Abstract
Human Enterovirus 71 (EV71) is an emerging pathogen of infectious disease and a serious threat to public health. Currently, there are no antivirals or vaccines to slow down or prevent EV71 infections, thus underscoring the urgency to better understand mechanisms of host-enterovirus interactions. EV71 uses a type I internal ribosome entry site (IRES) to recruit the 40S ribosomal subunit via a pathway that requires the cytoplasmic localization of hnRNP A1, which acts as an IRES trans-activating factor. The mechanism of how hnRNP A1 trans activates EV71 RNA translation is unknown, however. Here, we report that the UP1 domain of hnRNP A1 interacts specifically with stem loop II (SLII) of the IRES, via a thermodynamically well-defined biphasic transition that involves conserved bulge 5′-AYAGY-3′ and hairpin 5′-RY(U/A)CCA-3′ loops. Calorimetric titrations of wild-type and mutant SLII constructs reveal these structural elements are essential to form a high-affinity UP1-SLII complex. Mutations that alter the bulge and hairpin primary or secondary structures abrogate the biphasic transition and destabilize the complex. Notably, mutations within the bulge that destabilize the complex correlate with a large reduction in IRES-dependent translational activity and impair EV71 replication. Taken together, this study shows that a conserved SLII structure is necessary to form a functional hnRNP A1-IRES complex, suggesting that small molecules that target this stem loop may have novel antiviral properties.
Keywords: Enterovirus 71, Internal Ribosomal Entry Site, Protein-RNA interactions, RNA structure, hnRNP A1
Introduction
Enterovirus 71 (EV71), the major etiological agent of hand foot and mouth disease, is an emerging threat to public health.1-3 EV71 infections typically manifest with only mild illness; however, more serious infections can lead to severe neurological disorders, morbidity and death.1,2,4,5 In a recent outbreak, 78 Cambodian children were infected with EV71 of which 54 died from complications associated with the virus (WHO, Global Alert and Response report July 2012). There are currently no vaccines or antivirals to prevent EV71 outbreaks, thus underscoring the urgent need to better understand mechanisms of host-enterovirus interactions.
EV71 is a positive sense, single-stranded RNA Enterovirus that belongs to the Pircornaviridae family. Like other picornaviruses, infection proceeds via a cytoplasmic replication cycle where the approximately 7,500-nt RNA genome serves as template for negative strand amplification, viral protein synthesis and genomic RNA for new virions.6,7 Given their limited coding capacity, picornaviruses coordinate these different viral RNA functions through mechanisms that co-opt host proteins.6,8,9 Upon viral entry, EV71 initiates translation via a type I internal ribosomal entry site (IRES) located within the 5′-non-coding region (5′-NCR).10 The 5′-NCR is a highly structured multifunctional RNA element composed of six stem loops (SLI-SLVI; Fig. 1). Stem loop I facilitates viral genome replication, whereas stem loops II‒VI function as the basic IRES-active unit, recruiting ribosomal components in a cap-independent pathway.11 The molecular mechanism of EV71 RNA translation remains largely unknown; although, earlier studies have implicated several non-canonical host initiation proteins, collectively known as ITAFs, as key IRES trans acting factors in modulating internal ribosomal recruitment.12 The current dogma supports a model where ITAFs reshape and stabilize the IRES structure in a conformation that facilitates ribosome assembly.13-20 A molecular description of how RNA structure remodeling by ITAFs modulates internal translation efficiency is lacking, however.
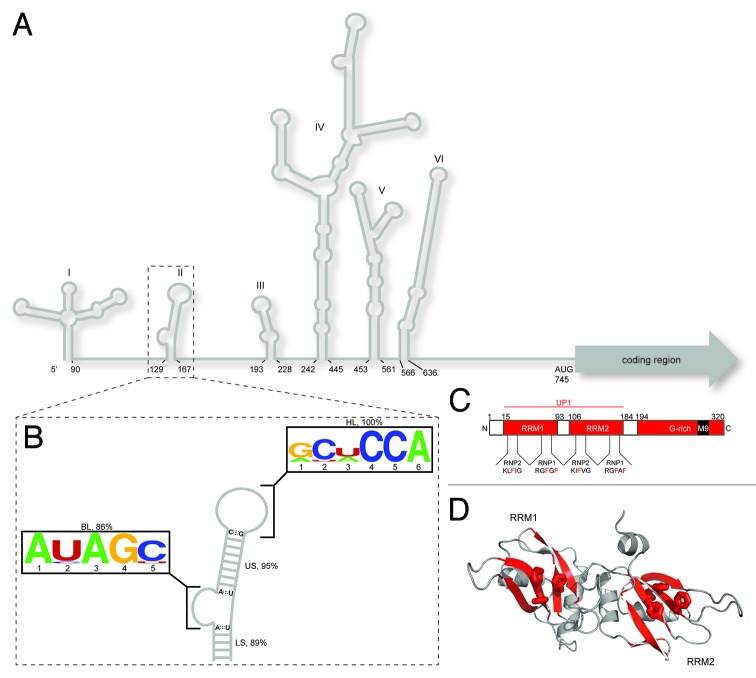
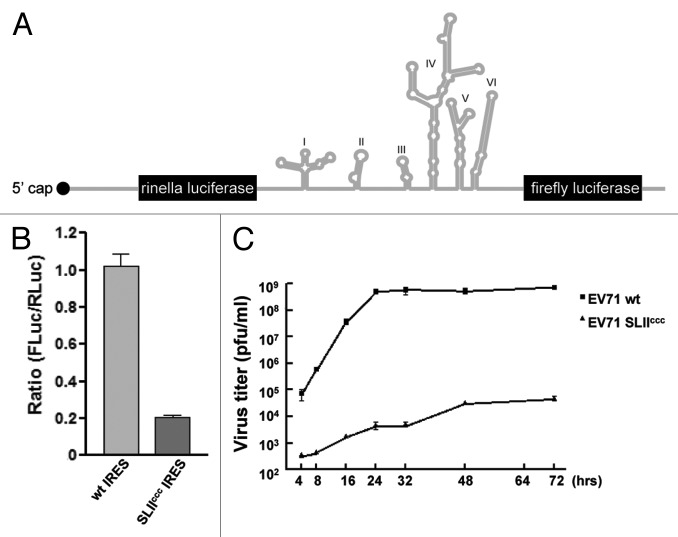
Figure 1. Overview of cis and trans acting factors that affect EV71 RNA translation. (A) Schematic representation of the RNA secondary structure of the EV71 5′ NCR. Stem loop I functions in negative strand synthesis, whereas stem loops II‒VI comprise the type I IRES element. (B) Consensus secondary structure of the SLII domain derived from sequence alignments (n = 147; see SI for accession codes) coupled with RNA structure prediction. SLII can be divided into four conserved elements of secondary structure: a five bp lower stem (LS, 89%); a five nt bulge loop (BL, 86%); a nine bp upper stem (US, 95%) and a six nt hairpin loop (HP, 100%), where percent values correspond to conservation of secondary structural elements not sequence. The BL and HL closing base pairs are invariably AU and CG, respectively. Consensus sequence motifs for the bulge and hairpin loops are depicted using the WebLogo format.55 (C) Domain organization of hnRNP A1 showing the N-terminal RNA recognition motifs (collectively known as UP1) and the C-terminal glycine rich domain. Signature RNP motifs are designated for RRM1 and RRM2 (D) Cartoon representation of the UP1 structure (PDB 2LYV) where aromatic residues from RNP1 and 2 are drawn as sticks.
