Abstract
The conserved human LIN28 RNA-binding proteins function in development, maintenance of pluripotency and oncogenesis. We used PAR-CLIP and a newly developed variant of this method, iDo-PAR-CLIP, to identify LIN28B targets as well as sites bound by the individual RNA-binding domains of LIN28B in the human transcriptome at nucleotide resolution. The position of target binding sites reflected the known structural relative orientation of individual LIN28B-binding domains, validating iDo-PAR-CLIP. Our data suggest that LIN28B directly interacts with most expressed mRNAs and members of the let-7 microRNA family. The Lin28-binding motif detected in pre-let-7 was enriched in mRNA sequences bound by LIN28B. Upon LIN28B knockdown, cell proliferation and the cell cycle were strongly impaired. Quantitative shotgun proteomics of LIN28B depleted cells revealed significant reduction of protein synthesis from its RNA targets. Computational analyses provided evidence that the strength of protein synthesis reduction correlated with the location of LIN28B binding sites within target transcripts.
Keywords: post-transcriptional regulation, RNA-binding protein, CLIP, stem cell
Introduction
Post-transcriptional gene regulation is elicited through a complex network of RNA-binding proteins (RBPs) and miRNA-containing ribonucleoprotein complexes that target defined sequence elements within mRNAs and thereby regulate all aspects of RNA metabolism.1 Individual RBPs can bind hundreds of RNAs and regulate their processing, cellular localization, translation and decay.2-5 Consequently, various RBPs have been implicated in human disease, including neurodegenerative disorders and cancer.6-8
The RNA-binding protein Lin28 was initially identified as a heterochronic gene, controlling developmental progression during the second larval stage (L2) in Caenorhabditis elegans (C. elegans).9 Vertebrates have two paralogs, LIN28A and LIN28B, which share a unique domain structure, consisting of a single cold-shock domain (CSD) and two CCHC-type zinc fingers that form a zinc knuckle domain (ZKD). Given its absence in non-vertebrates, it therefore appears likely that LIN28B originated from an early duplication event in vertebrate evolution. Strikingly LIN28A proteins in different species are more similar to each other than respective paralogs in the same species.10 In contrast to LIN28A, LIN28B encodes an extended C terminus and harbors nuclear and nucleolar localization signals (NLS, NLoS, respectively)11 that were shown to functionally impact the cellular localization of LIN28B proteins.12
In mammalian and nematode development, LIN28 proteins are highly expressed in undifferentiated cell types, while expression selectively declines during differentiation.13,14 In agreement with its stage-specific expression, LIN28A together with OCT4/POU5F1, SOX2 and NANOG overexpression was sufficient to reprogram adult human fibroblasts and reconstitute gene expression patterns of pluripotent stem cells.15 More recently, it was shown that perturbation of Lin28a levels lead to severe developmental defects in mammals. Transgenic mice expressing ectopic Lin28a showed increased body size and delayed onset of puberty, while Lin28a-deficient mice exhibited 20% less body weight at birth and died during early developmental stages.16 Interestingly, the human proteins LIN28B, and to a lesser extent LIN28A, were found to be reactivated in a variety of cancer cells and tumor tissues, including ovarian carcinoma and germ cell tumors, thus indicating the importance of tightly regulated LIN28 expression to maintain normal cell development and pluripotency.17,18
The molecular mechanisms underlying LIN28 function were initially suggested to largely revolve around its ability to block the processing of primary-let-7 (pri-let-7) and precursor-let-7 (pre-let-7) hairpins into mature let-7 miRNAs, another family of heterochronic gene products. Indeed, LIN28 and let-7 expression levels are reciprocal in early nematode and mammalian development and mature let-7 was shown to repress LIN28 translation as part of a negative feedback loop.13,19 The inhibition of let-7 miRNA biogenesis by Lin28 is required for normal development and contributes to maintenance of pluripotency by blocking let-7-induced differentiation in mouse embryonic stem cells.20 LIN28B binds to the terminal loop of pri- and pre-let-7, thereby preventing DROSHA and DICER cleavage. Interestingly LIN28A was suggested to partly act by an alternative mechanism, involving a terminal uridyl-transferase (TUT4/Zcchc11 in mammals, PUP-2 in C. elegans) that leads to LIN28A-promoted 3′-uridylation of pre-let-7.21,22 Uridylated pre-let-7 is a poor DICER substrate and subject to degradation. Independent biochemical studies identified a conserved GGAG motif within the terminal loop of pre-let-7 to be essential for LIN28 binding and uridylation.23
Recently published co-crystals of LIN28 and pre-let-7 revealed that this interaction is mediated in a sequence-specific manner by the ZKD of LIN28.24-26 Of note, the C-terminal region of LIN28 encoding the ZKD shows remarkable similarities to the HIV-1 protein NCp7 in terms of amino acid sequence and RNA-binding activity.27 Less is known about the nucleotide-binding preferences of the LIN28 CSD.28 Bacterial cold-shock domains can act as RNA chaperons and bind a variety of single-stranded nucleic acids.29,30 Mayr and colleagues reported the LIN28 CSD to be involved in remodeling of the terminal pre-let-7 stem loop,24 but to date, only low complexity-binding motifs have been suggested. However, both RNA-binding domains are essential for LIN28-mediated regulation of let-7 and the let-7 precursor contacting amino acids25 are highly conserved in the domain structure of both LIN28A and B.
Several studies indicate a let-7-independent mRNA-binding activity of Lin28. Notably, the gene expression profile of mouse embryonal carcinoma cells that constitutively express Lin28a is significantly changed during retinoic acid-induced cell differentiation before mature let-7 accumulates.31 In addition, Lin28 mediates translational activation of Igf2, Oct4, cyclin A, cyclin B, histone 2a and HMGA1 mRNAs.32-37 Two recent studies provide a transcriptome-wide view of the direct target sites of LIN28A in embryonic stem (ES) cells by carrying out RNA crosslinking-immunoprecipitation-sequencing (CLIP-seq).38,39 In addition to let-7 precursors, Lin28A seems to recognize the mRNA motifs AAGNNG, AAGNG and, less frequently, UGUG in mouse ES cells38 and GGAGA in human ES cells.39 In both cases, the consensus motifs were found to be preferentially located in terminal loops of hairpin structures. Furthermore, Cho and colleagues show that LIN28A decreases ribosome density on certain mRNAs associated with the endoplasmic reticulum,38 whereas Wilbert and colleagues indicate that LIN28A plays a role in mRNA processing by direct regulation of splicing factors leading to widespread changes in alternative splicing.39 A recent study by Hafner and colleagues performed Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP)40 of LIN28A and LIN28B and identified a largely overlapping set of around 3,000 mRNAs with about 9,500 target sites.41 LIN28 protein binding mildly stabilized target mRNAs and increased protein abundance.
To understand the biological function and differential activities of LIN28 proteins it is necessary to globally identify the RNA regions bound and regulated by the C-terminally extended LIN28 paralog LIN28B. We applied PAR-CLIP to generate a transcriptome-wide map of LIN28B interactions in human embryonic kidney (HEK) 293 cells. Identified targets include the let-7 precursors and a surprisingly abundant number of transcripts involved in protein translation, mRNA splicing and regulation of cell cycle. We further analyzed the nature of these interactions by computational means and report potential sequence preferences for binding of LIN28B. To further investigate how LIN28B interaction on mRNA is mediated by its RNA-binding domains, we developed individual domain PAR-CLIP (iDo-PAR-CLIP) and revealed a distinct orientation of the LIN28B CSD and ZKD on its target transcripts. Finally, we validated functionality of bound mRNA targets by using pulsed quantitative shotgun proteomics42-44 to detect changes in protein synthesis of target transcripts upon LIN28B depletion.
