additional CLN6 mutations will be uncovered. It is also possible that a common founder mutation—which may but need not be related to either the Costa Rican or the Venezuelan defect—will be found; for example, Indian, Pakistani, and Romany gypsy families with vLINCL, families that may share ancestry (Bennett and Hofmann 1999), may also possess a common mutation.
The spontaneous murine nclf mutation, currently the only CLN6 lesion known in the mouse, has revealed, in neurons and other cell types, early deposits that presage later retinal atrophy, reactive gliosis, and neuronal-cell degeneration (Bronson et al. 1998). These mice, therefore, offer an immediate resource for studies to discover very early pathological changes that can be investigated in human vLINCL tissue. Moreover, additional nonhuman CLN6 defects are likely to emerge from a variety of naturally occurring NCL animal models (Jolly et al. 1992), particularly that for the OCL6 defect in Southampshire sheep (Broom et al. 1998, 1999), which has revealed mitochondrial ATP synthase subunit c in NCL-storage bodies (Jolly 1997; Broom et al. 1998).
Knowledge of the fundamental CLN6 defects in animal models, together with mutations in man, will provide genotype-phenotype information that may be critical for unraveling of the function of the novel CLN6 protein. The CLN6-encoded protein, which we have dubbed “linclin” (for LINCL protein), is novel, without obvious relatives or protein domains that readily place it within a biochemical or functional pathway. The protein is, however, highly identical between man and mouse. The exception is the amino terminus, which may serve as a cleaved mitochondrion-targeting leader sequence, with subsequent glycine myristoylation perhaps anchoring the processed amino terminus in the membrane. The protein is predicted to form approximately six membrane-spanning domains, although it may lack N-glycosylation sites, suggesting that it may not mature via the secretory system. Another potential clue is a region between transmembrane segments, identical in man and mouse, that contains multiple histidine residues and an unusual tyrosine triplet, implying a domain that is essential to the protein's unknown functional activities.
The recessively inherited CLN6 mutations that we have identified cause identical disease in Costa Rican and Venezuelan sibships. The severely truncated mutant product, comprising the putative presequence and two transmembrane domains, predicted for the Costa Rican (and nclf) mutation implies a complete loss of functional activity; this loss apparently can be mirrored by the Tyr171 deletion. This strongly implies that Tyr171—or the surrounding putative domain—may be directly involved in the biochemical activity of the protein.
Linclin joins three unrelated transmembrane proteins—encoded by CLN3, CLN5, and CLN8—that, when disrupted, cause NCL symptoms that begin in school-age children. Moreover, CLN8, which causes EPMR in man (Ranta et al. 1999), is also mutated in the mnd mouse (Messer et al. 1992; Bronson et al. 1993; Ranta et al. 1999), producing disease that is identical to that caused by the truncating nclf mutation (Bronson et al. 1998). This close similarity in their “loss of function” phenotypes therefore suggests that the CLN6- and CLN8-gene products, as well as the protein products of CLN5 and CLN3, may either be in parallel biochemical pathways or, perhaps, comprise a functional complex.
Deficiencies for classic lysosomal enzymes cause severe (rapid-onset) disease in infants, and the presence of lysosomal inclusions had suggested classification of the NCLs as “lysosomal”-storage disorders (Bennett and Hofmann 1999). However, CLN3, CLN5, CLN8, and CLN6 all encode novel membrane-spanning proteins that may not be restricted to the lysosome (Janes et al. 1996; Pearce 2000). Defects in each of these novel loci cause onset of symptoms after infancy (at age 3–6 1/2 years), suggesting effects, on essential processes, that are less acute than the loss of lysosomal components. Thus, the novel linclin, battenin, and CLN5- and CLN8-encoded transmembrane proteins may comprise components of an “upstream” pathway that, if perturbed, eventually leads to lysosomal lipoprotein deposits.
Elucidation of the CLN6 gene will permit carrier testing and will allow accurate diagnosis of delayed infantile forms of NCL, offering improved disease management. Furthermore, knowledge of CLN6 will spur genotype-phenotype studies in patients with NCL and in spontaneous and created animal models, which promise to shed light on pathways critical for neurons. It is hoped that this knowledge will provide a complete understanding of the biochemical deficits that cause NCL, leading to the development of rational therapeutic interventions for these tragic disorders.
