Abstract
MiRNAs are short, non-coding RNAs that regulate gene expression post-transcriptionally through specific binding to mRNA. Deregulation of miRNAs is associated with various diseases and interference with miRNA function has proven therapeutic potential. Most mRNAs are thought to be regulated by multiple miRNAs and there is some evidence that such joint activity is enhanced if a short distance between sites allows for cooperative binding. Until now, however, the concept of cooperativity among miRNAs has not been addressed in a transcriptome-wide approach. Here, we computationally screened human mRNAs for distances between miRNA binding sites that are expected to promote cooperativity. We find that sites with a maximal spacing of 26 nucleotides are enriched for naturally occurring miRNAs compared with control sequences. Furthermore, miRNAs with similar characteristics as indicated by either co-expression within a specific tissue or co-regulation in a disease context are predicted to target a higher number of mRNAs cooperatively than unrelated miRNAs. These bioinformatic data were compared with genome-wide sets of biochemically validated miRNA targets derived by Argonaute crosslinking and immunoprecipitation (HITS-CLIP and PAR-CLIP). To ease further research into combined and cooperative miRNA function, we developed miRco, a database connecting miRNAs and respective targets involved in distance-defined cooperative regulation (mips.helmholtz-muenchen.de/mirco). In conclusion, our findings suggest that cooperativity of miRNA-target interaction is a widespread phenomenon that may play an important role in miRNA-mediated gene regulation.
Keywords: microRNA, target regulation, target prediction, cooperativity
Introduction
MicroRNAs (miRNAs) are small single-stranded non-coding RNAs, which are endogenously expressed and predominantly downregulate the expression of mRNA targets. They achieve post-transcriptional regulation of gene expression as part of the miRNA-induced silencing complex (miRISC), which consists of a miRNA and several proteins, including a member of the Argonaute (AGO) protein family. Binding of miRISC to its target sequence is guided by the miRNA and most commonly occurs within the 3′-untranslated region (3′-UTR) of the mRNA, thereby inducing translational repression or degradation of the mRNA (reviewed in refs. 1‒3).
It becomes increasingly apparent that deregulated expression of miRNAs is causally related to the development of various complex disorders. This includes cardiac disease,4,5 lung cancer,6 leukemia,7 neurological disorders such as Alzheimer disease,8 metabolic abnormalities like diabetes mellitus9 and rheumatoid arthritis.10
Unbiased approaches to miRNA function, for example, by application of synthetic miRNAs libraries to cells,11 indicated that cellular pathways are regulated by multiple miRNAs12,13 or are subject to regulation by a single miRNA acting on different levels.14 On the other hand, almost every miRNA investigated has been assigned several, often contradictory, physiological roles.15 Obviously, identifying the target mRNAs is crucial to understand the function of a disease-related miRNA and, consequently, to develop therapeutic approaches. To achieve this goal, we need to know the criteria according to which miRNAs (in the context of miRISCs) are guided to their respective targets and the principles leading to effective target regulation.
However, the mechanisms of miRNA-mRNA interactions are still about to be elucidated, and versatile, often contradictory modes of action have been reported.16-19 In metazoans, the suppressive effect of an individual miRNA on a target is often small,20 potentially due to the fact that miRNAs form only imperfect and thermodynamically unfavorable RNA-RNA hybrids with their targets over a short sequence (called the miRNA seed region nucleating the interaction).
A set of interaction rules has been formulated1 based on biochemical and bioinformatic analyses, but functional miRNA sites often show aberrant characteristics. In spite of these difficulties, there is good evidence that contiguous and perfect base pairing of nucleotide positions 2–8 of the miRNA (seed region) with the cognate mRNA sequence is predictive of true interactions between them.1,21
Therefore, one comparably successful approach to bioinformatically predict miRNA targets is to focus on the seed region in miRNA targets. The online tool TargetScan searches for conserved seed regions of 7 and 8 nucleotides in length as well as for 3′ compensated sites in 3′-UTRs. It ranks its predicted results based on further miRNA-mRNA binding properties summarized in a so called context+ score, including seed-pairing stability and target-site abundance.22-24 A similar tool to predict miRNA target sites, miRanda, scores and ranks its results based on a machine learning algorithm called mirSVR.25-27 The authors use support vector regression (SVR) to train on target site information as well as context features and calibrate their scores to correlate with observed downregulation of a published experimental data set.
