Abstract
Next-generation sequencing has been widely applied to understand the complexity of non-coding RNAs (ncRNAs) in a cost-effective way. In this study, we developed mirTools 2.0, an updated version of mirTools 1.0, which includes the following new features. (1) From miRNA discovery in mirTools 1.0, mirTools 2.0 allows users to detect and profile various types of ncRNAs, such as miRNA, tRNA, snRNA, snoRNA, rRNA, and piRNA. (2) From miRNA profiling in mirTools 1.0, mirTools 2.0 allows users to identify miRNA-targeted genes and performs detailed functional annotation of miRNA targets, including Gene Ontology, KEGG pathway and protein-protein interaction. (3) From comparison of two samples for differentially expressed miRNAs in mirTools 1.0, mirTools 2.0 allows users to detect differentially expressed ncRNAs between two experimental groups or among multiple samples. (4) Other significant improvements include strategies used to detect novel miRNAs and piRNAs, more taxonomy categories to discover more known miRNAs and a stand-alone version of mirTools 2.0. In conclusion, we believe that mirTools 2.0 (122.228.158.106/mr2_dev and centre.bioinformatics.zj.cn/mr2_dev) will provide researchers with more detailed insight into small RNA transcriptomes.
Keywords: ncRNA, miRNA, miRNA targets, web server, mirTools, next-generation sequencing
Introduction
Non-coding RNA (ncRNAs) has been increasingly recognized as an important molecular in the past few years.1 Among them, microRNA (miRNA) is small, approximately 19–25 nt RNA molecule, which is involved in post-transcriptional regulation of gene expression. It plays important roles in regulation of numerous biological processes, such as development, cell differentiation and proliferation, apoptosis and metabolism.2 Small nuclear RNA (snRNA) is primarily involved in RNA splicing and assists in the regulation of transcription factors and maintains telomeres.3 Small nucleolar RNA (snoRNA) plays a crucial role in modification of target RNAs and processing of rRNA during ribosome subunit synthesis.4 Piwi-interacting RNA (piRNA), an approximately 24–31 nt RNA molecule, plays an important role in regulation of cell division and maintenance of germline stem cells.5 A recent study showed that piRNA is also involved in epigenetic control of memory-related synaptic plasticity in neural cells.6
Next-generation sequencing (NGS) has been widely applied to characterize small RNA transcriptomes under various conditions. It provides an unprecedented opportunity to discover ncRNAs and identify differentially expressed ncRNA transcripts.7 However, the massive amount of data generated by NGS poses great bioinformatics challenges for detection and functional annotation of ncRNAs. Therefore, a number of computational methods have been developed for mining small RNA sequencing data. Among them, many tools have mainly focused on miRNA analysis, such as miRDeep,8-10 Mireval,11 miRFinder,12 miRNAkey,13 miRanalyzer,14 miRExpress,15 miRTRAP,16 DSAP17 and MIReNA.18 In addition, several integrated ncRNA analysis tools have been released, such as SeqCluster,19 DARIO,20 ncPRO-seq,21 CPSS,22 Shortran,23 NORAHDESK,24 APART25 and smyRNA.26
We previously developed a web service, mirTools 1.0, which provides annotation of miRNAs based on NGS and has been widely used.27 Comprehensive comparison and evaluation of bioinformatics tools for miRNA deep-sequencing has indicated that mirTools 1.0 has a good performance for miRNA coverage, accuracy and sensitivity, as well as computational time.28 However, in the past 2 y, we have received considerable feedback from users. These users expected us to update mirTools to include more versatile functions, such as miRNA-targeted genes and further functional annotation, other types of ncRNAs besides miRNAs and multiple sample comparison. Therefore, in this study, an integrated web server, mirTools 2.0, an updated version of mirTools 1.0, was developed to investigate ncRNA sequences, expression levels, differentially expressed ncRNAs and miRNA-targeted genes and their functional annotation, which will be valuable for deciphering the functional roles of ncRNAs hidden in the large amount of NGS data.