The multifunctional RNA-binding protein, hnRNP A1, was shown to stimulate EV71 IRES-mediated translation.21 HnRNP A1 consists of two N-terminal RNA recognition motifs (RRM1 and RRM2, collectively known as UP1) that interact specifically with nucleic acids and a C-terminal glycine-rich domain that contributes to protein-protein contacts, among other functions.22,23 Like most RRM-containing proteins, the UP1 subdomain adopts a mixed α/β fold where the four strand β sheet surfaces of RRM1 and RRM2 pack tightly against two pair of α helices, respectively (Fig. 1). Nucleic acid-binding specificity is conferred primarily through signature RNP motifs located in β strands 1 and 3, with additional RNA contacts directed through connecting loops.24 HnRNP A1 has been associated with a number of cellular functions, most notably as a regulator of alternative splicing.22,23 The nucleic acid binding properties of hnRNP A1 have been studied extensively, usually with model RNA or DNA oligomers that lacks well-defined secondary structure.25-31 However, hnRNP A1 is known to regulate RNA processes where the RNA targets contain elements of higher-order structure.12,21,32-38 Indeed, the EV71 IRES is a highly structured target for hnRNP A1 (Fig. 1), where an understanding of the influence of RNA structure to molecular recognition is lacking.
Lin et al. demonstrated that cells infected with EV71 show a cytoplasmic redistribution of hnRNP A1, and siRNA knockdowns of hnRNP A1 impair IRES mediated translation, RNA synthesis and markedly reduce enteroviral replication.21 IRES mapping studies localized the hnRNP A1-binding sites to stem loops II and VI; however, the specific binding surfaces, mechanisms of interaction and correlative structure-function properties remain unknown.21 To gain more insight into how hnRNP A1 trans activates EV71 RNA translation, we performed a systematic structural analysis of SLII and characterized its mode of binding to the UP1 domain of hnRNP A1. Here, we show that the ability to form a 40-nt SLII domain, interrupted by a 5-nt bulge and capped by a 6-nt hairpin, is a conserved structural feature among enteroviral IRES elements. The bulge and hairpin loops act synergistically to form a (UP1)2SLII complex, characterized by two distinct thermodynamic transitions. Notably, mutations that disrupt the apparent biphasic binding transition correlate with a significant decrease in IRES-dependent translation and impair EV71 replication. Taken together, this study shows that a conserved EV71 SLII structure actively defines a functional hnRNP A1-IRES complex, and further suggests that molecules that can disrupt this interaction may have novel anti-enteroviral properties.
Results
Stem loop II of the EV71 IRES folds into a phylogenetically conserved and independent structural domain
Phylogenetic alignments coupled with computational prediction of human EV71 sequences (n = 147; see SI for accession codes), corresponding to the SLII region, revealed a high level of RNA structural conservation. Overall, the consensus SLII secondary structure was conserved in 95% of the sequences analyzed (Fig. 1). Stem loop II can be sub-divided into four elements of secondary structure: a five base-paired lower stem (LS), a five nucleotide bulge loop (BL), a nine base-paired upper stem (US) and a six nucleotide hairpin loop (HL). Of the aligned sequences, the length of the US and HL were 95% and 100% conserved, respectively. The primary sequence of the HL also showed conserved features, allowing definition of a consensus motif: 5′-RY(U/A)CCA-3′. Using a smaller data set (n = 57) that included Enteroviridae and Rhinoviridae strains, Klinck et al. determined a similar HL consensus motif, 5′-ANNCCA-3′.39 Of note, the CCA motif was invariant using their smaller data set as well as in the much larger data set used here. Interestingly, the HL closing base pair for all aligned sequences corresponds to 5′-C:G-3′, which likely reflects favorable stacking energetics on the adjacent 5′-R:A-3′ mismatch.40 A similar high level of structural conservation was observed for the LS and BL. The lengths of the LS and BL were 89% and 86% conserved, respectively. The consensus motif for the bulge loop determined here is 5′-A(Y/A)AGY-3′. Moreover, the BL closing base pairs are invariably 5′-A:U-3′ (Fig. 1). Taken together, the EV71 stem loop II folds into a phylogenetically conserved structural domain that exposes invariant BL (AGY) and HL (CCA) sequence motifs.