More recently, computational methods were successfully combined with experimental miRISC-RNA crosslinking approaches to identify target mRNAs and characterize their miRNA binding sites: High-throughput sequencing of RNA isolated after UV crosslinking and immunoprecipitation (HITS-CLIP),28 photoactivatable ribonucleoside-enhanced CLIP (PAR-CLIP)29 and individual nucleotide resolution CLIP (iCLIP).30 These approaches are helpful to reduce the search space for miRNA targets since they select for RNA fragments that are bound to active miRISC complexes. By UV irradiation of living cells, native protein-RNA contacts will be covalently crosslinked and, thereby, the information about the binding region preserved for later miRNA binding site predictions. Next to CLIP methods, there is a range of approaches used for target identification that do not rely on crosslinking, such as pull down of biotinylated miRNAs.31
However, miRNA-target interactions are not only bidirectional but rather form complex networks.32,33 For the formation of a RISC on mRNA, seed pairing with as little as 6 or 7 nucleotides between miRNA and mRNA target seems sufficient (albeit thermodynamically unfavored and most likely dependent on further interaction between RISC components and the mRNA). Therefore, almost every miRNA known to date is computationally predicted to target more than one mRNA, and experimental evidence confirms this notion.17,22-24,34-36 Further, one mRNA is often controlled by multiple miRNAs. It has been shown that mRNAs with strong miRNA-mediated effects on their expression level typically contain multiple miRNA binding sites for the same or different miRNAs instead of a single one, which is therefore useful as a predictor of miRNA target regulation.35
The possibility that miRNAs could regulate their targets in a concerted—potentially cooperative—fashion has already been considered short after their identification, when the 3′-UTR of C. elegans lin-41 mRNA was shown to contain multiple targets sites (seven) for miRNA lin-4.37,38 Later, the integrity of more than one site for miRNA let-7 on the same target has been shown to be essential for efficient translational repression.39 Additional support comes from assays which showed that luciferase reporter mRNAs with two or four binding sites for an exogenous small RNA (CXCR4 siRNA) in their 3′-UTR were more efficiently repressed than single-site constructs.40
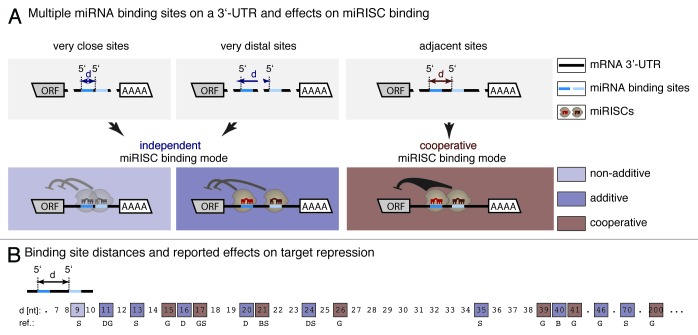
In principle, the combined regulation of a mRNA by several miRNAs could be achieved by (1) independent or (2) cooperative target interaction (Fig. 1A). Independent binding of several miRNAs (in the context of a miRISC) to the same mRNA may be presumed to confer additive regulatory effects, whereas cooperative binding enhances the individual regulatory potency of miRNAs. Depending on their experimental design, assays for translational repression of reporter constructs verified both independent23,41,42 and cooperative42,43 activities of small RNAs. According to these studies, additive effects on the same mRNA are, at best, moderate, whereas regulation by two or more sites within a certain distance amplified miRNA-mediated repression to an extent greater than expected from independent sites, supporting a concept of cooperative activity.
Figure 1. Distance between multiple miRNA binding sites as predictor of cooperative target regulation. (A) Concerted miRNA target regulation (in the context of miRISCs) may be described by independent or cooperative activities. An independent mode of repression has been described for very close and for very distant sites. Non-additive effects would be expected if overly close sites exclude simultaneous binding of miRNAs. Additive effects may occur when miRNAs occupy sites autonomously without activity-enhancing interactions between their miRISCs. In contrast, cooperative activity has been shown for miRNAs, whose binding sites on a specific mRNA are within a certain distance referred to as cooperativity range. The term cooperativity refers to a synergistic effect amplifying miRNA-mediated repression to an extent greater than expected from independent sites. (B) Summary of previous experimental studies investigating distance-dependency of cooperative target repression by multiple small RNA binding sites. Inter-site distances (d) tested in the reports (squares) are shown as the distance between adjacent miRNA 5′ ends on the respective mRNA target. Square colors, light and dark blue indicate that the repressive effect of multiple binding sites was weaker/similar or stronger than for a single target site. Brown squares emphasize 5′-to-5′ distances for which repression has been reported significantly greater than expected from additive effects (cooperative miRISC binding mode). The accumulation of cooperative regulation for distances between 15‒26 nucleotides indicates that directly adjacent miRISCs (with certain variations) have the highest potential to repress their target in a cooperative way. Cooperative regulation outside of the core cooperativity range might occur due to secondary structure formations of the target sequence. D, Doench et al.;43 S, Saetrom et al.;41 G, Grimson et al.;23 B, Broderick et al.42
However, only one of these studies42 characterized cooperative RISC binding in a quantitative way. The authors used siRNAs (i.e., small interfering RNAs of 18–21 nucleotides, which completely hybridize to their targets) instead of miRNAs and multiple binding sites on one reporter mRNA molecule. The Hill coefficient, a measure of cooperativity,44 was determined by fitting reporter repression as a function of the siRNA concentration to the Hill equation. Next to the identity of the involved Argonaute protein, Broderick et al.42 showed strong dependency of cooperative silencing on the distance between two adjacent binding sites. They could show that at least for AGO 1, 3 and 4, miRNA cooperativity is limited to directly adjacent binding sites. This is in accordance with previous approaches studying the spacing pattern between neighboring binding sites leading to cooperative repression. Although the conclusions drawn in these studies were not fully unanimous, they concurred that cooperative effects are facilitated when miRNA binding sites are directly adjoining40,42 (i.e., the distance from the 5′-end of one miRNA to the 5′-end of the next is 20‒22 nucleotides or the length of exactly one miRNA). Enhanced repression of mRNAs with two (vs. one) miRNA binding sites was also observed when sites partially overlapped (5′-end of downstream site moved four nucleotides into the accessory but not the seed region of the upstream site) or when they were separated by few additional nucleotides (5′-to-5′ distance of 25 nucleotides).43 It seems, however, unclear what mode of combined miRNA activity (independent or cooperative) underlies translational repression of these constructs. Broderick et al.42 found that cooperative effects are lost when miRNA sites (except bulged AGO 2 sites) were separated by 19 nucleotides (5′-to-5′ distance 40 nucleotides). These studies suggest that direct adjacency of binding sites promotes cooperative miRNA activities, whereas deviation from this rule may result in loss of combined effects or a shift toward independent activities. A schematic drawing of these correlations is shown in Figure 1B. Some outliers with larger 5′-to-5′ distance of cooperative binding sites may be explained by spatial proximity due to suitable secondary structure of the mRNA sequence.