Results and Discussion
Implementation
The web server mirTools 2.0 is constructed under the Apache/PHP/MySQL environment in the Linux system. The back-end pipeline is implemented in Perl language and the plots are generated by R packages (www.r-project.org). Compared with mirTools 1.0, the computational power of mirTools 2.0 has been enhanced and it is equipped with four Quad-Core AMD processors (2.2 GHz each) and 32 GB of RAM. It will only take approximately 30 min to detect and quantify ncRNAs for a given sample (~10 Mb size). Additionally, the queuing module can execute more jobs in parallel.
Data input
The web server mirTools 2.0 provides more functional modules than mirTools 1.0, including a single case, two cases, group cases and re-analysis. The single case module allows users to detect various types of known and novel ncRNAs, and performs functional annotation of the miRNA-targeted genes for a single sample. Two cases and group cases modules allow users to identify differentially expressed ncRNAs between or among samples. The re-analysis module is designed to allow users to run previously submitted data with adjustable parameters, which avoids resubmitting the sample data.
In single case and two case modules, similar to mirTools 1.0, the input of mirTools 2.0 is a trimmed FASTA file where all the identical raw reads are aggregated and cleaned into a non-redundant FASTA file to reduce the input size. To further reduce the input size, the FASTA file can be compressed in rar, zip or gz formats, with a maximum size of 30 Mb. In addition, mirTools 2.0 supports the input of original mapped reads in SAM/BAM format, which can be generated by many public alignment software, such as Bowtie (bowtie-bio.sourceforge.net) and BWA (bio-bwa.sourceforge.net). In the group case module, the expression table files of ncRNAs are required, which can be obtained from the single case and two case modules. Users can directly input a single case analysis job ID and the web server will retrieve the corresponding expression table file automatically. In all modules, the mail address is optional and the web server will give users a job ID, which can be used to retrieve the results once the job is finished or to reanalyze the data submitted previously.
Data output
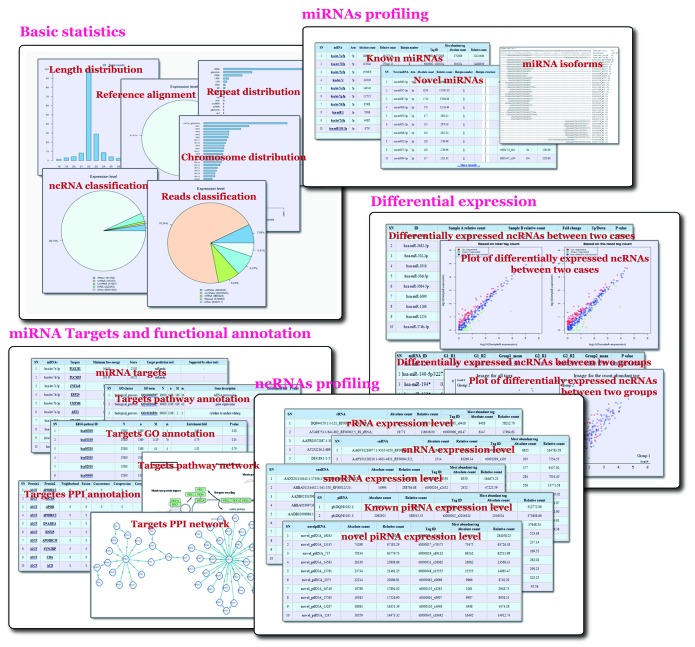
The mirTool 2.0 results are presented in intuitive HTML pages, of which a typical output consists of six parts: basic statistics, annotation, miRNA, miRNA targets, ncRNA and differential expression (Fig. 1). The basic statistics output contains length distribution charts of short reads, pie charts summary of reference genome mapping and the chromosome distribution. The annotation output includes pie charts of mapped reads with different functional categories, ncRNA distribution and repeat-associated RNA distribution. The web server mirTools 2.0 plots the unique read distribution and its expression levels (the number of reads for each tag reflects its relative abundance).