Results
PAR-CLIP reproducibly identifies thousands of human RNAs directly bound by LIN28B
To identify LIN28B-binding sites at high resolution, we applied PAR-CLIP in combination with next-generation sequencing.40 In PAR-CLIP experiments, nascent RNA is metabolically labeled with the photoreactive ribonucleosides 4-thiouridine (4SU) or 6-thioguanosine (6SG). Crosslinking of protein to 4SU or 6SG-labeled RNA leads to specific T to C or G to A transitions that occur at high-frequency in cDNA sequence reads and mark the protein crosslinking site on the target RNA.40 Briefly, HEK293 cells stably expressing inducible FLAG/HA-tagged LIN28B at physiological levels (Fig. S1A) were crosslinked after metabolic labeling of RNA with photoreactive nucleosides. Immunopurified, ribonuclease-treated and radiolabeled LIN28B-RNA complexes were separated by SDS-PAGE and bands migrating at the expected molecular weight of LIN28B protein were excised (Fig. 1A; Fig. S1B). Protein-protected RNA fragments were recovered and converted into a cDNA library amenable to Illumina sequencing.
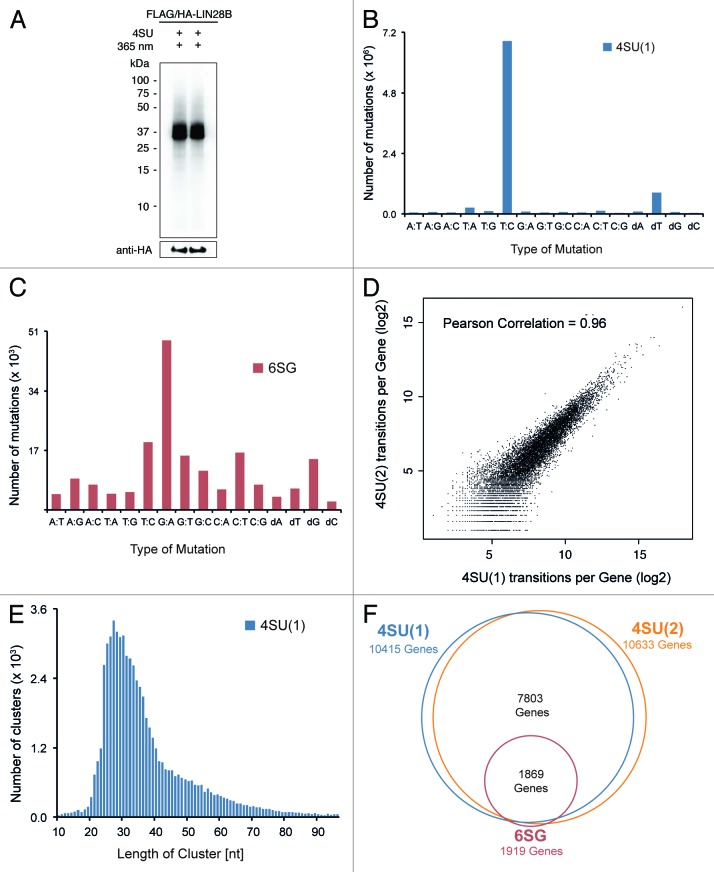
Figure 1. PAR-CLIP reproducibly identifies thousands of human mRNAs directly bound by LIN28B. (A) Autoradiogram of SDS-PAGE, transferred to nitrocellulose membrane. Crosslinked protein-RNA complexes migrating at 39 kDa correspond to epitope-tagged LIN28B. Anti-HA western blot confers expression of FLAG/HA-LIN28B. Lanes 1 and 2 show protein-RNA complexes used for generation of 4SU(1) and 4SU(2) PAR-CLIP libraries. (B and C) Frequency of nucleotide mutations detected in 4SU(1) and 6SG PAR-CLIP libraries after alignment to spliced human transcriptome. dA, dT, dG and dG indicate respective nucleotide deletions. (D) Number of diagnostic transitions per gene observed in 4SU(1) and 4SU(2) experiments. (E) Length distribution of 4SU(1) PAR-CLIP sequence clusters after quality filtering. (F) Scaled Venn diagram of target genes with at least two independent diagnostic transitions in indicated PAR-CLIP libraries.
In total, we performed three independent PAR-CLIP experiments (two biological replicates with 4SU and one experiment with 6SG; see Fig. S1C and Table S1). Sequence reads were aligned to the spliced human transcriptome and overlapping reads were used to build sequence read clusters. In PAR-CLIP experiments using 4SU, diagnostic T-C mutations were 30-fold more abundant than any other mutation within clustered sequence reads (Fig. 1B; Fig. S1D). Similarly, but less pronounced, the diagnostic G-A mutation was the most abundant mutation observed in sequence clusters from 6SG PAR-CLIP experiments (Fig. 1C). In addition to these diagnostic mutations and consistent with previous reports,42,45,46 we observed respective T or G deletions at crosslinking sites, however less frequently (Fig. 1B and C). We therefore considered the respective nucleotide mutations as well as nucleotide deletions as indicators for direct protein-RNA crosslinking events and refer to them as diagnostic transitions in what follows.
When comparing the number of diagnostic transitions per gene in the two 4SU experiments, we observed a high reproducibility between biological replicates (Pearson Correlation: 0.96) (Fig. 1D). Furthermore, crosslinking positions in one 4SU library were highly reproducible in the other 4SU replicate library (Fig. S1E and F). Comparing mRNA expression levels of genes covered by the top 1,000 4SU PAR-CLIP-binding sites to those of all transcribed genes indicated a good dynamic detection range (Fig. S1G). Figure 1E shows that the cluster length distribution peaked at a cluster length of ~27 nucleotides.