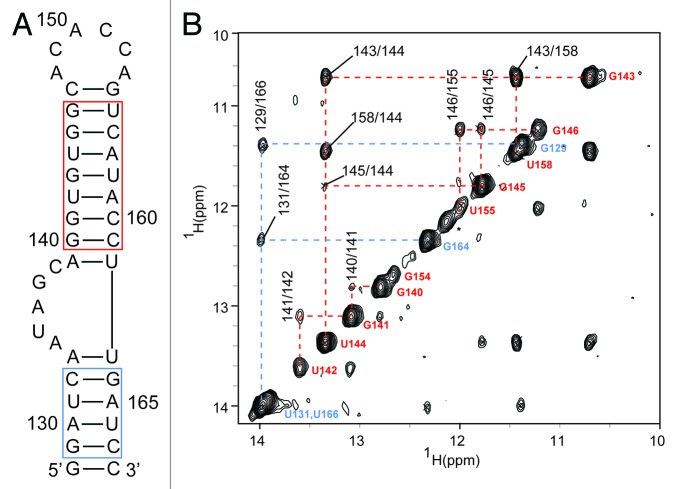
To determine if SLII is an independent folding IRES domain, we characterized a 41-nt construct (corresponding to strain 2231; Fig. 2) by two-dimensional NMR spectroscopy. The SLII2231 construct was prepared by in vitro transcription where a non-native terminal 5′-G:C-3′ pair was included to increase transcription yield.41 The oligomeric state of SLII2231 was analyzed under different solution conditions by resolving samples on non-denaturing PAGE (not shown). Under high monovalent cation concentration, SLII2231 migrated both as a fast and slow moving species, presumably representative of a hairpin and dimer conformation, respectively. Adjusting the buffer conditions (60 mM total K+) shifted the equilibrium primarily to the hairpin conformation (> 95%). Moreover, the line widths of the 1D H1 NMR signals at low ionic strength were sharp (not shown); indicating SLII2231 primarily adopts a single conformation under these conditions. The secondary structure of SLII2231 was confirmed using conventional 1H-1H nuclear Overhauser enhancement spectroscopy (NOESY) collected in water (283K; pH 6.5). As shown in Figure 2, sequential and long-range (G/U)NH-(G/U)NH nOe cross peaks can be traced for 7 out of 9-bps for US and 4 out of 6-bps for the LS. NOE cross peaks from the terminal 5′-G:C-3′ pair, and both BL closing 5′-A:U-3′ pairs were not observed due to rapid exchange with solvent. Strong nOe cross peaks were detected between the imino protons of G143/U158, indicating these nucleotides form a stable wobble pair. By comparison, the weaker nOe cross peaks between G146/U155 suggest these nucleotides form a less stable wobble pair, likely resulting from helical fraying at the adjacent HL junction (Fig. 2). Consistent with this conclusion, more intense nOe cross peaks are observed between G146/U155 for a SLII construct where the 6-nt HL loop was replaced with a GCGA tetraloop (Fig. S1C). This construct also gave well-defined stacking nOe interactions between G154 and the adjacent G146/U155 base pair. Collectively, the NMR chemical shifts and nOe cross peak patterns are consistent with the SLII secondary structure prediction and confirms that SLII is an independently folding IRES domain.
Figure 2. The EV71 stem loop II is an independent folding domain. (A) RNAStructure56 prediction of EV71 SLII from isolate 2231. The terminal GC pair is nonnative and was included to increase transcription yields. The blue and red boxes correspond to those helical stretches observed by NMR. (B) Six hundred megahertz 1H-1H NOESY spectrum of the imino protons of SLII2231 collected at 298K. Blue and red lines trace sequential imino assignments in the lower and upper helix, respectively.
RNA structural elements are necessary to form a high-affinity (UP1)2SLII2231 complex
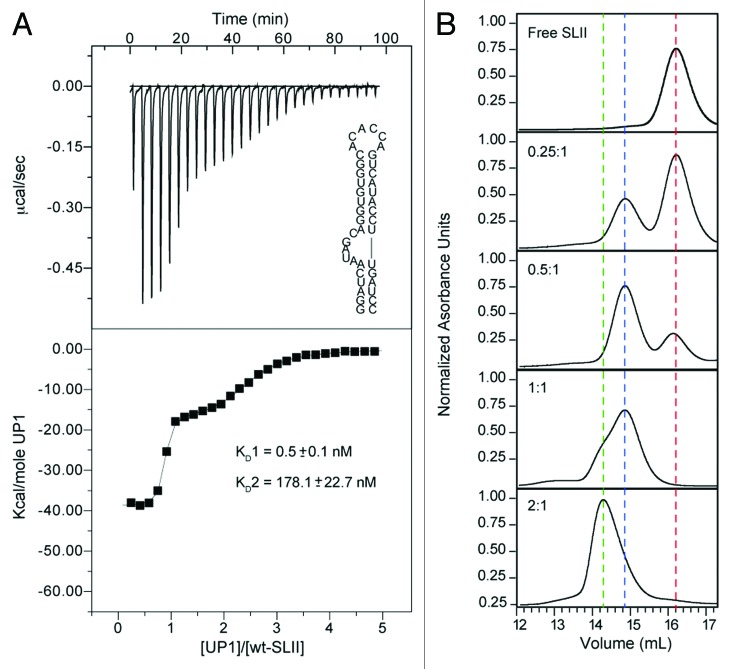
To gain insight into the thermodynamics of hnRNP A1-SLII2231 interactions, we performed calorimetric titrations with the UP1 sub-domain of hnRNP A1. UP1 is composed of two RRM domains connected through a 13 amino acid linker, and previously we showed that both RRMs interact cooperatively to form a high-affinity 1:1 complex with the HIV-1 exon splicing silencer 3 (ESS3) stem loop.33Figure 3A shows the results of the UP1-SLII2231 calorimetric titration. Under the conditions used here (see Methods), the titration profile reproducibly displayed a biphasic transition. Fitting of the normalized data to a non-sequential (independent sites) model isotherm provided the best physical description of the binding event. Both transitions are characterized by favorable changes in enthalpy and opposed by unfavorable changes in entropy (Table 1). The thermodynamic signatures are consistent with recent calorimetric studies of other RRM-RNA systems, which show large exothermic enthalpies associated with complex formation.42 Control reverse titrations of SLII2231 into UP1 gave a thermodynamic signature in general agreement with the forward titration (Fig. S2); however, the binding profile cannot be directly compared due to differences in the number of apparent receptor sites of each titrand (presumably two for SLII2231 and one for UP1). Nevertheless, both the forward and reverse titrations are consistent with a binding event that is more complex than a simple one-to-one stoichiometry (Fig. 3A; Fig. S2).
Figure 3. UP1 interacts with SLII2231 through distinct modes of recognition. (A) Representative calorimetric titration of (His)6-UP1 into SLII2231 at 298K. The upper panel shows the raw thermogram along with the SLII2231 secondary structure. The processed isotherm is shown in the lower panel along with the nonlinear least squares fit of the data to a two-independent sites binding model. Average values and standard deviations of the dissociation constants (two replicates) are provided. (B) Analytical size exclusion chromatographic titration of (His)6-UP1 into SLII2231. Titrations were performed by pre-incubating a fixed amount (5 uM) of SLII2231 with increasing amounts of (His)6-UP1 until a final molar ratio of 2:1 (taken as concentration of binding sites on SLII2231) was reached. Complexes were resolved on a Superdex 200 10/300 GL column (GE Healthcare Life Sciences). Vertical dashed lines are drawn to help visualize the different molecular species present during the titration: red, free SLII2231; blue, UP1-SLII2231 and green, (UP1)2SLII2231.