Mechanistically, distance constraints between miRNA binding sites have been suggested to result from interactions between adjacent RNA-induced silencing complexes which stabilize target mRNA binding and increase the probability of occupancy (binding cooperativity).41,42 Another possibility would be a cooperative influence on the recruitment or effectiveness of further proteins leading to enhanced target degradation or repression (functional cooperativity42). Too close sites might be underrepresented due to steric hindrance of neighboring RISCs resulting in reduced effectiveness, possibly even lower than for a single site. On the other hand, if binding sites are too distant, the RNA-protein complexes might not be able to positively interact. However, it has to be elucidated if cooperative target regulation reflects a general concept of miRNA mediated mRNA regulation.
Here we present a systematic distance analysis of predicted miRNA target sites in human 3′-UTRs. Compared with randomized controls, distances shown by experimental studies to generate cooperative effects were enriched for naturally occurring miRNAs and miRNA binding sites. Further, functionally related miRNAs tend to bind more distance-defined cooperative targets, as the number increases for groups of miRNAs co-expressed in the same tissue or co-regulated in specific disease contexts. Our results, which are based on binding sites predicted by TargetScan are in good agreement with both a second computational target site predictor (miRanda/mirSVR) and experimentally verified miRNA interaction sites derived from HITS-CLIP or PAR-CLIP experiments.
Our findings support the importance of inter-site distance as a parameter defining miRNA-mediated repression. The comprehensive analysis of multiple miRNAs per target rather than miRNA-mRNA pairs appears essential to exploit disease-associated miRNAs and respective targets suitable for therapeutic purposes. To facilitate further research in miRNA cooperativity we developed miRco, a public web application that predicts cooperative miRNA-target interactions based on inter-site distance constrains: mips.helmholtz-muenchen.de/mirco.
Results
MiRNA target sites are enriched within cooperativity-promoting distance
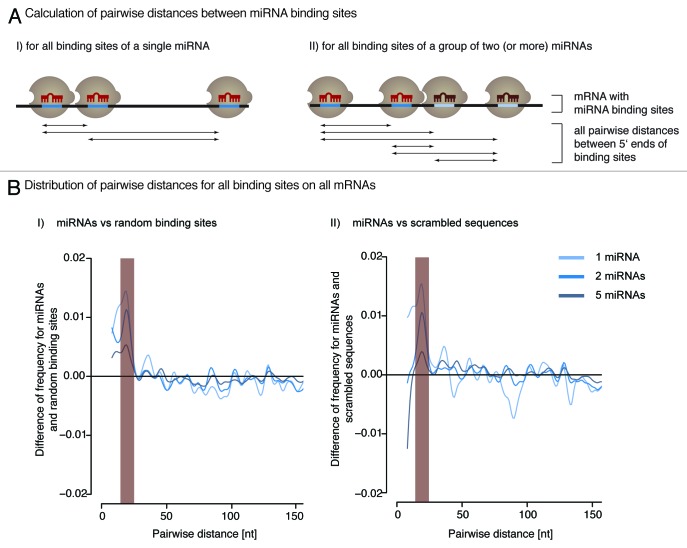
If a cooperative mode of action was functionally relevant, then cooperativity-promoting distance between target sites should be statistically overrepresented for intact binding sites of miRNAs. To test this, we computed the distribution of pairwise distances between predicted binding sites of evolutionary conserved human miRNAs. MiRNA targets were predicted using TargetScan, version 6.2.45 The data set contained 1,537 conserved human miRNAs. We calculated the distribution of distances between all binding sites of each miRNA individually. In addition, we determined distances for all binding sites of groups of two and five miRNAs (Fig. 2A). These group sizes have been defined in order to analyze combinatory effects. They were sampled 1,000 times from the complete set of miRNAs.