Figure 1. Output screenshots of mirTools 2.0. The output includes: (1) basic statistics, such as length distribution, percentage of reads aligned to the reference genome, chromosome distribution, functional categories of reads and ncRNA distribution; (2) known miRNA, putative novel miRNA, miRNA isoforms and modification; (3) miRNA-targeted genes and functional annotation based on GO, the KEGG pathway and the PPI network; (4) expression information of other types of ncRNA, such as rRNA, tRNA, snRNA, snoRNA and piRNA; and (5) differentially expressed ncRNAs between two cases, two experimental groups or among multiple samples.
The miRNA output consists of known miRNAs and putative novel miRNAs. The detailed annotation of each miRNA contains the miRNA name, arm on the hairpin, absolute count, relative count, pre-miRNA number (hairpin secondary structure for novel miRNAs) and related expression information of the most abundant tag. In addition, users can view the read mapping information and miRNA isoforms in a pop-up webpage by clicking the “pre-miRNA” link.
The miRNA targets output contains the predicted miRNA-targeted genes and their functional annotation with GO, the KEGG pathway and the PPI network. The miRNA-targeted genes tables display the miRNA name, the targeted gene name, the minimum free energy, the score value (P value for RNAhybrid) and target prediction tool used. The known miRNA-targeted genes tables also contain the “other tools” column to indicate whether the targets are supported by other tools. The GO and KEGG pathway annotation tables illustrate the enriched GO terms and pathway terms of targeted genes predicted, respectively, which can be sorted by enrichment fold and P value. The PPI annotation tables depict the protein interaction information of miRNA targets in STRING databases. Users can visualize the interaction intuitively in the implemented Cytoscape Web, which supports node dragging and searching, by clicking the “show the network in Cytoscape” link.
The ncRNA output shows information of other ncRNAs, except for miRNAs and their expression level. Information on the identified known piRNAs and novel piRNAs are also included in this output. The detailed annotation of each ncRNA contains the ncRNA name, absolute count, relative count, hairpin number and related expression information of the most abundant tag.
In a two-sample study, the differential expression output contains expression correlation dot charts and differentially expressed ncRNAs lists between the two samples. The annotation information of the differential expression list contains the ncRNA name, sample “a” relative count, sample “b” relative count, the fold change, the up/down tag and the P value. In group case results, differentially expressed ncRNAs between two groups are listed. The annotation information of the group expression list also contains the expression value of each sample, the median expression value of the group, the up/down tag and the statistical P value. All these components are well organized with examples to facilitate users with correct input and expected results.
Discussion
NGS has greatly facilitated RNA transcriptome studies, among which small RNA sequencing offers a cost-effective and in-depth method to comprehensively investigate ncRNAs in a genome-wide manner.7 However, one of the main challenges lies with the analysis of miRNAs and other ncRNAs from the large amount of sequencing data. The web server mirTools 2.0 was developed for research communities toward a fully automated and easy to use web service suitable for ncRNA discovery, profiling and functional annotation based on high-throughput sequencing.
The web server mirTools 2.0 is freely available for non-commercial use and will be updated regularly to keep up with the latest annotation information of the implemented databases. In mirTools 1.0, we received a lot of valuable feedback and suggestions from users worldwide, and this feedback has been helpful for developing mirTools 2.0. Therefore, we sincerely welcome questions, comments and suggestions, which will be useful for feedback for the enhanced function of mirTools 2.0. Currently, mirTools 2.0 can only detect the known ncRNAs, novel miRNAs and novel piRNAs. In the future, we will develop or incorporate a tool to predict other type of novel ncRNAs. In the meantime, phylogenetic conservation analysis of ncRNAs across different species will be provided. Considering the network limits, currently, the maximum file upload size is 30 Mb, regardless of whether compression is involved. Therefore, we have developed a stand-alone version of mirTools 2.0 to allow users to run it on their own server. In the future, we will design an FTP module to allow users to submit larger data to enhance the usability of web server. In conclusion, we believe that mirTools 2.0 will provide the scientific community with an integrated web server to assist research for identifying various types of ncRNA, profiling expression levels, predicting miRNA targeted genes and functional annotation based on the large amount of data generated from NGS.