Table 1. Thermodynamic parameters for the binding of UP1 to wild-type SLII and structural mutants.
| SLII construct | Kd (nM) | n | ΔG° | ΔH° | -TΔS° |
|---|---|---|---|---|---|
| WT | |||||
| site 1 | 0.5 ± 0.1 | 0.7 ± 0.1 | -12.7 ± 0.1 | -38.3 ± 0.5 | 25.6 ± 0.6 |
| site 2 | 178.1 ± 22.7 | 1.5 ± 0.2 | -9.2 ± 0.1 | -17.6 ± 0.3 | 8.4 ± 0.4 |
| SLIIΔL ΔBL | 28.4 ± 2.0 | 0.87 ± 0.01 | -10.2 ± 0.1 | -28.2 ± 0.2 | 17.9 ± 0.2 |
| SLIIΔBL | 239.0 ± 9.0 | 0.95 ± 0.3 | -9.01 ± 0.2 | -30.22 ± 5.3 | 21.14 ± 5.5 |
| SLIIGNRA | 967.3 ± 159.2 | 1.10 ± 0.05 | -8.2 ± 0.04 | -56.5 ± 1.3 | 48.3 ± 1.3 |
| SLIICCC | 138.5 ± 14.8 | 1.4 ± 0.4 | -9.4 ± 0.1 | -35.9 ± 3.4 | 26.5 ± 3.5 |
a Measurements were made at 298K in 40 mM KCl and 5mM K2PHO4, pH 6.5; bThermodynamic parameters presented as avg +/− std taken from two replicates.
As a further probe of the solution properties and stoichiometry of the UP1-SLII2231 complex, analytical gel filtration chromatography was used to monitor the interaction. Figure 3B shows the chromatographic trace of UP1-SLII2231 prepared at different molar ratios, where the SLII2231 concentration was held constant at 5 μM (note concentration of binding sites is 10 μM). Consistent with the calorimetric titrations, the SLII2231 chromatographic trace shifted to lower elution volumes as a function of increasing UP1 amount (Fig. 3). At a molar ratio of 0.5, a new peak corresponding to a 1:1 UP1-SLII2231 complex is observed well-resolved from the free SLII2231 peak. Increasing the UP1 concentration to 5 μM causes a reduction in the free SLII2231 peak along with a proportionate increase in the peak for the 1:1 complex. At a molar ratio of 2, SLII2231 is completely bound; however, a second earlier eluting component is observed as a shoulder on the peak corresponding to the 1:1 complex. This leading shoulder is further resolved into a distinct peak at higher UP1-SLII2231 molar ratio. The slight asymmetry observed in this new peak indicates a mixture of protein-RNA species, corresponding to the 1:1 and perhaps a 2:1 complex.
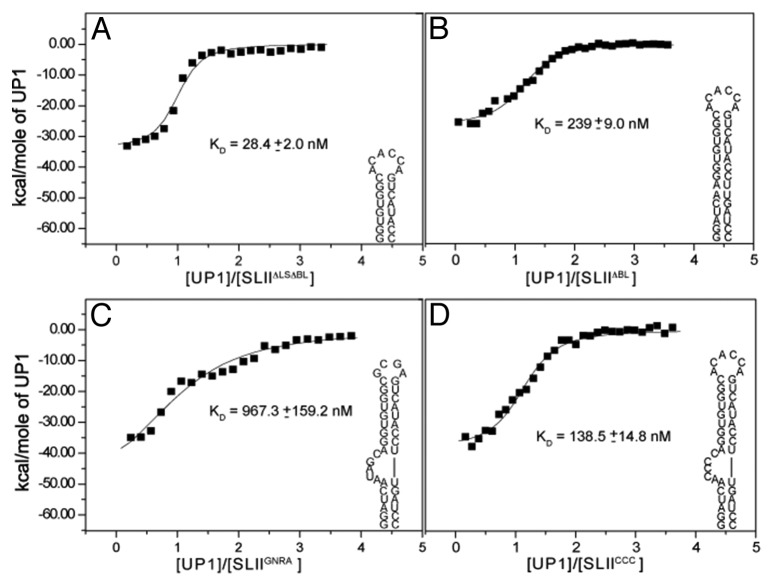
To gain insight into the mode of recognition of the UP1-SLII2231 complex, calorimetric titrations were performed with a series of SLII2231 structural mutants (Fig. 4), strategically designed to probe elements of secondary structure: (1) BL deletion (SLIIΔBL); (2) LS + BL deletion (SLIIΔLSΔBL); (3) HL to GNRA substitution (SLIIGNRA) and (4) BL, UAG to CCC substitution (SLIICCC). Two-dimensional 1H-1H NOESY spectra were collected for the mutant constructs to verify the RNAs fold as predicted (Fig. S1). Representative isotherms for each titration are shown in Figure 4 and a summary of the thermodynamic parameters is provided in Table 1. Since hnRNP A1 is known to bind single strand 5′-UAG-3′ stretches with high affinity and specificity,27 we first tested binding with the SLIIΔLS ΔBL construct, which lacks the only exposed 5′-UAG-3′ motif and lower base paired stem. Surprisingly, titration of UP1 into SLIIΔLS ΔBL resulted in a well-defined monophasic transition (Fig. 4). The binding event is driven by a large favorable change in enthalpy (ΔH = -28.2 ± 0.2 kcal/mol) and opposed by an unfavorable change in entropy (-TΔS = 17.9 ± 0.2 kcal/mol). This enthalpy-entropy signature leads to a stable 1:1 complex, characterized by a nanomolar binding affinity (Kd = 28.4 ± 2.0 nM). To test if the lower stem contributes to binding, the titration was repeated with the SLIIΔBL construct. Like SLIIΔLS ΔBL, the titration displayed a monophasic transition that was fit to a single-site isotherm with similar thermodynamic parameters (Table 1). Given the observation that UP1 binds SLIIΔLS ΔBL (and SLIIΔBL) as a stable 1:1 complex, calorimetric titrations were repeated with the SLIIGNRA construct. SLIIGNRA retains the 5-nt BL (and 5′-UAG-3′ site); however, the 6-nt HL is replaced with a GCGA tetraloop. Relative to SLII2231, UP1 associates with SLIIGNRA weakly and with a single transition (Fig. 4). The titration profile and thermodynamics suggest the 5-nt BL is a second, albeit weaker, site of interaction for UP1. Lastly, calorimetric titrations were performed using the SLIICCC construct, where CCC replaces the BL 5‘-UAG-3′ site. As shown in Figure 4, UP1 binds SLIICCC with moderate affinity, consistent with the SLIIΔLS ΔBL and SLII ΔBL titrations.