Figure 2. Naturally occurring miRNA binding sites are more frequently spaced within the cooperativity range (15‒26 nucleotides) than expected by chance. (A) The pairwise distance between all binding sites of a single (I) or multiple (II) miRNAs is calculated for each mRNA 3′-UTR. (B) The distribution of pairwise distances shows an enrichment of the cooperativity permitting distance for miRNAs compared with randomly picked sites (I) and predicted binding sites of scrambled sequences (II) (p < 2.2 × 10−16, Wilcoxon Rank Sum test).
To test for statistical significance, the results were compared with two different null models: (1) randomly chosen binding sites and (2) predicted target sites for scrambled miRNA-like sequences. For the first null model, we randomly selected target sites in a sequence-independent manner and, thus, generated artificial target sets with random binding positions. We picked random positions from the complete set of real human 3′-UTRs. The number of sites was normalized to predictions for human miRNAs. For the second, we designed arbitrary sequences of 22 nucleotides with the constraint that they are not similar to known miRNAs. We predicted targets with TargetScan and kept only those results that have a similar number of targets than native human miRNAs.
The distribution of all pairwise distances significantly differed for endogenous miRNAs and randomly selected binding sites in the range of 15‒26 nucleotides (Fig. 2B, p value < 2.2 × 10−16). This is in accordance with experimental findings (Fig. 1B) and is hence referred to as cooperativity range. This holds true for individual as well as for combinations of two and five different miRNAs. The enrichment of miRNA binding sites shows a peak for an inter-site distance of ~21 nucleotides (i.e., when two miRNAs bind in immediate vicinity). The distance distribution of predicted binding sites for scrambled sequences was also found to be different from miRNAs, again with significant underrepresentation within the cooperativity range (Fig. 2B).
In summary, when only small distances are considered (< 3 miRNA lengths), predictions for randomly picked sites and scrambled sequences produced similar results, while predicted target sites for human miRNAs displayed significant enrichment of inter-site spacing between 15‒26 nucleotides. These findings correlate with previous studies (Fig. 1B) and, thus, we used this window of inter-site spacing in subsequent analyses to determine cooperatively regulated miRNA targets.
HITS-CLIP and PAR-CLIP data sets show cooperativity
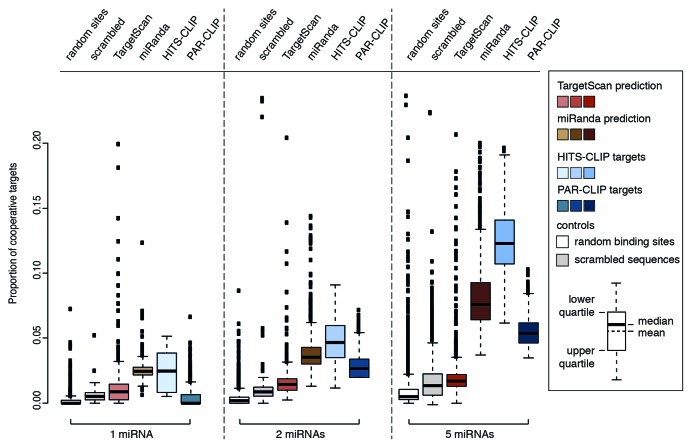
We calculated the fraction of targets that are potentially regulated in a cooperative manner for four distinct sets of miRNA targets: (1) TargetScan predictions for human miRNAs,44 (2) miRanda/mirSVR predictions for human miRNAs as a second target prediction tool,26,27 (3) experimentally validated data from a HITS-CLIP28 and (4) from a PAR-CLIP study.29 All data sets were compared with random target sites and random sequences. As above, we analyzed single as well as groups of two and five miRNAs to take combined activity into account.
Looking at target prediction, both tools show significantly more cooperative targets than random binding sites and random sequences (p-values < 2.2 × 10−16). This holds true for single and groups of two and five miRNAs. Interestingly, in all cases, miRanda/mirSVR has a higher percentage of cooperative targets. We find a mean of 2%, 4% and 8% for miRanda/mirSVR and a mean of 1%, 1.7% and 2.2% for TargetScan.
The difference between miRNAs and controls increased with the number of miRNAs and we found the largest difference for groups of five naturally occurring miRNAs. This indicates that targets controlled by multiple different miRNAs are more frequently regulated in a cooperative fashion than mRNAs with multiple binding sites for an identical miRNA species.
Recently, biochemical methods to identify miRNA binding sites on a genome-wide scale have been developed. For an experimental validation of our in silico results, we analyzed the published HITS-CLIP and PAR-CLIP data sets. The former contains mRNA binding sites for the 20 most abundant miRNAs from mouse brain while the latter contains 47 human miRNAs. Both HITS-CLIP and PAR-CLIP identifie similar numbers of targets as TargetScan and miRanda/mirSVR and, thus, allow for comparison with our findings for computational prediction.
We retrieved all binding sites for both data sets and calculated the proportion of cooperative targets (Fig. 3, brown and blue boxes). For a single miRNA, only HITS-CLIP shows significantly more cooperative targets than controls with a mean of 2.5% compared with 0.5% and 0.2% for random sequences and random sites. PAR-CLIP data shows a mean of 0.4% cooperative targets and thus is not different from controls.