Materials and Methods
Overview of the workflow of mirTools 2.0
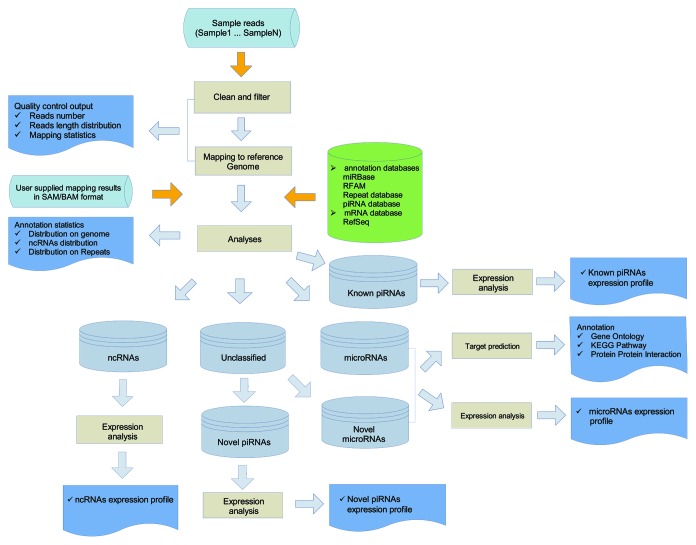
The overall workflow of mirTools 2.0 is shown in Figure 2. Briefly, mirTools 2.0 filters out raw reads to exclude low quality and 3/5′ adaptor sequences and trim them into clean reads. Clean reads are then mapped onto the reference genome and mapping results are converted into the SAM/BAM format with SAMtools (samtools.sourceforge.net) to serve as a generic alignment format compatible with different alignment tools. Based on public resources, the mapped reads are annotated and classified into known ncRNAs. Novel miRNAs and piRNAs will be predicted from unclassified aligned reads. miRNA-targeted genes and further functional annotations are conducted for both known and novel miRNAs based on a number of implemented tools. Finally, all the results are shown in different types of tables and figures on an HTML page, and these are available for downloading intermediate annotation results and the final results.
Figure 2. The overall workflow of mirTools 2.0. The workflow includes clean and filtering raw reads, alignment of them to the reference genome, classification of aligned reads, detection of expression levels of various types of ncRNAs and the differentially expressed ncRNAs between two cases/two experiment groups or among multiple samples, prediction of novel miRNAs and piRNAs, identification of miRNA targeted genes and further functional annotation based on GO, the KEGG pathway and the PPI network.
Discovery and profiling of known and novel ncRNAs
To identify known and novel ncRNAs, sequence reads are first aligned to the reference genome using the SOAP program.29 Subsequently, aligned reads are associated with the annotation information of several public databases. In addition to miRBase (www.mirbase.org), Rfam (rfam.sanger.ac.uk), repeat database produced by RepeatMasker (www.repeatmasker.org) and coding genes of the reference genome, piRNA from the piRNABank database (pirnabank.ibab.ac.in) is also incorporated to identify known piRNAs. Currently, mirTools 2.0 is compatible for use with 32 reference genomes across vertebrates, insects, deuterostomes, nematodes and plants. The aligned reads are classified into known miRNAs, other types of ncRNAs, known piRNAs, repeat-associated RNA and mRNAs. miRNA isoforms and modification can be obtained through changing the mismatch number in the SOAP program. The ncRNAs reads annotated by Rfam are further classified into sRNA, tRNA, snRNAs and snoRNA.