Figure 4. The bulge and hairpin loops of SLII contribute to high affinity UP1 recognition. Calorimetric titrations of (His)6-UP1 into various SLII structural mutants: (A) A SLIIΔLSΔBL; (B) SLIIΔBL; (C) SLIIGNRA and (D) SLIICCC. Titrations were performed at 298K. Secondary structures for each structural mutant are drawn. Processed isotherms shown along with non-linear least squares fits to a single site binding model. Average values and standard deviations of the dissociation constants (two replicates) are provided.
Altogether, the calorimetric and chromatographic titrations show that UP1 interacts with SLII2231 through multiple modes that involve conserved elements of RNA secondary structure. Additional NMR and biophysical studies are underway to determine the mechanism of interaction between UP1 and SLII2231.
The SLII bulge loop is necessary for IRES activity and EV71 replication
To assess the functional role of high-affinity hnRNP A1 assembly, we introduce the BL, CCC substitution into an in vivo translation construct and tested IRES activity in HeLa cells. A bicistronic reporter plasmid, pRHF-EV71-5′UTR, was used as template for the synthesis of CMV-RLuc-EV71-5′UTR-FLuc RNA transcript. As hnRNP A1 could be possibly involved in nuclear export of mRNA, to avoid this confounding issue, the CMV-RLuc-EV71-5′UTR-FLuc RNA transcripts, wild-type or with CCC substitution was transfected into cells to measure the IRES activity of EV71. The bicistronic transcript contained the EV71 5′UTR flanked by the Renilla luciferase (RLuc) and Firefly luciferase (FLuc) open reading frames (Fig. 5A). Translation of RLuc is cap-dependent, whereas translation of FLuc is EV71 IRES-dependent. The RLuc and FLuc expression levels were measured 2 d after transfection. The relative translation efficiency of EV71-IRES was determined by comparison of the level of downstream reporter gene product with the level of upstream reporter gene product. A bicistronic transcript with the anti-sense of the EV71-IRES (pRHF-EV71-5′UTR-AS) was used as a control to confirm that the EV71-IRES is actually functional in these assays.
Figure 5. BL 5′-UAG-3′ element is important for EV71 RNA translation and replication. (A) Diagram of the bicistronic construct used in the dual luciferase assay. (B) Effect of CCC substitution on EV71 IRES activity. Cells were transfected with CMV-RLuc-EV71-5′UTR-FLuc or CMV-RLuc-EV71-5′UTR-CCC-FLuc RNA transcript as described in Materials and Methods. Relative luciferase activity was measured 2 d after transfection. Mean values and standard errors from triplicate samples are shown in the bar graph. (C) Replication of CCC mutant EV71 in RD cells. RD cells were infected with wild tpe or CCC mutant EV71 at an m.o.i. of 0.01 pfu/cell and incubated at 37°C. Medium was harvested at various times and assayed for infectious virus by plaque formation on RD cells. Mean values and standard errors from triplicate samples are shown.
As shown in Figure 5B, the EV71 IRES activity was reduced dramatically in cells transfected with CMV-RLuc-EV71-5′UTR-CCC-FLuc RNA compared with that in cells transfected wild-type RNA transcript. The result suggested that the UAG element is critical for EV71 RNA translation. To investigate effects on virus replication, the CCC mutation was introduced into the infectious clone of EV71 and the full-length viral RNA was synthesized and transfected into RD cells. Virus released into medium was harvested 2 d after transfection and the titer was determined by plaque assay on RD cells. To determine the effect of the CCC mutation on virus replication, equal moi of wild-type and CCC mutant viruses were used to infect RD cells. Medium was collected at various times and the virus titer was determined by plaque forming on RD cells. As shown in Figure 5C, the CCC mutant virus replicates slower than the wild-type throughout the infection. The titer of mutant virus is about four logs less than that of the wild-type. Taken together these results suggest that the BL 5′-UAG-3′ element is important for virus translation and replication.
Discussion
Translation events in Enterovirus 71 are driven through the synergistic action of a type I IRES element and several ITAFs.8-10 Like other members of the Picornaviridae family, the mechanism of ITAF-mediated protein synthesis remains unknown, partly due to a paucity of structural and mechanistic descriptions of protein-RNA interactions that facilitate 40S ribosomal assembly. The work presented here offers some insight into the molecular determinants of functional hnRNP A1-IRES interactions, and provides a gateway to better understand the contribution of IRES RNA structure to EV71 replication fidelity.
Bioinformatic analysis of the EV71 stem loop II domain revealed that it is composed of highly conserved RNA structural and sequence elements. The consensus structure of SLII (Fig. 1) was derived from folding 147 aligned Enteroviral 71 sequences, and the predicted secondary structure was further validated by solution NMR (Fig. 2). The lower and upper stems are connected through a purine-rich bulge loop. Positions 1, 3 and 4 are invariably A, A and G, respectively. Pyrimidine residues occupy the remaining two positions, with a preference for U at position 2 and C at position 5. Searches of the RNA CoSSMos database43 for structures that contain the bulge loop consensus motif (AYAGY) yielded zero hits. Using a more relaxed search (ANAGN), an AAAGA bulge loop (nucleotides 2812–2816) was identified in domain VI of the H. marismortui 23S rRNA subunit.44 Interestingly, the AAAGA bulge loop protrudes out from helix 94, where ribosomal proteins L3 and L13 contact the base edge of G2815. Additional long-range interactions are made between A2814 and C533, located in domain II. Apparently, the AAAGA bulge loop forms an important site of molecular recognition within the context of the ribosome. Structural studies of the EV71 SLII bulge loop domain will provide insight into its free form conformation and facilitate comparisons with the ribosomal AAAGA loop.