Figure 3. The fraction of cooperative targets per total targets grows for increasing numbers of miRNAs. Analysis of cooperative targets was performed with computationally predicted (TargetScan, red; miRanda, brown) and experimentally identified (HITS-CLIP, light blue; PAR-CLIP, dark blue) target sets. The proportion of cooperative targets is plotted for single miRNAs and sampled groups of two and five miRNAs. The mean is always higher for existing miRNAs than either of the controls (p < 2.2 × 10−16, tested with Wilcoxon Rank Sum test), except PAR-CLIP data for single miRNAs. This indicates that they are more often located in a potential cooperative distance than expected by chance.
This picture changed when groups of two or five miRNAs were taken into account. While both data sets retrieved by experimental methods exhibited the same tendency as the target prediction tools, both HITS-CLIP and PAR-CLIP showed a stronger gain. The mean fraction of cooperative targets increased to 4.7% and 12.5% for HITS-CLIP and to 3.4% and 5.7% for PAR-CLIP. The mean percentage of coperative targets for the controls increased only to 0.4%/0.8% for random positions and 1%/1.3% for random sequences. In general, miRanda/mirSVR resembled HITS-CLIP and PAR-CLIP more closely while for TargetScan the number of cooperative targets was slightly lower.
These results show that cooperative regulation is likely to involve different miRNAs. Most importantly, the data for computational target prediction was confirmed with two independent sets of experimentally validated miRNA targets.
Functionally related miRNAs show an enrichment of target sites within cooperativity permitting distance
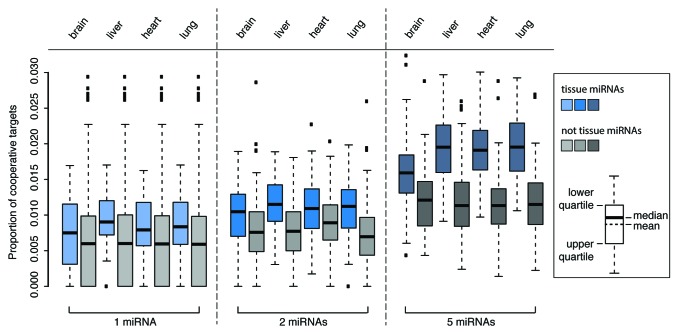
As shown above, endogenous miRNAs are more likely to posess target sites in a cooperativity range than randomly picked sites or scrambled sequences. If cooperativity is relevant in miRNA-mediated gene regulation, then functionally related miRNAs may share more cooperative targets than others. As most miRNAs are not comprehensively understood with respect to function, the field widely relies on two criteria that may be indicative of functional relation: (1) Co-expression of miRNAs within a particular tissue and (2) co-regulation in a common disease context. To put the first criterion to the test, we used the miRNA expression profiling database mimiRNA.46 For the second, we employed PhenomiR,47 a database of differentially regulated miRNA expression in diseases. For all miRNAs in both databases, targets were retrieved from TargetScan as described before.
The mimiRNA database employs normalized human miRNA expression profiles from four different sources: Sequencing data from the miRNA Atlas,48 quantitative real-time PCR data from Gaur et al.49 and Lee et al.50 and microarray and deep sequencing data from the Gene Expression Omnibus (GEO).51 The complete data set for 188 different tissues was used to calculate the proportion of cooperative targets among all targets for single and groups of two and five miRNAs. Co-expressed miRNAs were compared with all non-expressed miRNAs as a control. As shown exemplary for brain, liver, heart and lung (Fig. 4), miRNAs that are co-expressed in a tissue target more mRNAs in a potentially cooperative manner than miRNAs that are not co-expressed in a particular tissue (one-sided Wilcoxon Rank Sum test with p < 2.2 × 10−16). Moreover, the difference increases for groups of two and five co-expressed miRNAs, suggesting that these co-expressed miRNAs are in a functional relation with each other.
Figure 4. Functionally related miRNAs show an enrichment of target sites within the cooperativity range. Fraction of potentially cooperative targets for miRNAs in four exemplary tissues (blue) compared with a control set of miRNAs not expressed within the respective tissue (gray). Targeting within the cooperativity permitting distance is over-represented for co-expressed miRNAs (one-sided Wilcoxon Rank Sum test with p < 2.2 × 10−16).