The total number of target genes identified in each PAR-CLIP experiment was strongly dependent on the photoreactive nucleosides used, likely reflecting different crosslinking efficiencies.40 While the two 4SU experiments identified target transcripts of 10,415 and 10,633 genes, respectively, only 1,919 genes were detected in the 6SG PAR-CLIP. Despite obvious differences in the total number of target transcripts captured in 4SU or 6SG experiments, the identity of target genes was largely overlapping (Fig. 1F). The surprisingly large number of bound transcripts detected in both 4SU experiments points toward an unusual widespread mode of LIN28B target interaction that encloses the majority of all expressed transcripts. A similar observation was recently described for LIN28A.38
LIN28B binds to let-7 precursors and protein-coding transcripts
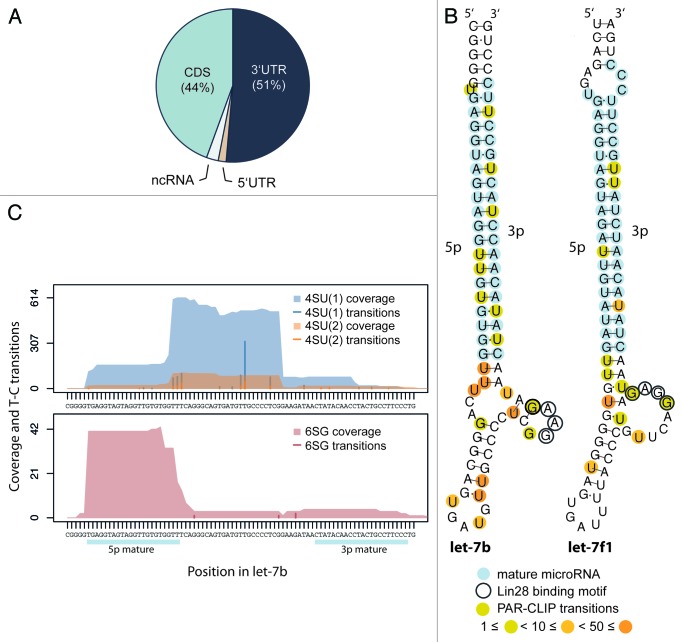
For further analysis, we defined a conservative set of sequence clusters that showed at least two independent diagnostic transitions in overlapping reads from 4SU and 6SG PAR-CLIP libraries. Applying these criteria with a flank of 30 nt, we retained 2,540 conservative sequence clusters mapping to transcripts of 1,527 protein-coding genes. Almost all of LIN28B-binding sites were detected within 3′UTRs (51%) and CDS (44%) of mRNAs (Fig. 2A). While early studies on the mRNA-binding activity of LIN28 focused on binding elements in 3′UTRs,32-34 the high frequency of CDS targeting is surprising, but not unreported for other RNA-binding proteins.7
Figure 2. LIN28B binds to 3′UTRs and CDS of protein-coding genes and interacts with let-7 precursors. (A) Distribution of LIN28B-binding sites in conservative sequence clusters to non-coding RNAs and different transcript regions (5′UTR, CDS and 3′UTR) of protein-coding genes. (B) Identified LIN28B-binding sites in let-7b and let-7f1 precursors. Mature microRNA sequences (light-blue), biochemically identified GGAG motif (encircled) and weighted PAR-CLIP transition sites (yellow-orange) are indicated. Structures are adapted from RNAfold (ViennaRNA). (C) Alignment of sequence coverage signal and diagnostic nucleotide transitions observed in 4SU (1) (blue), 4SU (2) (orange) and 6SG (dark red) libraries to the genomic region encoding let-7b precursor.
Consistent with previous in vitro experiments, we found pre-let-7b and pre-let-7f to be directly contacted by LIN28B in loop and hairpin regions in all three PAR-CLIP experiments, while pre-let-7d was detected in 4SU experiments only (Fig. 2B and C; Fig. S2A and B). Since the let-7 family of miRNAs represents the best-studied group of functionally regulated LIN28 targets, we considered them as important internal controls. Diagnostic transitions within the loop regions of pre-let-7b and pre-let-7f precisely occurred in the previously described GGAG-binding motif (Fig. 2B and C), thus validating that our approach captures functional LIN28B target interactions at high resolution. Interestingly, Figure 2C shows extensive sequence coverage of 4SU experiments in the pre-let-7b loop region, while 6SG preferentially captured the 5p stem region of the same precursor. Apart from the let-7 family we found only three other miRNA precursors (pre-miR-19b-1, pre-miR-663 and pre-miR-16-2) being bound by LIN28B, underlining the specificity of our approach (Fig. S2C).
Target transcripts are enriched for a RGGSWG consensus motif
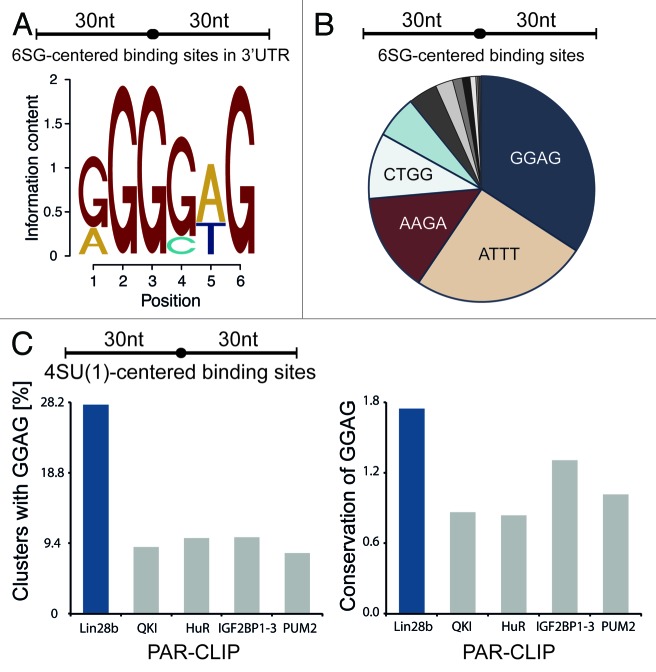
To enable identification of sequence motifs responsible for LIN28B mRNA binding, we generated crosslink centered regions (30 nt upstream and downstream of crosslinking sites) from the conservative set of sequence clusters. We applied MEME motif finding algorithm47 on the top 300 conservative 6SG-centered target regions in 3′UTRs and identified RGGSWG (R = G or A, S = G or C, W = A or T) as the most enriched motif (E = 0.14, 74 sites) (Fig. 3A). Consistently, GGAG was the most frequently observed tetramer in all 6SG-centered binding sites within our conservative target transcripts (Fig. 3B). Reducing the window size from 60 to 10 nts around crosslinked sites left the results largely unchanged, indicating that the GGAG motif is mostly observed in the vicinity of 6SG crosslinks (Fig. S3A). On the other hand, when applying our analysis to random G-centered sequences derived from the same transcript set, AGAA was the most frequently observed 4mer (Fig. S3B). Interestingly, a motif search in CDS clusters yielded AAGRWG (R = A or G), which is highly similar to the LIN28A consensus sequence reported by Cho et al. (Fig. S3C). To exclude that a technical bias leads to an enrichment of GGAG in our PAR-CLIP data, we compared the occurrence of the GGAG motif in LIN28B PAR-CLIP clusters to the presence of the same motif in PAR-CLIP data from previously studied RBPs. Figure 3C shows that GGAG-containing clusters were at least 2-fold more enriched in LIN28B PAR-CLIP data. At the same time, the evolutionary conservation of the GGAG motif in LIN28B clusters exceeded the conservation in other PAR-CLIP clusters by a factor of 2 (Fig. 3C). In conclusion, the GGAG motif appears to be a crucial determinant for LIN28B binding, not only in let-7 precursor interaction, but also in recognition of target mRNAs. While co-crystals of LIN28 and let-7 revealed that GGAG is contacted by the ZKD of LIN28, evidence for a distinct binding motif or region contacted by the CSD is less clear. Nam et al. proposed NGNGAYNNN within a closed loop as a consensus for CSD binding,25 whereas Mayr et al. identified a GUNNUNN motif.24 However, neither of these motifs is enriched in our data set.