Contrast to the bulge, the apical hairpin shows a stronger preference for pyrimidine residues. Positions 4 and 5 are invariably cytosines, whereas position 2 is either C or U, and position 3 shows a slightly higher preference for U over A (Fig. 1). Purines occupy the remaining two positions, with an immutable A at position 6. Klink and coworkers determined the solution NMR structure of an upper hairpin analog of SLIII derived from poliovirus type II Lansing (note, numbering system of stem loops off by +1 in early PV secondary structures).45 The sequence of their hairpin construct was AAUCCA, which shows good agreement with the consensus sequence determined here for all available EV71 strains (Fig. 1). The Lansing hairpin loop adopts a two-sided structure where the first two bases are exposed to the major groove, the third base represents the turning nucleotide and the remaining three bases form a continuous stack along the closing 5′-C:G-3′ base pair.45 Klink et al. postulated the hairpin might function as a site of recognition for trans factors or form long-range interactions with a distant site within the IRES. Both postulates were in agreement with the contemporary virological and phylogenetic studies on polioviruses.46,47 Indeed, Le and coworkers used sequence alignments to propose that stem loop VI, which has a similar ANCCA motif as stem loop III, participates in a pseudoknot structure with complementary residues downstream.46 We also observed a high-level of conservation of the AANCCA motif in stem loop V using our Enterovirus 71 alignments (not shown). Additional structural studies on larger EV71 IRES constructs will shed light on potential tertiary interactions between these highly conserved hairpin loops.
Using RNA pull downs, Lin et al. identified hnRNP A1 as one of 15 proteins that interact with the EV71 IRES element.21 The biological significance of this interaction, including global hnRNP A1-binding sites along the IRES, were later determined. The work presented here clearly shows the UP1 domain of hnRNP A1 binds SLII2231 via a well-defined biphasic transition (Fig. 3). Each binding event is driven by favorable changes in enthalpy and opposed by unfavorable changes in entropy (Table 1). Titrations of UP1 into various SLII structural mutants revealed the hairpin and bulge loops function synergistically to form a high-affinity complex (Fig. 4 and Table 1). Deletion or substitution mutations within the bulge loop in either SLIIΔLS ΔBL or SLIICCC reduced the UP1 binding affinity between ~280‒480-fold (Table 1), relative to the first transition of wild type SLII2231. Furthermore, substitution of the hairpin loop with a GNRA tetraloop reduced the apparent binding affinity by approximately three orders of magnitude relative to the first transition. Taken together, these results clearly indicate the bulge and hairpin loops act synergistically to assemble a stable UP1-SLII2231 complex. Titrations from size exclusion chromatography further support the calorimetric data and help resolve the stoichiometry. At this time, the mechanism of this interaction is unknown and will require additional structural/biophysical studies to fully elucidate; however, we hypothesize that the native bulge sequence induces a kink in the RNA backbone that likely facilitates stable protein-protein and protein-RNA contacts. The bulge loop in the SLIICCC constructs likely adopts a different tertiary conformation that does not support the biphasic binding transition.
The thermodynamic results also show the apical hairpin loop retains significant binding affinity for UP1 (Fig. 4 and Table 1). Indeed, calorimetric titrations with the SLIIGNRA construct had the most severe impact on UP1 recognition. This observation is very intriguing since the 6-nt hairpin loop does not contain a 5′-UAG-3′ motif, which is the central element of the hnRNP A1 high-affinity “winner” sequence.27 Rather, the SLII2231 hairpin loop is AC rich and the consensus motif determined here for all aligned EV71 strains is RY(U/A)CCA. Thus, hnRNP A1 interacts with the SLII hairpin loop using a mode of recognition likely distinct from its established mode of binding single strand nucleic acids.28,30,48 This observation suggests that hnRNP A1 can accommodate different RNA substrates using sequence and shape-specific recognition principles. The results obtained here are in good agreement with our previous study that showed UP1 binds the HIV-1 ESS3 hairpin loop as a high-affinity 1:1 complex; however, the sequence of the ESS3 apical loop (5′-GAUUAGU-3′) more closely matches the hnRNP A1 “winner.”33 Similar plasticity has been observed for the PTB and La proteins, which both use RRM domains to interact with RNA substrates that contain various secondary structures, including IRES elements.49,50 Indeed, it was suggested that the adaptability of PTB-RNA interactions modulate the structural composition of the RNP complex, which might influence RNA regulatory processes.50 We propose that hnRNP A1 also executes its many functions through substrate-specific adaptive mechanisms. Therefore, attempts to define the biological functions of hnRNP A1 using only simple oligonucleotide substrates might lead to restrictive models.51
Of important interest, we demonstrate that the ability to disrupt the UP1-SLII2231-binding transition in vitro correlates robustly with impaired EV71 RNA translation and replication (Fig. 5). Introduction of the bulge loop 5′-UAG-3′ to 5′-CCC-3′ mutation into the bicistronic pRHF-EV71-5′UTR reporter construct reduced IRES driven translation to within ~20% of wild-type levels, supporting previous work by Lin et al. showing that hnRNP A1 stimulates IRES activity through interactions with SLII and SLVI.21 The observed reduction in translation activity also correlates nicely with an impaired EV71 growth phenotype (Fig. 5). The question remains, how does hnRNP A1 trans activate the EV71 IRES? Taking into consideration earlier phylogenetic and structural work on poliovirus, which provided evidence that the apical hairpins of SLIII and SLVI may form pseudoknot structures,46 we propose that hnRNP A1 binds the apical loop in the hairpin (exposed) conformation, thus preventing pseudoknot (sequestered) formation (Fig. 6). In this context, the hnRNP A1-bound IRES associates with translational activities whereas the predicted pseudoknot conformation functions in some other RNA process, such as strand synthesis or genome packaging. While additional structural and virological studies are needed to test this postulate the work presented here clearly shows a functional link between high-affinity UP1-SLII binding and EV71 replication. Binding of hnRNP A1 to the SLII apical loop would compete with it forming a long-range pseudoknot structure. Lastly, the proposed model differs subtly from other predictions of ITAF functions, which posits ITAFs act to chaperone or stabilize the RNA in a conformation supported for 40S assembly.
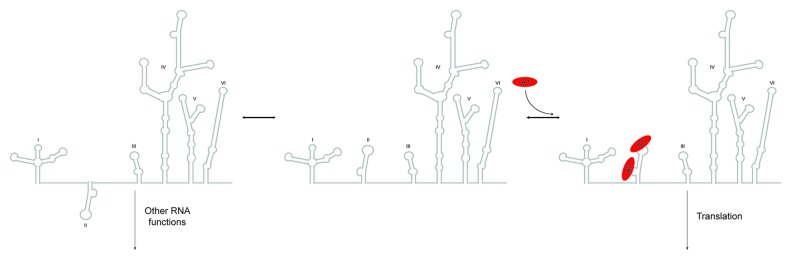
Figure 6. A model of how hnRNP A1 activates EV71 RNA translation. In this model, stem loop II exists in two conformations, sequestered and exposed. The exposed conformation supports hnRNP A1 high-affinity assembly and favors translation, whereas the sequestered state (possible pseudoknot as predicted in ref. 46) promotes other RNA functions.