To test for second presumed indicator of functional interrelation, i.e., co-regulation in disease as another indicator for functional relationship, we applied the latest release of PhenomiR, a database, which comprises 126 diseases and 615 associated miRNAs. Again, the fraction of cooperative targets was determined for single and sampled combinations of two and five miRNAs. The complete set of non-regulated miRNAs was used as control for each disease. Targeting with at least two binding sites within the cooperativity range was more often found for co-regulated miRNAs than for control groups not associated with a particular disease (one-sided Wilcoxon Rank Sum test with p < 2.2 × 10−16) (data not shown). Notably, this holds true for all diseases covered by PhenomiR. Similar to co-expressed miRNAs, we found an increase of the difference between disease-associated miRNAs and controls for groups of two and five miRNAs.
miRco: A tool to predict miRNA targets with binding sites in a cooperativity-permitting distance
We have shown that miRNA binding sites are more often located in the cooperativity range than expected by chance. Additionally, functionally related miRNAs show an enrichment of cooperative binding sites. Still, the biological relevance of cooperativity in miRNA function has to be shown experimentally. To support further research in this topic, we developed the web application miRco, a tool to predict potentially cooperative miRNA interactions and their mRNA targets. Upon user input of miRNAs and distance allowance between miRNA binding sites, miRco identifies mRNAs that may be controlled by cooperative miRNA activities. Additionally, miRco can find all miRNAs that bind cooperatively to a given list of genes or mRNAs (Fig. 5A and B). To predict mRNAs that are cooperatively regulated, miRco searches by default for target sites within a distance of 15−26 nucleotides between two consecutive miRNA 5′ ends. As described above, this setting was chosen based on our findings and reported experimental data.23,41-43 Alternatively, the user may define a custom lower and upper limit of the distance. The tool includes miRNAs and mRNAs from human, mouse and rat. Target predictions are obtained from the current release of TargetScan (version 6.2).
Figure 5. Functionality of the miRco web application. (A) A search for cooperative miRNA-target interactions is performed by selecting miRNA candidates, relevant target genes or both. The user is able to specify parameters for the range in which the spacing between two adjacent miRISC binding sites (d) is assumed to lead to cooperative target repression. Default values are 15‒26 nucleotides. Predictions for three species are available: human, mouse and rat. (B) Screenshot of the user interface of the online tool.
First, the user is asked to choose the species for which the search is to be performed. Then, a list of miRNAs, or genes, or both may be submitted. If either miRNAs or genes are left blank, the complete data set is used for analysis. Our tool is connected to the PhenomiR database. The user can select a disease annotated in PhenomiR and input a set of disease-associated miRNAs (Fig. 5B). The output of miRco is presented as a list of target genes with corresponding binding sites in the aforementioned cooperativity range. Data are initially sorted based on the context+ score calculated by TargetScan and can subsequently be listed by target gene symbol and average distance between the binding sites. Furthermore, the result table can be filtered for the occurrence of one or multiple miRNAs within the list of candidate mRNAs.
The improved data set of the latest TargetScan release is a solid fundament for prediction of cooperative targets for three major model organisms used for medical research. miRco may serve as a hypothesis-generator to aid further research on the mechanisms underlying concerted miRNA-mediated target regulation.
Discussion
The study presented here addresses a largely unresolved question in miRNA research: Do miRNAs confer physiological effects on their own, or do they function in a concerted, possibly cooperative manner?
Literature provides certain evidence: Experiments in which a single small regulatory RNA binds to a single site within a mRNA often fail to show effects (e.g. refs. 23 and 40). On the other hand, studies indicated that miRNA-mediated target regulation is particularly effective if several miRNAs bind within a close distance.23,41-43 However, these results rely on expression of artificial reporter constructs and do not provide comprehensive evidence that cooperativity is a general principle of miRNA-mediated target regulation.
In principle, one way to explain the basic concept of miRNA cooperativity is that proximity of binding sites on mRNAs stabilizes miRISCs’ binding to their mRNA targets, leading to an increased silencing effect. This proximity concept has already been discussed in literature and several of our observations provide further support for it on a genome-wide scale: First, we showed that mRNAs with more than one miRNA site are more likely to have these sites placed in cooperativity-promoting distance (15−26 nucleotides, 5′-to-5′) than randomized controls. Interestingly, the peak distance of ~21 nucleotides reflects binding of two miRNAs in direct neighborhood. Second, by in silico prediction (TargetScan, miRanda/mirSVR), as well as experimentally supported (HITS-CLIP, PAR-CLIP), we retrieved more mRNAs with miRNA sites in cooperativity range than from control setups. Third, the higher proportion of such mRNAs goes along with the co-regulation of miRNAs in tissue as well as similar disease context, underscoring the suspected functional interplay of these miRNAs on the respective mRNAs. The enrichment of miRNA binding sites in cooperativity-promoting distance speaks for a prevalently concerted, maybe cooperative way of miRNA target regulation.
However, the mechanisms of targeting are complex. Apparently, the type of Argonaute protein involved in a particular silencing complex has great influence on the nature of target regulation.42 For example AGO 1 and AGO 2 show distinct characteristics with respect to the distance requirement between binding sites leading to cooperative targeting.42 While bulged binding of miRNAs within AGO 1-complexes shows cooperativity only for adjacent binding sites, bulged sites of AGO 2-containing RICSs can act cooperatively in adjacent as well as in extended compositions. Consequently, the cell-specific proportion of the different AGO subtypes as well as the concentration of other potential effector proteins may be important. Furthermore, the sequence context around miRNA sites might affect cooperative actions of miRISCs, with other protein binding sites and secondary structure as the most likely determinants.