Figure 3. LIN28B target transcripts are enriched for GGAG consensus motif. (A) Top sequence motif identified by MEME in top 300 6SG centered 3′UTR-binding sites (extended by 30 nt upstream and downstream) within the conservative sequence cluster set. (B) Most frequent tetramers within 6SG centered conservative clusters, extended by 30 nt upstream and downstream of 6SG-crosslinks. (C) Frequencies of GGAG in the top 1,000 sequence clusters of indicated PAR-CLIP data sets (left) and mean vertebrate conservation (phyloP) of GGAG within those sequence clusters (right).
iDo-PAR-CLIP (individual domain PAR-CLIP) enables characterization of domain specific target interactions
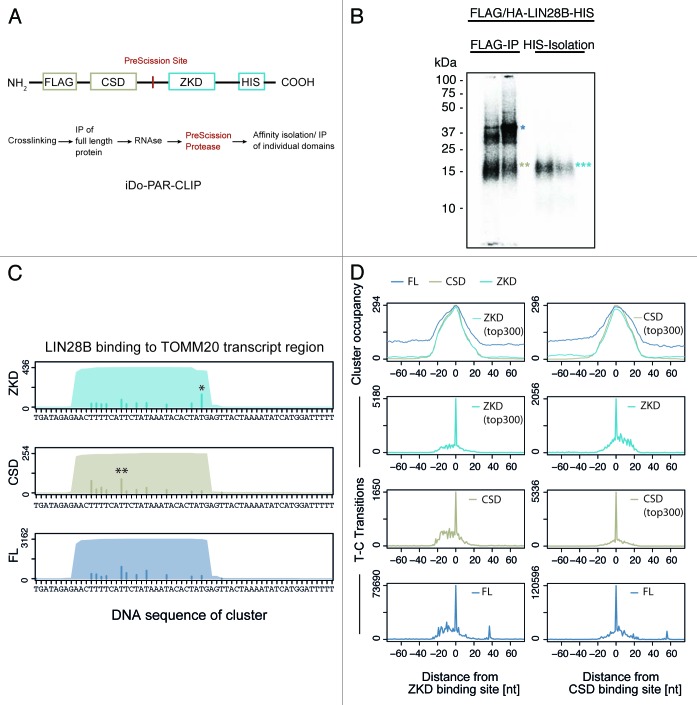
To further explore the contribution of CSD and ZKD binding to LIN28 target recognition, we generated a stable cell line, expressing FLAG/HA-LIN28B-HIS protein that contains a PreScission protease cleavage site between the two RNA binding domains at amino acids 108–114 (Fig. 4A). Following crosslinking and RNase digest, the N-terminal FLAG-tag was used to immunopurify the full-length protein. We then used PreScission protease to cleave crosslinked LIN28B protein between CSD and ZKD. Following cleavage of full-length LIN28B, the C-terminal HIS-tag enabled purification of the ZKD fragment allowing us to perform individual domain PAR-CLIP (iDo-PAR-CLIP). Resulting domain fragments were separated on SDS-PAGE (Fig. 4B), and excised from the gel. Crosslinked RNA fragments were converted into a cDNA library amenable for Illumina sequencing. After aligning the sequence reads to the spliced human transcriptome, we detected characteristic PAR-CLIP nucleotide transitions (Fig. S4A and B). Surprisingly, we observed differences in CSD and ZKD crosslinking patterns on individual target transcripts. Figure 4C exemplifies LIN28B domain interactions on TOMM20 mRNAs. A global analysis revealed that both domains bound to largely overlapping regions with highly similar cluster occupancy profiles (determined as the number of clusters mapping to the respective region) (Fig. 4D top panel). However, centering sequence clusters on the strongest local transition sites observed in the ZKD PAR-CLIP showed increased CSD crosslinking in a 5′-proximal region of ZKD binding sites (Fig. 4D, left column). Consistently, elevated ZKD crosslinking was observed 3′ of CSD binding sites (Fig. 4D, right column). Comparing the number of diagnostic transitions observed 5′ and 3′ of the respective preferred crosslinking site, we found highly significant differences in CSD and ZKD crosslinking patterns (ZKD p = 4.7e-15; CSD p = 4.9e-06) (Fig. 4D). Together these results indicate that both RNA-binding domains interact with the same RNA region and bind in close proximity of each other, suggesting a defined 5′ to 3′ domain orientation of LIN28B CSD and ZKD on target RNAs.
Figure 4. iDo-PAR-CLIP (individual Domain PAR-CLIP) enables characterization of domain-specific target interactions. (A) Domain structure of FLAG/HA-LIN28B-HIS protein harboring a PreScission protease cleavage that replaces interdomain amino acids 108–114 in LIN28B. Flowchart of iDo-PAR-CLIP approach. (B) Autoradiogram of SDS-PAGE, transferred to nitrocellulose membrane. Crosslinked protein-RNA complex migrating at 39 kDa (blue single asterisk) corresponds to full-length LIN28B protein. Two beige asterisks indicate N-terminally FLAG/HA-tagged LIN28B CSD fragment after PreScission protease cleavage. Three cyan asterisks indicate C-terminally HIS-tagged LIN28B ZKD fragment. (C) Full-length LIN28B and individual domain binding sites in TOMM20 transcript region. Sequence coverage and number of crosslinks derived from 4SU PAR-CLIPs of ZKD, CSD and LIN28B full-length (FL) protein are shown. Asterisks indicate preferred local transition site. (D) Global analysis of CSD and ZKD crosslinking patterns in iDo-PAR-CLIP data. Top panel: comparison of cluster occupancy (number of cluster at respective position). Lower panels: diagnostic transitions observed in CSD, ZKD and FL PAR-CLIP clusters. Left column: crosslinking signal in ZKD, CSD and FL PAR-CLIPs, centered on strongest local crosslinking site in ZKD PAR-CLIP data. Right column: crosslinking signal in ZKD, CSD and full-length iDo-PAR-CLIPs, centered on preferred crosslinking site in CSD iDo-PAR-CLIP data.
Next we used the full-length LIN28B 4SU PAR-CLIP library to overlap the top 300 CSD or ZKD-binding sites and deduce RNA-binding motifs that might be specific to CSD or ZKD target interactions. We found DGGGAG (D = A, T, or G) to be the best scoring motif in the top 300 ZKD-overlapping 4SU-binding sites. Conversely, the best scoring motif observed in the top 300 CSD-overlapping 4SU-binding sites was UUUUCC and rather distinct from the top scoring ZKD motif. Although we detect the domain-specific motifs with low frequency, our findings are consistent with biochemical efforts, elucidating LIN28B-binding preferences on let-7 precursors.24-27,48
LIN28B enhances protein production of mRNA target transcripts
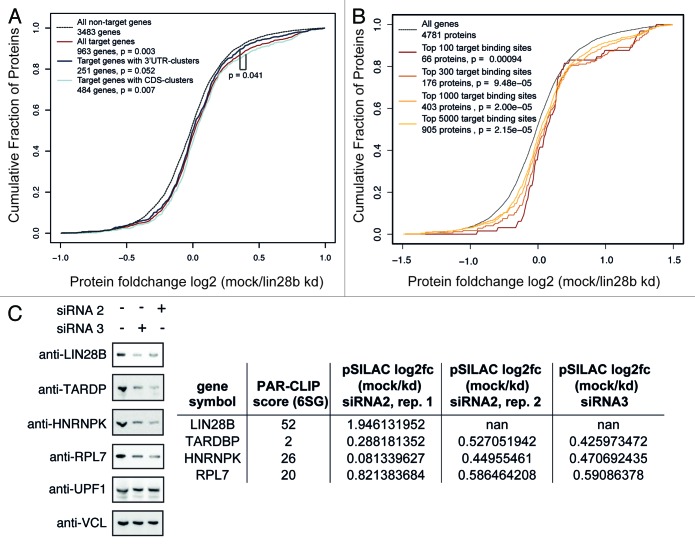
To examine the effect LIN28B exerts on expression of its mRNA target transcripts, we performed pulsed SILAC proteomics measurements upon LIN28B knockdown. Pulsed stable isotope labeling by amino acids in cell culture (pSILAC) was essentially performed as described before.42-44 Briefly, cells were grown in medium supplemented with “light” stable isotope-labeled amino acids. Upon knockdown of endogenous LIN28B, siRNA transfected cells were cultured for 24 h in medium containing “medium-heavy” stable isotope-labeled amino acids, while mock-treated cells were grown in medium containing “heavy” stable isotope-labeled amino acids (Fig. S1C). The labeled amino acids are incorporated into newly synthesized proteins, leading to a mass shift of proteins derived from LIN28B knock down (“medium-heavy”) and mock-treated (“heavy”) cells, allowing the quantification of changes in newly synthesized protein levels independent of the pool of “light” labeled pre-existing proteins. We used two different siRNAs in independent experiments and achieved 80–90% decrease in LIN28B mRNA levels, resulting in a significant reduction of