Materials and Methods
RNA preparation
The SLII constructs used in this study were prepared by in vitro transcription using recombinant T7 RNA polymerase that was overexpressed and purified from BL21 (DE3) cells. Synthetic DNA templates, corresponding to the EV71 2231 isolate, were purchased from Integrated DNA Technologies. Transcription reactions were performed using standard procedures,41 and consisted of 15‒30 ml reaction volumes containing unlabeled ribonucleotide triphosphates (rNTPs; Acros Organics). Following synthesis, the SLII constructs were purified to homogeneity by 12‒20% denaturing PAGE, excised from the gels, electroeluted and desalted using GE illustra™ NAP™-25 columns. Additional rounds of desalting were performed by exhaustively washing the sample with RNase-free water using a Millipore Amicon® Ultra-4 centrifugal filter device. Purified constructs were dried under vacuum and stored at -20°C until further use.
The solution properties of the SLII constructs were probed by annealing the RNA under different solute conditions, followed by native PAGE analysis. Typical RNA folding reactions were performed by heating the sample to 95°C for 2 min followed by flash cooling on ice. The samples were prepared with buffer salts only (5 mM K2HPO4 and 0.5 mM sodium EDTA, pH 6.5) or with buffer salts plus the addition of 40 mM KCl (NMR buffer). Independent of the method of preparation, the predominant (> 95%) SLess3 species migrated as a hairpin on native PAGE; however, a small population (< 5%) of duplex or dimer was observed at NMR concentrations. Final NMR samples were prepared in NMR buffer, dried under vacuum and resuspended in 90% H2O/10% D2O. The NMR samples were subsequently annealed by heating at 95°C for 2 min and flashed cooled on ice. Concentrations were determined using theoretical molar extinction coefficients (Ambion Inc.), and typical NMR samples ranged from 0.1‒0.5 mM at 250 uL.
NMR spectroscopy
NMR spectra were recorded on a Bruker Avance (600 MHz) spectrometer equipped with room temperature HCN triple resonance probes and a z-axis pulsed field gradient accessory. All NMR data were processed with nmrPipe/nmrDraw52 and analyzed using Sparky.53 Exchangeable 1H spectra were measured at 283 K with the Watergate NOESY (τm = 250 ms) pulse sequence.54
Preparation and calorimetric titrations of UP1
The UP1 construct was prepared as previously described33 with the exception that a cysteine-less mutant was used in these studies. To avoid complications of using reducing agents in the ITC, we mutated cysteine residues 43 and 175 to serines. NMR 1H-15N HSQC spectra (not shown) collected on the cysteine-less construct showed that it folds identical to wild-type UP1.
All calorimetric titrations were performed at 25°C on a VP-ITC calorimeter (MicroCal, LLC) as described previously.33 UP1 was exchanged into titration buffer (5 mM K2HPO4, 40 mM KCl, pH 6.5) by gel filtration. The SLII constructs were prepared in titration buffer using a Millipore Amicon® Ultra-4 centrifugal filter device and subsequently annealed by heating at 95°C for 2 min, followed by flash cooling on ice. (His)6-UP1 at 50 uM was titrated into ~1.4 mL of 2.0‒2.5 uM SLII constructs over 35 injections of 8 uL each. Prior to non-linear least squares fitting in Origin v7.0, the raw data were corrected for dilution by subtracting the average heats from the saturated upper asymptotes.
Analytical gel filtration (FPLC)
Analytical gel filtration was done with Superdex 200 10/300 GL column (GE Healthcare Life Sciences). Samples were manually loaded into 100 µL loop and run through column at 0.5 mL/min, with elution volume starting at point of column loading. Samples were run at 5 µM RNA and UP1-SLII2231 ratios of 0, 0.5, 1.0, 2.0 and 4.0. For UP1-SLII2231 complexes, the samples were mixed at dilute volume (4 mL) and then concentrated down to 250 µL volume with Amicon Ultra centrifugal filters. For molecular weight determination, elution volumes were divided by the void volume and the resulting ratios were compared with a standard curve (Fig. S3). The void volume and the standard curve were determined using Sigma Gel Filtration Molecular Weight Markers (MW-GF-70).
Cells
HeLa cells were cultured at 37°C in Eagle’s minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS) (Mediatech). RD (human embryonal rhabdomyosarcoma) cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FCS.
Plasmid construction and in vitro transcription
The RHF plasmid was constructed as described previously.21 The bicistronic reporter plasmid pRHF-EV71-5′UTR, which contains EV71 IRES between Renilla and Firefly luciferase, was constructed by ligating a NotI-EV71-5′UTR-NotI fragment into pRHF. The pRHF-EV71-5′UTR-AS was constructed by inserting the reverse sense of NotI-EV71-5′ UTR-NotI fragment into pRHF. pRHF-EV71-5′UTR-CCC has CCC substitution of TAG at nts 135–137 of EV71 IRES.
pRHF-EV71-5′UTR, pRHF-EV71-5′UTR-CCC or pRHF-EV71-5′UTR-AS was linearized by Drd I digestion and used as template for the synthesis of CMV-RLuc-EV71-5′UTR-FLuc RNA, CMV-RLuc-EV71-5′UTR-CCC-FLuc RNA or CMV-RLuc-EV71-5′UTR-AS-FLuc RNA using the MAXIscript kit (Life Technologies).