Acknowledgments
We are extremely grateful to the families with vLINCL families, to their physicians (Drs. David Luna and Joachin Pena and the Costa Rican Parent Association), and to Jamileth Chavez, Dehuel Jimenez, Elimar Rojas, and many others, for participating in this research. We acknowledge the services of the Massachusetts General Hospital Genomics Core Facility, and we are grateful to Mary Anne Anderson, Patricia Crawford, Francesca Puglisi, Anuli Ajene, and Karida Shook for technical assistance. This work was supported by National Institutes of Health grants NS33648 (to M.E.M.) and NS30170 (to R.-M.N.B.) and by grants from the Betsy Campbell Trust of the Children’s Brain Disease Foundation (to T.J.L. and R.-M.N.B.) and the Batten Disease Support and Research Association (to T.J.L. and R.-M.N.B.). S.L.C. receives a postdoctoral fellowship from the Batten Disease Support and Research Association.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST 2 Sequences, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html (for nucleic-acid and amino acid alignments)
- CBS TargetP, http://www.cbs.dtu.dk/services/TargetP/ (for subcellular targeting predictions)
- Celera, http://celera.com (for mouse FLJ20561 homologue [mCG9290 ; sequence 55053941–55061138] and for location relative to the order of mouse chromosome 9 genes in the region: Fem1b [mCG9291; sequence 55005793–55020906], Itga11 [mCG10167–mCG10169; sequence 54887579–54992508], Ddxbp1 [mCG9281; sequence 55084931–55182946], and Map2k5 [mCG9283; sequence 55363607–55576330])
- Foundation Jean Daussett CEPH, http://www.cephb.fr/ (for marker-allele sizes and frequencies)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for mRNAs and ESTs)
- MITOPROT, http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter (for subcellular targeting predictions)
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for accession numbers and homology searches)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for INCL [CLN1] [MIM 256730], LINCL [CLN2] [MIM 204500], JNCL [CLN3] [MIM 204200], vLINCL [CLN5] [MIM 256731], vLINCL [CLN6] [MIM 601780], and EPMR [CLN8] [MIM 600143])
- PredictProtein, http://cubic.bioc.columbia.edu/predictprotein/ (for protein predictions of transmembrane domains, functional motifs, and modifications deduced from the primary amino acid sequence)
- PSORT, http://psort.nibb.ac.jp/ (for protein predictions of transmembrane domains, functional motifs, and modifications deduced from the primary amino acid sequence)
- UCSC Human Genome Project Working Draft, http://genome.ucsc.edu (for human genomic-DNA-sequence matches, identity and location of annotated genes, and predicted exon-intron structure)
- Whitehead Institute/MIT Center for Genome Research, http://carbon.wi.mit.edu (for DNA-marker PCR primers and amplification conditions)
- Weizmann Institute of Science Genome and Bioinformatics, Protein Hydrophilicity/Hydrophobicity Search and Comparison, http://bioinformatics.weizmann.ac.il/hydroph/hydroph_help.html (for hydrophobicity comparisons)
References
- Anderson MA, Gusella JF (1984) Use of cyclosporin in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro 20:856–858 [DOI] [PubMed] [Google Scholar]
- Auger KJ, Ajene A, Boustany R-M, Lerner T (1999) Progress toward the cloning of CLN6, the gene underlying a variant LINCL. Mol Genet Metab 66:332–336 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Hofmann SL (1999) The neuronal ceroid-lipofuscinoses (Batten disease): a new class of lysosomal storage diseases. J Inherit Metab Dis 22:535–544 [DOI] [PubMed] [Google Scholar]