LIN28B protein level (Fig. S5A). In measurements of two biological replicates (Pearson Correlation = 0.71; Fig. S5B) we were able to quantify changes in protein synthesis for about 4,500 proteins. Interestingly, mRNA transcripts bound by LIN28B showed significantly higher protein expression levels in mock-treated cells when compared with LIN28B knockdown cells (p < 0.003) (Fig. 5A; Fig. S5C). Next, we subdivided the mRNA targets into different groups based on the location of LIN28B-binding sites and found that targets bound within the CDS showed a mild, but significantly higher change in protein synthesis when compared with 3′UTR-bound targets (p < 0.041) (Fig. 5A; Fig. S5C). Interestingly, we observed increasing changes in protein synthesis when considering only the top 5,000, 1,000, 300 or 100 binding sites in our conservative set of target clusters (Fig. 5B; Fig. S5D). This observation was confirmed for the group of genes mapping to the top 100 binding sites in 4SU and 6SG PAR-CLIP experiments (Fig. S5E). We next focused on the 100 lowest ranked binding sites in 4SU and 6SG PAR-CLIPs and did not observe significant changes in protein synthesis for the corresponding genes (Fig. S5F). Thus, we hypothesize that protein production from highly ranked PAR-CLIP targets is more likely to be regulated by LIN28B. This effect is independent of let-7, as PicTar49,50 predicted let-7 targets, as a group, do not show a significant change in protein production upon LIN28 knockdown (Fig. S5G). The observation that CDS bound targets show enhanced protein production when compared with 3′UTR-bound targets suggests a previously unappreciated aspect of LIN28B regulation and points towards a functional relevance of LIN28B-binding events in the CDS.
Figure 5. LIN28B globally enhances protein synthesis of target mRNAs. (A) Cumulative density of log2 transformed changes in newly synthesized protein levels, measured by pulsed SILAC upon LIN28B knock down (pSILAC data using siRNA2, replicate1 is shown). PAR-CLIP targets with 3′UTR-binding sites only and targets with CDS-binding sites are compared with all targets and non-targets. (B) Cumulative density of log2 transformed changes in newly synthesized protein levels upon LIN28B knockdown. Genes covered by the top 5,000, 1,000, 300 and 100 conservative binding sites are shown (pSILAC data from siRNA2, replicate 1 is shown). All indicated p values are based on Wilcoxon rank sum test to test whether the two distributions significantly differ by a non-zero shift. (C) Western analysis of target transcript encoded proteins upon LIN28B knockdown using siRNA2 and siRNA3. UPF1 and vinculin (VCL) served as controls. Table indicates log2 fold changes in protein synthesis upon LIN28 knockdown as determined by pSILAC experiments.
We validated the changes in protein synthesis as observed in pSILAC experiments in LIN28B-knockdown cells by western analysis of target transcript-encoded proteins (Fig. 5C). A reduction in protein levels was observed for the LIN28-targets TARDP, HNRNPK and RPL7 upon LIN28 depletion, whereas no significant protein changes could be detected for non-targets UPF1 and vinculin (VCL).
LIN28B controls core cell cycle regulators
Gene Ontology analysis of LIN28B-targeted transcripts revealed a highly significant enrichment of genes involved in ribosome (p = 3.0E-120), cell cycle (p = 1.3E-36), spliceosome (p = 4.0E-36) and pathways in cancer (p = 6.7E-30) (Table S3). Importantly, genes belonging to the most significantly enriched GO-term “ribosome” also represent the strongest LIN28B PAR-CLIP targets and exhibit highest log2 fold changes in pSILAC experiments upon LIN28B knockdown (Table S2 and Fig. S6). LIN28B binding and regulation of mRNA targets involved in cell cycle control and gene regulation is consistent with its well-established role in stem cell differentiation and oncogenesis.17 In agreement with these findings, we observed a strong reduction of cell proliferation in LIN28B-knockdown cells (Fig. 6A). Accordingly, cell cycle analysis by DNA content (propidium iodide staining) revealed a substantially higher percentage of cells residing in the G2/M phase under LIN28B-knockdown conditions (Fig. 6B), further supporting the importance of LIN28B-mRNA interactions in cell cycle control.
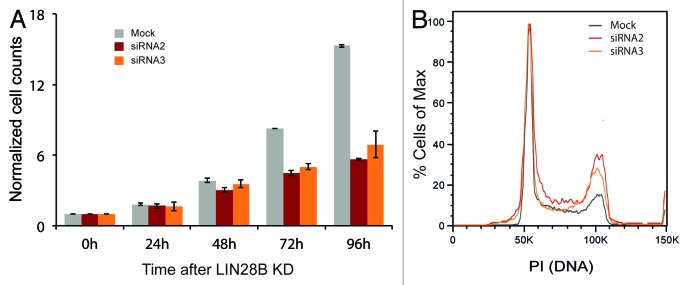
Figure 6. LIN28B controls cell growth and regulates cell cycle. (A) Normalized cell numbers following LIN28B knockdown using two different siRNAs over a period of 96 h. (B) Flow cytometry plot of cell cycle staining by propidium iodide after 72 h of LIN28B knockdown. Results are representative for three independent experiments using two different siRNAs.
Discussion
Understanding LIN28 biology at both the molecular and functional level is as fascinating as complex. The complexity of LIN28 mediated regulation manifests in our finding, consistent with other studies (ref. 38), that LIN28A can bind to most expressed mRNAs in the cell. While the LIN28-let-7 axis clearly represents the most intensely studied aspect of LIN28 function we provide insights into the mRNA binding and regulatory activity of this RBP. Application of PAR-CLIP enabled us to generate a high-resolution map of LIN28B-RNA interactions and revealed that the most abundantly bound class of RNAs are protein coding transcripts rather than miRNA precursors. In addition to LIN28B binding to precursors of the let-7 family as well as pre-miRNA-663, pre-miR-19b and pre-miR-16, we observed LIN28B crosslinking to about 10.000 protein-coding transcripts.
Among the top LIN28B targets is the LIN28B message itself. Autoregulation of their own mRNA is a commonly observed feature of many RBPs, including SR proteins SRSF1 and SRSF2, hnRNP members HNRNPD, PTB, and HNRNPL, ELAVL1 and TARDBP/TDP-43.51-61 In the case of LIN28, this is particular interesting as it suggests the existence of a second and let-7-independent feed-forward mechanism to maintain high levels of LIN28B in undifferentiated, highly proliferative cell types. Conversely, low LIN28B levels allow accumulation of mature let-7 that was, in turn, shown to repress LIN28B translation as part of a negative feedback loop, thereby promoting cell differentiation.