Therefore, the next step in studying miRNA cooperativity will be to more comprehensively analyze it in a biological context. Analysis of several instead of single miRNAs and their potential cooperativity could lead to a better understanding of the complex interplay of miRNAs and genetic networks in health and disease.
Cooperativity as a moderator of strongly increased effects would be interesting for the therapeutic use of miRNAs: If two miRNAs downregulate an mRNA target cooperatively, a lower level of expression might suffice to exert the designated effect. This would potentially decrease side effects of the miRNAs or miRNA mimics and, thereby, lead to a more tolerable treatment.
In addition, combinations of miRNAs could be employed to improve experimental protocols. Similar to the idea of decreased side effects of therapeutic miRNAs, the combination of different miRNAs might increase the specific effect on the targets of interest. Interestingly, the combined activity of multiple miRNAs has recently been reported to facilitate the reprogramming of fibroblasts to cardiomyocyte-like cells52 as well as the induction of pluripotent stem cells (iPSCs).53,54
Recently, several studies highlighted the interaction of AGO/miRNAs with other RNA binding proteins (RBP).55-57 In the future, the concept of cooperativity may extend to all RBPs in order to better predict mRNA regulation.
In the context of this work, we also developed miRco (mips.helmholtz-muenchen.de/mirco), a web application meant to aid experimental research into the cooperative action of miRNAs. It predicts potentially cooperatively targeted mRNAs based on binding site distances and, thus, might help to identify key regulatory miRNA-mRNA networks. miRco serves as a starting point for wet lab scientists: It allows one to input miRNAs and search for cooperative targets. In addition, the user can specify a set of genes and find all miRNAs that target these genes in a cooperative fashion. This dual approach helps to narrow down lists of candidate genes and miRNAs and makes it more feasible to test cooperativity in a complex biological context.
Taken together, our data indicate that cooperativity of miRNA-target interaction is a wide-spread phenomenon that may play an important role in miRNA-mediated gene regulation.
Materials and Methods
Criteria for the prediction of cooperativity
Cooperativity of two miRNAs is defined by the distance between the 5′-starts of their binding sites. We used 15 nucleotides as the lower and 26 nucleotides as the upper limit of the cooperative distance, following experimental studies of distance-dependent cooperative effects and our data showing an enrichment of binding sites for human miRNAs in this window.
To determine whether a mRNA may be cooperatively regulated, we take a single gene, acquire all binding sites of a given set of miRNAs on this mRNA and cluster them in groups where the distance between two adjacent sites lies within the cooperativity interval. If at least two binding sites fulfill this criterion, a mRNA is considered to be potentially regulated in a cooperative manner.
All data sets are stored in a MySQL database containing tables for genes, miRNAs and binding sites as well as their relations. Analyses are performed with Python programs combined with data plotting using R.
MiRNA target prediction
We used computational target prediction of human miRNAs from TargetScan22,45 release 6.2 and miRanda/mirSVR release August 2010.26,27 For TargetScan, we used the predictions for conserved miRNAs and targets. Scores of target sites are the context+ scores calculated by TargetScan. The release 6.2 contains 1,536 conserved human miRNAs and prediction is performed on a multiple sequence alignment of 18413 3′-UTRs from 23 species. For miRanda/mirSVR, we used the predictions for conserved miRNAs with a good mirSVR score. The release contains 249 human miRNAs.
Random distribution of target sites
Randomly distributed target positions were used as a null model for cooperativity. We picked random positions within the real set of human 3′-UTRs. UTR data for all human genes (assembly GRCh37.p10) was downloaded from ENSEMBL BioMart (www.ensembl.org/). The number of positions per UTR was normalized to lie within the range of TargetScan predictions. This approach is completely independent of miRNAs, their sequences and pairing determinants. Thus, this represents the most basic null model for binding site allocation and does not rely on any prior knowledge.
Random miRNA-like sequences
To augment the basic random position control, we generated 1,000 completely random 22 nucleotides long sequences. We only used sequences which are not known human miRNAs and do not contain seeds (nucleotide 2−8) of known human miRNAs. We predicted targets for these seeds with the TargetScan 6.2 software and the UTR data provided by TargetScan. For the subsequent analyses, only random sequences that produce the same numbers of targets (i.e., between 10−2,719) as human miRNAs were taken into account.
Sampling of groups
For analyses using single miRNAs, the complete data set was considered. Groups of two and five miRNAs or controls were sampled randomly 1,000 times from the complete set with no recurrence.
HITS-CLIP data set
The data set of Chi et al.28 is available at ago.rockefeller.edu, including mapping of miRNA binding sites onto genomic positions. The authors of this study used neocortex of P13 mouse brain, crosslinked RNA binding proteins and RNA with UV irradiation and immunoprecipitated AGO-RNA complexes. Subsequently, RNA was purified and sequenced. Computational analysis produced a miRNA-mRNA interaction map. We used the mapping on mouse genome assembly mm9.
PAR-CLIP data set
The data set of Hafner et al.29 is available through starBase (starbase.sysu.edu.cn), a database providing gene mappings for a wide range of CLIP experiments.58 We used the “target site interaction” tool of starBase with settings for at least one microRNA read and “stringent miRNA targets” as described in the starBase publication.