The unclassified aligned reads termed as “unclassified” are used to detect novel miRNAs and piRNAs. In mirTools 1.0, we used the miRDeep program to predict novel miRNAs. In mirTools 2.0, we implemented a new version of miRDeep9 to discover novel miRNAs. We also implemented another broadly used program, Mireap (sourceforge.net/projects/mireap), which combines secondary structure, minimum free energy, Dicer cleavage site, small RNA position and depth, to discover novel miRNAs from NGS.28,30 During this process, the secondary structures are predicted using the RNAfold program in Vienna RNA package (www.tbi.univie.ac.at/RNA). The remaining unclassified reads are used to detect novel piRNAs using a k-mer scheme, which has been indicated to be high accuracy and specificity for predicting novel piRNAs.31
Identification of miRNA-targeted genes and functional annotation
To identify known and novel miRNA targeted genes, mirTools 2.0 implements two widely used tools miRanda (www.microrna.org) and RNAhybrid (bibiserv.techfak.uni-bielefeld.de/rnahybrid/). In addition, miRNA-targeted gene results from another six tools or databases are also integrated, including TargetScan (www.targetscan.org), TargetSpy (www.targetscan.org), miRNAMap (mirnamap.mbc.nctu.edu.tw), microT v4.0 (diana.cslab.ece.ntua.gr/microT/), MicroCosm (www.ebi.ac.uk/enright-srv/microcosm) and MirTarget2 (mirdb.org).
To explore the potential biological function of predicted miRNA-targeted genes, we annotated them with Gene Ontology (GO), the KEGG pathway and the protein-protein interaction (PPI) network. For GO analysis, the predicted targets are mapped to the GO annotation data set to extract their GO annotation,32 and then Fisher’s exact test is used to perform GO enrichment analysis (enrichment ratio > 2 and P value < 0.01 at default). Pathway assignment information of miRNA-targeted genes is extracted from the KEGG pathway database33 and corresponding enrichment analysis is performed using the hypergeometric test (enrichment ratio > 2 and P value < 0.01 at default). Moreover, PPI annotation of miRNA-targeted genes is retrieved from the STRING database.34 Visualization of the PPI network can be conducted using the implemented Cytoscape Web tool, which is an interactive web-based network browser that allows easy displaying of graphs.35
Detection of differentially expressed ncRNAs
To determine the relative ncRNA expression level and its abundance, each identified ncRNA read count is normalized to the total read count of its belonging type of ncRNA to obtain reads per million (RPM) value. Similar to mirTools 1.0, mirTools 2.0 has two strategies to estimate the expression level of a given ncRNA: the relative total read count and the most abundant read (often considered as mature miRNA). To detect differentially expressed ncRNAs between two samples, the Bayesian method is used to calculate the statistical significance (P value) based on the relative total read count and most abundant read count.36
In addition, we developed a group case module, which can compare the difference within and between experimental groups with multiple replicates or samples. If a specific experimental group has two conditions, the Wilcoxon Rank-sum test is applied to infer the statistical significant difference. If a specific experimental group has more than two conditions, the Kruskal-Wallis H test is applied to infer the statistical significant difference. The Wilcoxon Rank-sum test is used to identify the differentially expressed ncRNAs between experimental groups. In all conditions, at default, a specific ncRNA is considered to be differentially expressed with a P value < 0.01 and a fold change > 2.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 31171236), the National High Technology Research and Development Program of China (2012AA02A201), the Key Science and Technology Innovation Team of Zhejiang Province (2012R10048-05) and the International S&T Cooperation Program of China (2011DFA30670).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25193
References
- 1.David R. Non-coding RNAS: A new member of the family. Nat Rev Mol Cell Biol. 2012;13:686. doi: 10.1038/nrm3449. [DOI] [PubMed] [Google Scholar]