We identified RGGSWG as the most enriched motif in our conservative set of LIN28B PAR-CLIP-binding sites. A highly similar motif was previously shown to be sufficient for LIN28 binding to the terminal pre-let-7 loop. The RGGSWG motif is highly similar to the most frequently observed pentamer GGAGA and hexamer AAGGAG in Lin28a HITS-CLIP experiments,38,39 suggesting LIN28A and B contact highly similar recognition elements. However, when comparing the sequence regions reported to be bound by LIN28A in HEK293 cells39 to LIN28B-binding sites identified in this study, we detected only a limited overlap (Fig. S1H). Thus, this observation could be interpreted as evidence for largely distinct LIN28A and B-binding sites. On the other hand, Hafner and colleagues showed that LIN28A and LIN28B proteins bound largely the same target sites, as indicated by a 60% overlap of binding sites.41
Two recent studies provided biochemical evidence linking the GGAG consensus to binding of the LIN28 ZKD,24,25 while no consistent sequence motif could be identified to explain CSD binding. Several studies suggested low sequence specificity for RNA binding by CSD. Furthermore, Mayr and colleagues showed that the LIN28 CSD is involved in remodeling the terminal pre-let-7 loop.24 Thus, it appears conceivable that yet-to-be-identified structural elements contribute to CSD target recognition. Using N- and C-terminally tagged LIN28B that harbors a protease cleavage site between CSD and ZKD, we were able to develop iDo-PAR-CLIP (individual Domain PAR-CLIP) a method to study the binding preferences of different RNA-binding domains encoded by a single RBP in isolation. As full-length LIN28B proteins are crosslinked to their natural targets prior to protease cleavage, physiological binding preferences should be retained. Our data suggest binding of both domains in close proximity to largely overlapping sequence clusters with a maximal binding distance of 20 nts between the domain specific interaction sites (Fig. 4D). Consistent with earlier studies, these findings indicate that both RNA-binding domains interact with the same RNA molecule, pointing toward a 1:1 ratio between LIN28 protein and its bound RNAs.62 Furthermore, we provide evidence that the CSD and ZKD of LIN28B are arranged in a 5′ to 3′ orientation on RNA. Similarly, Nam and colleagues concluded based on structural and biochemical observations that the ZKD interacts with a G-rich region downstream of the CSD-binding site.25 Consistent with this finding, we report a G-rich binding motif in ZKD-bound sequence clusters. In conclusion, iDo-PAR-CLIP provides many valuable insights into LIN28B CSD and ZKD interaction on binding targets and confirms important biochemical and structural observations. iDo-PAR-CLIP is readily applicable to any RBP that harbors different RNA-binding domains spaced by linker regions. We envision our approach to enhance our understanding of complex molecular mechanisms underlying RBP target recognition.
Most importantly, our study revealed that LIN28B globally enhances protein production of its target mRNAs independent of let-7 regulation. Strikingly, efficiently crosslinked transcripts showed most robust changes in protein synthesis upon LIN28B knockdown and CDS-bound targets were slightly more enhanced than 3′UTR-bound targets. While extensive binding of RBPs in coding sequences of target transcripts has been observed previously, we show that LIN28B-binding sites within the CDS are functionally equally relevant to 3′UTR-binding sites, thus revealing an unexpected aspect of LIN28B regulation on mRNA. This is especially intriguing as 3′UTRs are largely considered to be the major region of post-transcriptional gene regulation.1,2 Several studies provide compelling evidence that LIN28 acts as a translational activator on individual mRNAs,32-37 but until now no global effect on enhancing protein production of target mRNAs was described. Contrary to these studies and our data Cho et al. reported that LIN28A is a suppressor of ER-associated translation in embryonic stem cells,38 underlining the importance to comprehensively investigate the regulatory effect of LIN28 on protein synthesis.
LIN28-dependent translational enhancement of target transcripts involves recruitment of auxiliary factors, such as RNA helicase A (RHA/DHX9).48,63 RHA interaction is mediated by the C-terminal region of LIN28 and mutation or deletion of this region alleviates translational stimulation of LIN28-binding targets.48 Interestingly, C-terminally truncated LIN28B does not induce cancer cell proliferation.27 In this context, it appears conceivable that the variety of regulatory effects exerted by LIN28B is influenced by additional factors interacting with its C-terminal regions.48
Looking more closely into genes bound and potentially regulated at the level of protein synthesis by LIN28B, it is striking that the genes associated with the ribosome and cell cycle pathways are significantly overrepresented. Consistent with strong proliferative defects observed upon LIN28B knockdown, prominent genes such as CDK1, NRAS, RAN and ERK, controlling core signaling pathways, were shown to be directly bound by LIN28B. The ERK signaling cascade participates in the regulation of a large variety of processes including cell cycle progression, differentiation, tumorigenesis and cell death.64 Strikingly, ribosomal proteins stand out both in being among the top LIN28B targets and showing strong changes in protein levels upon LIN28B knockdown. The latter observation is of particular interest given that LIN28 mutants showed strong phenotypes in growth and metabolism.16 Thus, our transcriptome-wide study on LIN28B-binding preferences might provide a molecular link connecting LIN28B phenotypes with specific cellular pathways.
Finally, the type-2 diabetes-associated genes HMGA2 and IGF2BP2 were also found among direct LIN28B-binding targets, and showed a up to 2-fold decrease in protein production after LIN28B knockdown. Interestingly, a recent study in mice correlated impaired glucose tolerance and insulin resistance with muscle-specific loss of Lin28 and regulation of the insulin-PI3K-mTOR pathway, partly as a result of let-7 activity.65 Consistent with these findings, the identification of genes belonging to the insulin-PI3K-mTOR pathway as direct LIN28B targets suggest an even more prominent role of LIN28B in directly regulating insulin-PI3K-mTOR signaling through its mRNA-binding function.
Materials and Methods
Antibodies
anti-HA.11 (COVANCE, 16B12), anti-FLAG (SIGMA, F1804), anti-LIN28B (Cell Signaling, 4196) anti-TARDP (Abcam, ab57105), anti-HNRNPK (Abcam, ab52600), anti-RPL7 (Abcam, ab72550), anti-UPF1 (Bethyl, A300-036A), anti-VCL (Sigma, V4505), anti-tubulin (Sigma, T4026).
Oligonucleotides
Small RNA cloning adapters
5′adapter
rGrUrUrCrArGrArGrUrUrCrUrArCrArGrUrCrCrGrArCrGrArUrC
3′ barcoded adapters (barcode is underlined)
NBC3: AppTCTGGGATCGTATGCCGTCTTCTGCTTG-InvdT
NBC4: AppTCTTTTATCGTATGCCGTCTTCTGCTTG-InvdT
NBC6: AppTCTCCATTCGTATGCCGTCTTCTGCTTG-InvdT
NBC7: AppTCTCGTATCGTATGCCGTCTTCTGCTTG-InvdT
NBC8: AppTCTCTGCTCGTATGCCGTCTTCTGCTTG-InvdT
siRNAs
siRNA 2: GGAAGGAUUUAGAAGCCUA
siRNA 3: GGGAAGACAGGAAGCAGAA.
Plasmids
pENTR constructs were generated by PCR amplification of the LIN28B coding sequences (CDS) from cDNA followed by restriction digest and ligation into pENTR4 (Invitrogen) backbone. The PreScission site (amino acids LEVLFQGPT) was inserted in pENTR4/LIN28B to replace the interdomain region between amino acids 108 und 114. In addition, a C-terminal HIS-tag was added. pENTR4/LIN28B and pENTR/LIN28BPrescission-HIS was recombined into pFRT/TO/FLAG/HA-DEST destination vector7 using GATEWAY LR recombinase (Invitrogen) according to manufacturer's protocol to allow for doxycycline-inducible expression of stably transfected FLAG/HA-tagged LIN28B and F:AG/HA-LIN28BPrescission-HIS protein in Flp-In 293 T-REx (Invitrogen) from the inducible TO/CMV promoter. The plasmids described in this study can be obtained from Addgene (www.addgene.org).