Statistics
The distributions of pairwise distances (in a given distance window) as well as the percentage of cooperative targets were tested for a significant difference between miRNAs and controls with a one-sided Wilcoxon Rank Sum test.59 We used the wilcox.test function in the “stats” package of the R statistical computing software with a confidence interval of 0.95 to calculate p values. P values < 2.2 × 10−16 occur due to the limits in floating point precision in R.
miRco web application
The miRco web tool is implemented as a JAVA EE application running on a Tomcat 6 servlet engine, using the same MySQL database described above. It employs TargetScan release 6.2. For a given set of mRNAs and miRNAs, miRco produces groups of miRNA binding sites that fulfill the user set distance criteria. Whenever two binding sites are at the exact same position or overlap (i.e., their distance is smaller than the lower limit), the binding site with the best context+ score calculated by TargetScan is used.
Acknowledgments
We would like to thank Nikola Müller, Carsten Marr and Dominik Lutter for helpful comments and proofreading of the manuscript. This work was supported by the Bundesministerium für Bildung und Forschung (MedSys project “LungSys” and programmes m4, Personalized Medicine and ANR-BMBF-01KU0902A). This work was further supported in part by grants from the Fondation Leducq (to S.E.); the Bavarian Ministry of Sciences, Research and the Arts in the framework of the Bavarian Molecular Biosystems Research Network (to S.E. and F.T.), the Helmholtz Alliance on Systems Biology (project “CoReNe”), the German Science Foundation (DFG) within the SPP 1395 (InKoMBio) and the Studienstiftung des Deutschen Volkes (to A.R.).
Glossary
Abbreviations:
- miRNA
microRNA
- miRISC
miRNA-induced silencing complex
- AGO
Argonaute protein
- 3′-UTR
3′ untranslated region
- siRNAs
short interfering RNAs
- iPSCs
induced pluripotent stem cells
- RBP
RNA-binding protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24955
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–8. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 3.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 4.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 6.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon Y-J, Ngankeu A, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2012;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–63. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–30. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan SD, Krüger M, Willmes DM, Redemann N, Wunderlich FT, Brönneke HS, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13:434–46. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 10.Baxter D, McInnes IB, Kurowska-Stolarska M. Novel regulatory mechanisms in inflammatory arthritis: a role for microRNA. Immunol Cell Biol. 2012;90:288–92. doi: 10.1038/icb.2011.114. [DOI] [PubMed] [Google Scholar]
- 11.Jentzsch C, Leierseder S, Loyer X, Flohrschütz I, Sassi Y, Hartmann D, et al. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2012;52:13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Mestdagh P, Boström A-K, Impens F, Fredlund E, Van Peer G, De Antonellis P, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol Cell. 2010;40:762–73. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell. 2010;38:140–53. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, et al. MiR-378 Controls Cardiac Hypertrophy by Combined Repression of MAP Kinase Pathway Factors. Circulation. 2013 doi: 10.1161/CIRCULATIONAHA.112.000882. In press. [DOI] [PubMed] [Google Scholar]
- 15.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–74. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 21.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lal A, Thomas MP, Altschuler G, Navarro F, O’Day E, Li XL, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowarsch A, Marr C, Schmidl D, Ruepp A, Theis FJ. Tissue-Specific Target Analysis of Disease-Associated MicroRNAs in Human Signaling Pathways. Morris RJ, ed. PLoS ONE 2010;5(6):e11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowarsch A, Preusse M, Marr C, Theis FJ. miTALOS: analyzing the tissue-specific regulation of signaling pathways by human and mouse microRNAs. RNA. 2011;17:809–19. doi: 10.1261/rna.2474511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 35.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 36.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;267:529–35. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 38.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 39.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–91. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–42. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saetrom P, Heale BSE, Snøve O, Jr., Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–42. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–69. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill AV. A new mathematical treatment of changes of ionic concentration in muscle and nerve under the action of electric currents, with a theory as to their mode of excitation. J Physiol. 1910;40:190–224. doi: 10.1113/jphysiol.1910.sp001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–46. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie W, Flamant S, Rasko JEJ. mimiRNA: a microRNA expression profiler and classification resource designed to identify functional correlations between microRNAs and their targets. Bioinformatics. 2010;26:223–7. doi: 10.1093/bioinformatics/btp649. [DOI] [PubMed] [Google Scholar]
- 47.Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, et al. PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 2010;11:R6. doi: 10.1186/gb-2010-11-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–68. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 50.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–69. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–8. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Jiang P, Coller H. Functional Interactions Between microRNAs and RNA Binding Proteins. MicroRNA. 2012;1:70–9. doi: 10.2174/2211536611201010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci. 2012;13:372–9. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–9. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J-H, Li J-H, Shao P, Zhou H, Chen Y-Q, Qu L-H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bauer DF. Constructing Confidence Sets Using Rank Statistics. J Am Stat Assoc. 1972;67:687–90. doi: 10.1080/01621459.1972.10481279. [DOI] [Google Scholar]