- 2.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7:192–204. doi: 10.4161/rna.7.2.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–69. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L, Li X, Liu Q, Zhao F, Wu J. Small RNA transcriptome investigation based on next-generation sequencing technology. J Genet Genomics. 2011;38:505–13. doi: 10.1016/j.jgg.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 9.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011;27:2614–5. doi: 10.1093/bioinformatics/btr430. [DOI] [PubMed] [Google Scholar]
- 11.Ritchie W, Théodule FX, Gautheret D. Mireval: a web tool for simple microRNA prediction in genome sequences. Bioinformatics. 2008;24:1394–6. doi: 10.1093/bioinformatics/btn137. [DOI] [PubMed] [Google Scholar]
- 12.Huang TH, Fan B, Rothschild MF, Hu ZL, Li K, Zhao SH. MiRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinformatics. 2007;8:341. doi: 10.1186/1471-2105-8-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronen R, Gan I, Modai S, Sukacheov A, Dror G, Halperin E, et al. miRNAkey: a software for microRNA deep sequencing analysis. Bioinformatics. 2010;26:2615–6. doi: 10.1093/bioinformatics/btq493. [DOI] [PubMed] [Google Scholar]
- 14.Hackenberg M, Sturm M, Langenberger D, Falcón-Pérez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37(Web Server issue):W68-76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrix D, Levine M, Shi W. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 2010;11:R39. doi: 10.1186/gb-2010-11-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, et al. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res. 2010;38(Web Server issue):W385-91. doi: 10.1093/nar/gkq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathelier A, Carbone A. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics. 2010;26:2226–34. doi: 10.1093/bioinformatics/btq329. [DOI] [PubMed] [Google Scholar]
- 19.Pantano L, Estivill X, Martí E. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics. 2011;27:3202–3. doi: 10.1093/bioinformatics/btr527. [DOI] [PubMed] [Google Scholar]
- 20.Fasold M, Langenberger D, Binder H, Stadler PF, Hoffmann S. DARIO: a ncRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2011;39(Web Server issue):W112-7. doi: 10.1093/nar/gkr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CJ, Servant N, Toedling J, Sarazin A, Marchais A, Duvernois-Berthet E, et al. ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics. 2012;28:3147–9. doi: 10.1093/bioinformatics/bts587. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Xu B, Yang Y, Ban R, Zhang H, Jiang X, et al. CPSS: a computational platform for the analysis of small RNA deep sequencing data. Bioinformatics. 2012;28:1925–7. doi: 10.1093/bioinformatics/bts282. [DOI] [PubMed] [Google Scholar]
- 23.Gupta V, Markmann K, Pedersen CN, Stougaard J, Andersen SU. shortran: a pipeline for small RNA-seq data analysis. Bioinformatics. 2012;28:2698–700. doi: 10.1093/bioinformatics/bts496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragan C, Mowry BJ, Bauer DC. Hybridization-based reconstruction of small non-coding RNA transcripts from deep sequencing data. Nucleic Acids Res. 2012;40:7633–43. doi: 10.1093/nar/gks505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4013–24. doi: 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salari R, Aksay C, Karakoc E, Unrau PJ, Hajirasouliha I, Sahinalp SC. smyRNA: a novel Ab initio ncRNA gene finder. PLoS One. 2009;4:e5433. doi: 10.1371/journal.pone.0005433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, et al. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38(Web Server issue):W392-7. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4298–305. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 30.Cheng WC, Chung IF, Huang TS, Chang ST, Sun HJ, Tsai CF, et al. YM500: a small RNA sequencing (smRNA-seq) database for microRNA research. Nucleic Acids Res. 2013;41(Database issue):D285–94. doi: 10.1093/nar/gks1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Wang X, Kang L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics. 2011;27:771–6. doi: 10.1093/bioinformatics/btr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–60. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–8. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–95. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]