Cell lines and culture conditions
Flp-In 293 T-REx cells (Invitrogen) were grown in D-MEM high glucose with 10% (v/v) fetal bovine serum, 1% (v/v) 2 mM L-glutamine, 1% (v/v) 10,000 U/ml penicillin 10,000 µg/ml streptomycin, 100 µg/ml zeocin and 15 µg/ml blasticidin.
Cell lines stably expressing FLAG/HA-tagged Lin28B FLAG/HA-LIN28B-prescissionHIS protein were generated by co-transfection of pFRT/TO/FLAG/HA constructs with pOG44 (Invitrogen). Cells were selected by exchanging zeocin with 100 µg/ml hygromycin (Invivogen). Expression of epitope-tagged proteins was induced by addition of 1 µg/ml doxycycline 15–20 h before crosslinking. The expression of FLAG/HA-tagged LIN28B protein was assessed by western analysis using mouse anti-HA.11 monoclonal antibody (Covance).
For quantitative proteomics, cells were grown in SILAC medium as described in references 44 and 66. Briefly, Dulbecco’s modified Eagle’s medium (DMEM) Glutamax lacking arginine and lysine (PAA) supplemented with 10% dialyzed fetal bovine serum (dFBS, Gibco) was used. Amino acids (84 mg/l 13C615N4 l-arginine plus 146 mg/l 13C615N2 l-lysine or 84 mg/l 13C6-l-arginine plus 146 mg/l D4-l-lysine) or the corresponding non-labeled amino acids (Sigma), were added to obtain “heavy,” “medium-heavy” or “light” cell culture medium, respectively. Labeled amino acids were purchased from Sigma Isotec.
PAR-CLIP
Stably transfected and inducible LIN28B-expressing cells were labeled with 100 µM 4-thiouridine (4SU) or 6-thioguanosine (6SG) for 12 h. After labeling the cells, PAR-CLIP was performed as described in reference 40. Briefly, UV-irradiated cells were lysed in NP-40 lysis buffer [50 mM HEPES-KOH at pH 7.4, 150 mM KCl, 2 mM EDTA, 0.5% (v/v) NP40, 0.5 mM DTT, complete EDTA-free protease inhibitor cocktail]. Immunoprecipitation was performed with protein G magnetic beads (Invitrogen) coupled to anti-FLAG M2 antibody (SIGMA) from extracts of FLAG/HA-LIN28B expressing and 4SU-labeled HEK 293 cells for 1 h at 4°C. Following RNase T1 (Fermentas) treatment, beads were incubated with calf intestinal phosphatase (NEB) and RNA fragments were radioactively end-labeled using T4 polynucleotide kinase (Fermentas). The crosslinked protein-RNA complexes were resolved on a 12% NuPAGE gel (Invitrogen). The SDS-PAGE gel was transferred to a nitrocellulose membrane (Whatman) and the protein-RNA complex migrating at a molecular weight of ~40 kDa was excised. RNA was isolated by Proteinase K (Roche) treatment and phenol-chloroform extraction, reverse transcribed and PCR-amplified. The amplified cDNA was sequenced on a HighSeq2000 (Illumina) with a 1 × 50 nt cycle.
iDo-PAR-CLIP
We generated a recombinant FLAG/HA-LIN28B-HIS construct encoding a PreScission protease cleavage site between the two RNA-binding domains at amino acids 108–114. Stably transfected and inducible FLAG/HA-LIN28B-HIS-expressing cells were labeled with 100 µM 4-thiouridine (4SU) for 12 h. Initial RNase treatment and FLAG immunoprecipitation, dephosphorylation and 5′end radiolabeling of crosslinked full-length LIN28B-RNA complexes was performed as described for PAR-CLIP experiments. PreScission protease was used at a final concentration of 0.45 µg/ml for 1h at 4°C to cleave immunopurified LIN28B protein between CSD and ZKD. Following incubation, supernatant was removed and incubated with HIS-tag isolation dynabeads (Invitrogen) for 30 min at room temperature. Remaining FLAG beads were washed three times in IP wash buffer and resuspended in SDS-loading buffer to elute full-length LIN28B and N-terminal CSD fragments. Following incubation, HIS-tag beads were washed three times in IP wash buffer and C-terminal ZKD fragments were eluted by boiling beads for three min in SDS-PAGE sample loading buffer. LIN28B fragments were separated on SDS-PAGE and transferred to a nitrocellulose membrane (Whatman) (1 h 20V). Radiolabeled N- and C-terminal LIN28B fragments were excised from the membrane and crosslinked RNA fragments were converted into a cDNA library as described for PAR-CLIP experiments.
siRNA knockdown and pSILAC
FLAG/HA-LIN28B-HEK293 cells were grown in SILAC medium supplemented with “light” labeled amino acids prior to siRNA-knockdown experiments. siRNAs were transfected at a final concentration of 60 nM using Lipofectamine RNAiMAX (Invitrogen). Controls (mock) were treated with transfection reagent only. Following 24 h of incubation, siRNA transfected cells were switched to “medium-heavy”-labeled SILAC medium, while mock control cells were switched to “heavy”-labeled SILAC medium. After another 24 h of labeling, cells were harvested and equal amounts of siRNA- and mock-transfected cells were combined. Proteins were extracted and disulfide bridges reduced and alkylated with iodoacetamide. After overnight digestion with LysC and trypsin, the peptide mixture was desalted and fractionated by isoelectric focusing on a microrotofor Cell device (Biorad).
Peptides from each fraction were desalted using STAGE Tips67 and analyzed by LC-MS/MS on a Thermo LTQ Velos mass spectrometer. Raw data were analyzed using MaxQuant software for peptide/protein identification (1% false discovery rate) and for quantification.
Western blot
Total cell lysates were prepared in 1x SDS-PAGE sample loading buffer (50 mM Tris pH7.5, mercaptoethanol, 1% SDS, 0.01% bromophenol blue, 10% glycerol) and resolved on a 12% SDS-PAGE gel. Proteins were transferred to nitrocellulose membrane (Whatman) using a semi-dry blotting apparatus (BioRad) at 2 mA/cm2. The membrane was blocked in 5% non-fat milk and incubated with primary antibody from 1 h to overnight. Following incubation, membranes were washed three times in TBST and incubated with HRP-conjugated secondary antibody for 1 h. Following three additional TBST washes, protein bands were visualized using ECL detection reagent (GE Healthcare, RPN2106) and a LAS-4000 imaging system (GE-Healthcare).
Quantitative PCR
Following siRNA knockdown, cells were harvested and RNA was isolated using Trizol (Invitrogen). CDNA (cDNA) synthesis was performed after DNase (Invitrogen) treatment using Superscript III (Invitrogen) with Oligo(dT18–20) primers. qPCR analysis was performed with SYBR Green PCR Master Mix and ABI light cycler as described in the manufacturers’ instructions.