- Boustany RM (1997) Batten disease or neuronal ceroid lipofuscinosis. In: Moser H (ed) Handbook of clinical neurology. Vol 22: Neurodystrophies and lipodosis. Elsevier, Amsterdam, pp 671–700 [Google Scholar]
- Bronson RT, Donahue LR, Johnson KR, Tanner A, Lane PW, Faust JR (1998) Neuronal ceroid lipofuscinosis (nclf), a new disorder of the mouse linked to chromosome 9. Am J Med Genet 77:289–297 [DOI] [PubMed] [Google Scholar]
- Bronson RT, Lake BD, Cook S, Taylor S, Davisson MT (1993) Motor neuron degeneration of mice is a model of neuronal ceroid lipofuscinosis (Batten's disease). Ann Neurol 33:381–385 [DOI] [PubMed] [Google Scholar]
- Broom MF, Zhou C, Broom JE, Barwell KJ, Jolly RD, Hill DF (1998) Ovine neuronal ceroid lipofuscinosis: a large animal model syntenic with the human neuronal ceroid lipofuscinosis variant CLN6. J Med Genet 35:717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broom MF, Zhou C, Hill DF (1999) Progress toward positional cloning of ovine neuronal ceroid lipofuscinosis, a model of the human late-infantile variant CLN6. Mol Genet Metab 66:373–375 [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786 [DOI] [PubMed] [Google Scholar]
- Dyken PR (1989) The neuronal ceroid lipofuscinoses. J Child Neurol 4:165–174 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016 [DOI] [PubMed] [Google Scholar]
- Gardiner RM (2000) The molecular genetic basis of the neuronal ceroid lipofuscinoses. Neurol Sci 21:S15–S19 [DOI] [PubMed] [Google Scholar]
- Haines JL, Boustany RM, Alroy J, Auger KJ, Shook KS, Terwedow H, Lerner TJ (1998) Chromosomal localization of two genes underlying late-infantile neuronal ceroid lipofuscinosis. Neurogenetics 1:217–222 [DOI] [PubMed] [Google Scholar]
- Haltia M, Tyynela J, Hirvasniemi A, Ylitalo MS, Herva R (1999) Northern epilepsy: a novel form of neuronal ceroid-lipofuscinosis. In: Bennett MJ (ed) Seventh international congress on neuronal ceroid-lipofuscinoses. Mol Genet Metab 66(4) [Google Scholar]
- International Batten Disease Consortium, The (1995) Isolation of a novel gene underlying Batten disease, CLN3. Cell 82:949–957 [DOI] [PubMed] [Google Scholar]
- Janes RW, Munroe PB, Mitchison HM, Gardiner RM, Mole SE, Wallace BA (1996) A model for Batten disease protein CLN3: functional implications from homology and mutations. FEBS Lett 399:75–77 [DOI] [PubMed] [Google Scholar]
- Jolly RD (1997) The ovine model of neuronal ceroid lipofuscinosis (NCL): its contribution to understanding the pathogenesis of Batten disease. Neuropediatrics 28:60–62 [DOI] [PubMed] [Google Scholar]
- Jolly RD, Martinus RD, Palmer DN (1992) Sheep and other animals with ceroid-lipofuscinoses: their relevance to Batten disease. Am J Med Genet 42:609–614 [DOI] [PubMed] [Google Scholar]
- Kohlschutter A, Gardiner RM, Goebel HH (1993) Human forms of neuronal ceroid-lipofuscinosis (Batten disease): consensus on diagnostic criteria, Hamburg 1992. J Inherit Metab Dis 16:241–244 [DOI] [PubMed] [Google Scholar]
- Messer A, Plummer J, Maskin P, Coffin JM, Frankel WN (1992) Mapping of the motor neuron degeneration (Mnd) gene, a mouse model of amyotrophic lateral sclerosis (ALS). Genomics 13:797–802 [DOI] [PubMed] [Google Scholar]
- Mitchell WA, Wheeler RB, Sharp JD, Bate SL, Gardiner RM, Ranta US, Lonka L, Williams RE, Lehesjoki AE, Mole SE (2001) Turkish variant late infantile neuronal ceroid lipofuscinosis (CLN7) may be allelic to CLN8. Eur J Pediatr Neurol 5 Suppl A:21–27 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8:581–599 [DOI] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 86:2766–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce DA (2000) Localization and processing of CLN3, the protein associated to Batten disease: where is it and what does it do? J Neurosci Res 59:19–23 [PubMed] [Google Scholar]