Determination of EV71 IRES activity
HeLa cells were seeded in 12-well plates in antibiotic-free MEM. Two hundred nanograms of CMV-RLuc-EV71-5′UTR-FLuc, CMV-RLuc-EV71-5′UTR-CCC-FLuc or CMV-RLuc-EV71-5′UTR-AS-FLuc capped RNA templates were transfected along with 5 µl SuperFect transfection reagent (Qiagen) in 400 µl MEM supplemented with 10% FCS following manufacturer's directions. The IRES activity was determined 2 d after transfection by measuring the Renilla luciferase (RLuc) and Firefly luciferase (FLuc) activity in a 20/20 luminometer (Turner Biosystems) using a dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
Viral growth and plaque assay
Nucleotide 134 CCC mutations were introduced into the infectious clone of EV71 by site-directed mutagenesis according to the manufacturer’s instruction (Strategene). Full-length viral RNA was synthesized by in vitro transcription using infectious cDNA as template and transfected into RD cells. Supernatant was collected 2 d after transfection. Virus titer was determined by plaque forming on RD cells. To determine the viral growth curve, RD cells were infected with wild-type or mutant virus at an MOI of 0.01 PFU/cell. Medium was harvested at various time points and virus titer was determined by plaque assay on RD cells.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health (GM101979A to .BS.T.).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25107
References
- 1.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. 2010;10:778–90. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 2.Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 3.McMinn PC. Recent advances in the molecular epidemiology and control of human enterovirus 71 infection. Curr Opin Virol. 2012;2:199–205. doi: 10.1016/j.coviro.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Chang LY, Lin TY, Huang YC, Tsao KC, Shih SR, Kuo ML, et al. Comparison of enterovirus 71 and coxsackie-virus A16 clinical illnesses during the Taiwan enterovirus epidemic, 1998. Pediatr Infect Dis J. 1999;18:1092–6. doi: 10.1097/00006454-199912000-00013. [DOI] [PubMed] [Google Scholar]
- 5.McMinn PC. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev. 2002;26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 6.Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–6. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang HI, Weng KF, Shih SR. Viral and host factors that contribute to pathogenicity of enterovirus 71. Future Microbiol. 2012;7:467–79. doi: 10.2217/fmb.12.22. [DOI] [PubMed] [Google Scholar]
- 9.Shih SR, Stollar V, Li ML. Host factors in enterovirus 71 replication. J Virol. 2011;85:9658–66. doi: 10.1128/JVI.05063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson SR, Sarnow P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003;315:259–66. doi: 10.1016/S0042-6822(03)00544-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J Gen Virol. 2008;89:2540–9. doi: 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789:518–28. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–7. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–76. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plank TD, Kieft JS. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA. 2012;3:195–212. doi: 10.1002/wrna.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011;39:4851–65. doi: 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011;30:4423–36. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kafasla P, Morgner N, Pöyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell. 2009;34:556–68. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Kafasla P, Morgner N, Robinson CV, Jackson RJ. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–22. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin JY, Shih SR, Pan M, Li C, Lue CF, Stollar V, et al. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J Virol. 2009;83:6106–14. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chabot B, LeBel C, Hutchison S, Nasim FH, Simard MJ. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog Mol Subcell Biol. 2003;31:59–88. doi: 10.1007/978-3-662-09728-1_3. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–47. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 24.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 25.Abdul-Manan N, O’Malley SM, Williams KR. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry. 1996;35:3545–54. doi: 10.1021/bi952298p. [DOI] [PubMed] [Google Scholar]
- 26.Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–70. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–15. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayeda A, Munroe SH, Xu RM, Krainer AR. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–23. doi: 10.1017/S135583829898089X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers JC, Shamoo Y. Human UP1 as a model for understanding purine recognition in the family of proteins containing the RNA recognition motif (RRM) J Mol Biol. 2004;342:743–56. doi: 10.1016/j.jmb.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Shamoo Y, Abdul-Manan N, Patten AM, Crawford JK, Pellegrini MC, Williams KR. Both RNA-binding domains in heterogenous nuclear ribonucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry. 1994;33:8272–81. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- 32.Damgaard CK, Tange TO, Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA. 2002;8:1401–15. doi: 10.1017/S1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levengood JD, Rollins C, Mishler CH, Johnson CA, Miner G, Rajan P, et al. Solution structure of the HIV-1 exon splicing silencer 3. J Mol Biol. 2012;415:680–98. doi: 10.1016/j.jmb.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 34.Marchand V, Méreau A, Jacquenet S, Thomas D, Mougin A, Gattoni R, et al. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J Mol Biol. 2002;323:629–52. doi: 10.1016/S0022-2836(02)00967-1. [DOI] [PubMed] [Google Scholar]
- 35.Tange TO, Damgaard CK, Guth S, Valcárcel J, Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 2001;20:5748–58. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–6. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 37.Guil S, Long JC, Cáceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–58. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–8. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klinck R, Sprules T, Gehring K. Structural conservation in RNA loops III and VI of the internal ribosome entry sites of enteroviruses and rhinoviruses. Biochem Biophys Res Commun. 1998;247:876–81. doi: 10.1006/bbrc.1998.8852. [DOI] [PubMed] [Google Scholar]
- 40.Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of terminal mismatches in ribooligonucleotides. Biochemistry. 1987;26:4559–62. doi: 10.1021/bi00388a059. [DOI] [PubMed] [Google Scholar]
- 41.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–98. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaughlin KJ, Jenkins JL, Kielkopf CL. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–31. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanegas PL, Hudson GA, Davis AR, Kelly SC, Kirkpatrick CC, Znosko BM. RNA CoSSMos: Characterization of Secondary Structure Motifs--a searchable database of secondary structure motifs in RNA three-dimensional structures. Nucleic Acids Res. 2012;40(Database issue):D439–44. doi: 10.1093/nar/gkr943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 45.Klinck R, Sprules T, Gehring K. Structural characterization of three RNA hexanucleotide loops from the internal ribosome entry site of polioviruses. Nucleic Acids Res. 1997;25:2129–37. doi: 10.1093/nar/25.11.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le SY, Chen JH, Sonenberg N, Maizel JV. Conserved tertiary structure elements in the 5′ untranslated region of human enteroviruses and rhinoviruses. Virology. 1992;191:858–66. doi: 10.1016/0042-6822(92)90261-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholson R, Pelletier J, Le SY, Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol. 1991;65:5886–94. doi: 10.1128/jvi.65.11.5886-5894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers JC, Moore SA, Shamoo Y. Structure-based incorporation of 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine into the human telomeric repeat DNA as a probe for UP1 binding and destabilization of G-tetrad structures. J Biol Chem. 2003;278:42300–6. doi: 10.1074/jbc.M306147200. [DOI] [PubMed] [Google Scholar]
- 49.Martino L, Pennell S, Kelly G, Bui TT, Kotik-Kogan O, Smerdon SJ, et al. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res. 2012;40:1381–94. doi: 10.1093/nar/gkr890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clerte C, Hall KB. The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry. 2009;48:2063–74. doi: 10.1021/bi8016872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barraud P, Allain FH. Solution structure of the two RNA recognition motifs of hnRNP A1 using segmental isotope labeling: how the relative orientation between RRMs influences the nucleic acid binding topology. J Biomol NMR. 2013;55:119–38. doi: 10.1007/s10858-012-9696-4. [DOI] [PubMed] [Google Scholar]
- 52.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Goddard TD, Kneller DG, Sparky 3. University of California, San Francisco
- 54.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–5. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 55.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.