- Pena JA, Cardozo JJ, Montiel CM, Molina OM, Boustany R (2001) Serial MRI findings in the Costa Rican variant of neuronal ceroid-lipofuscinosis. Pediatr Neurol 25:78–80 [DOI] [PubMed] [Google Scholar]
- Prasad A, Kaye EM, Alroy J (1996) Electron microscopic examination of skin biopsy as a cost-effective tool in the diagnosis of lysosomal storage diseases. J Child Neurol 11:301–308 [DOI] [PubMed] [Google Scholar]
- Ranta S, Zhang Y, Ross B, Lonka L, Takkunen E, Messer A, Sharp J, Wheeler R, Kusumi K, Mole S, Liu W, Soares MB, Bonaldo MF, Hirvasniemi A, de la Chapelle A, Gilliam TC, Lehesjoki AE (1999) The neuronal ceroid lipofuscinoses in human EPMR and mnd mutant mice are associated with mutations in CLN8. Nat Genet 23:233–236 [DOI] [PubMed] [Google Scholar]
- Rider JA, Rider DL (1988) Batten disease: past, present, and future. Am J Med Genet Suppl 5:21–26 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Thirty years of Batten disease research: present status and future goals. Mol Genet Metab 66:231–233 [DOI] [PubMed] [Google Scholar]
- Savukoski M, Kestila M, Williams R, Jarvela I, Sharp J, Harris J, Santavuori P, Gardiner M, Peltonen L (1994) Defined chromosomal assignment of CLN5 demonstrates that at least four genetic loci are involved in the pathogenesis of human ceroid lipofuscinoses. Am J Hum Genet 55:695–701 [PMC free article] [PubMed] [Google Scholar]
- Savukoski M, Klockars T, Holmberg V, Santavuori P, Lander ES, Peltonen L (1998) CLN5, a novel gene encoding a putative transmembrane protein mutated in Finnish variant late infantile neuronal ceroid lipofuscinosis. Nat Genet 19:286–288 [DOI] [PubMed] [Google Scholar]
- Sharp JD, Wheeler RB, Lake BD, Fox M, Gardiner RM, Williams RE (1999) Genetic and physical mapping of the CLN6 gene on chromosome 15q21-23. Mol Genet Metab 66:329–331 [DOI] [PubMed] [Google Scholar]
- Sharp JD, Wheeler RB, Lake BD, Savukoski M, Jarvela IE, Peltonen L, Gardiner RM, Williams RE (1997) Loci for classical and a variant late infantile neuronal ceroid lipofuscinosis map to chromosomes 11p15 and 15q21-23. Hum Mol Genet 6:591–595 [DOI] [PubMed] [Google Scholar]
- Sleat DE, Donnelly RJ, Lackland H, Liu CG, Sohar I, Pullarkat RK, Lobel P (1997) Association of mutations in a lysosomal protein with classical late- infantile neuronal ceroid lipofuscinosis. Science 277:1802–805 [DOI] [PubMed] [Google Scholar]
- Vesa J, Hellsten E, Verkruyse LA, Camp LA, Rapola J, Santavuori P, Hofmann SL, Peltonen L (1995) Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376:584–587 [DOI] [PubMed] [Google Scholar]
- Wheeler JE, Sharp JD, Williams RE, Lake BD, Gardiner RM (19991125) Molecular genetic analysis of late infantile and adult onset NCL. In: Bennett MJ (ed) Seventh international congress on neuronal ceroid-lipofuscinoses. Mol Genet Metab 66(4) [Google Scholar]
- Wheeler RB, Sharp JD, Mitchell WA, Bate SL, Williams RE, Lake BD, Gardiner RM (1999) A new locus for variant late infantile neuronal ceroid lipofuscinosis—CLN7. Mol Genet Metab 66:337–338 [DOI] [PubMed] [Google Scholar]
- Wisniewski KE, Rapin I, Heaney-Kieras J (1988) Clinico-pathological variability in the childhood neuronal ceroid- lipofuscinoses and new observations on glycoprotein abnormalities. Am J Med Genet Suppl 5:27–46 [DOI] [PubMed] [Google Scholar]