Cell cycle analysis
Briefly, cells were harvested 48 h or 72 h after knockdown using 0.05% Trypsin (PAA) to bring cells into single cell suspension. To exclude secondary effects on the cell cycle due to proliferation-based depletion of medium-nutrients, medium was replaced daily and in all conditions by fresh medium. Cells were then counted, diluted to same concentrations, fixed by pure ethanol and labeled by 10 µg/ml propidium-iodide (Sigma-Aldrich) after digestion of RNA through 0.5 mg/ml RNase (Roche). Finally, cells were acquired by FACS at 488 nm (BD Fortessa) and single cells were analyzed using FlowJo 8.8.6 (Tree Star).
Computational analysis
Cluster definition
Deep sequencing reads were quality trimmed to at least three subsequent nucleotides (nts) at their 3′-end with a minimal Sanger quality score of 25. Adaptor sequences with a minimal overlap of 7 nts were clipped and a minimal final read length of 15 nts was required to avoid ambiguous mapping. Processed reads were mapped to the human genome (hg18), precursor RNAs and spliced RNAs using BWA (version 0.5.8c,68) and read-length and library-dependent mapping distances to optimize the signal-to-noise ratio (4SU: 15–23 nt 1, > 23 nt 2; 6SG: 15–22 nt 1, > 22 nt 2). Reads mapping to chromosome Y and antisense to spliced RNAs were used for noise estimation. Alignments mapping to antisense-spliced RNA or to multiple loci of different genes were discarded. The alignments were further processed using samtools (version 0.1.8). Clusters were extracted from the pileup files and scored for signal transitions (T to C mutations and T deletions or G to A mutations and G deletions), mutations, coverage and entropy. These clusters were filtered to have at most 20% of all nucleotides mutated or only signal transitions otherwise, at least 50% of non-repetitive contribution, a non-zero entropy and a minimal signal to coverage ratio of 1% (4SU) or 0.1% (6SG). These parameters were defined to exclude low-quality clusters and potential low-affinity binding sites. For further analysis, only clusters with two independent transitions (i.e., two different transition types or positions) were considered to counteract false-positive signals, such as SNPs. These filtered clusters were then intersected to a “conservative cluster set,” consisting only of binding sites with a cluster in the 6SG library and an overlapping cluster within one of the 4SU libraries, allowing for a flank of 30 nts to retain binding sites in close proximity that potentially may only be detected by one or the other type of nucleoside used in the libraries.
Annotation of clusters
Binding sites were annotated using customized scripts and based on hg18 refseq tables. Binding sites with ambiguously mapping, e.g., due to different isoforms or overlapping annotation boundaries, were subjected to a priority classification if unique annotation was needed. Priority was given to CDS, 3′UTR, 5′UTR and intron in this order as the coding sequence is annotated with high confidence.
Generation of control clusters
Different isoforms were projected to a single transcript per gene, such that the coverage of the annotated regions is maximized in the priority given above. This was done to maximize the homogeneity of sequence features that serve as a control. For example, a region used as a CDS in one isoform likely shows dominant CDS features and should not be subjected to a possible control for a 3′UTR sequence. Control clusters for binding sites were selected from the same gene out of a random region with the same annotation. When control sites for transition-centered binding sites were generated, these were forced to have the same nucleotide in their center (e.g., a T or a G), to minimize possible biases.
Motif analysis
Motifs in different subsets and of different lengths were searched using MEME,69 mostly using slightly modified standard parameters (-dna-mod zoops-minsites 20). In general, this method was complemented by extensive analysis based on customized scripts, especially focusing on the frequencies of specific motifs and their conservation both in binding sites and control sites. Motifs were centered and subjected to binding site analysis.
Binding site analysis
Vertebrate conservation scores were retrieved from UCSC genome browser (hg18, vertebrate, phyloP44way) and averaged if multiple binding sites were overlaid. Binding site probabilities were computed using a customized script42 that implements binding probability matrices of the VIENNA package.
Quantification of protein expression
Proteins were quantified based on pSILAC ratios computed by MaxQuant.70 For stringency, a minimal ratio count of three was used as a threshold and known contaminants were excluded. The shown protein changes represent the log2 transformed normalized H/M values, therefore indicating changes in protein production in mock-treated cells compared with LIN28B-knockdown cells.
Supplementary Material
Acknowledgments
R.G. and M.M. contributed equally and are listed in alphabetical order. R.G. performed the computational analysis, cell cycle experiments and designed figures under the supervision of N.R. M.M. performed all other experiments and developed iDo-PAR-CLIP under the supervision of M.L. G.M. performed pSILAC measurements supervised by S.K. F.M. contributed FLAG-LIN28B-HIS plasmid supervised by U.H. We thank all members of the Landthaler and N. Rajewsky labs for discussions and comments. We would like to express our gratitude to Marvin Jens for computational advice, Claudia Langnick and Mirjam Feldkamp from the Wei Chen lab (MDC) for sequencing and Ouidad Benlasfer (Landthaler lab) for generation of cell lines. We acknowledge the following funding sources: International PhD program of the Max-Delbrück-Center (MDC) (R.G.); the MDC-NYU exchange program (M.M.); BMBF support for the DZHK (N.R.); Funding for the group of M.L. is supported by BMBF-funding for the Berlin Institute for Medical Systems Biology (0315362C). R.G. thanks Klaus Rajewsky for support.
Accession Number
The sequencing data have been deposited in the GEO database under GSE46908.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25194
References
- 1.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 2.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 3.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–30. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–25. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 8.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–16. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Ng SB, Chng WJ. LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells. Int J Biochem Cell Biol. 2013;45:973–8. doi: 10.1016/j.biocel.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–79. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 14.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3:719–26. doi: 10.1016/S1567-133X(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–30. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–82. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 20.Martinez NJ, Gregory RI. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell. 2010;7:31–5. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–20. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Mayr F, Schütz A, Döge N, Heinemann U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 2012;40:7492–506. doi: 10.1093/nar/gks355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–91. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, Allain FH. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol. 2012;19:84–9. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 27.Desjardins A, Yang A, Bouvette J, Omichinski JG, Legault P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2012;40:1767–77. doi: 10.1093/nar/gkr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. Bioessays. 2010;32:109–18. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 29.Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J Mol Biol. 2006;360:702–14. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 30.Zeeb M, Balbach J. Single-stranded DNA binding of the cold-shock protein CspB from Bacillus subtilis: NMR mapping and mutational characterization. Protein Sci. 2003;12:112–23. doi: 10.1110/ps.0219703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 32.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–38. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 34.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–61. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang YC, Chen YL, Yuan RH, Pan HW, Yang WC, Hsu HC, et al. Lin-28B expression promotes transformation and invasion in human hepatocellular carcinoma. Carcinogenesis. 2010;31:1516–22. doi: 10.1093/carcin/bgq107. [DOI] [PubMed] [Google Scholar]
- 36.Xu B, Huang Y. Histone H2a mRNA interacts with Lin28 and contains a Lin28-dependent posttranscriptional regulatory element. Nucleic Acids Res. 2009;37:4256–63. doi: 10.1093/nar/gkp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–8. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–77. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 39.Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, et al. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19:613–26. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 44.Schwanhäusser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–9. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- 45.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–64. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 46.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202-8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei XX, Xu J, Ma W, Qiao C, Newman MA, Hammond SM, et al. Determinants of mRNA recognition and translation regulation by Lin28. Nucleic Acids Res. 2012;40:3574–84. doi: 10.1093/nar/gkr1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 50.Anders G, Mackowiak SD, Jens M, Maaskola J, Kuntzagk A, Rajewsky N, et al. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2012;40(Database issue):D180–6. doi: 10.1093/nar/gkr1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pullmann R, Jr., Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, et al. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–78. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–30. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, et al. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29:1442–51. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–96. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–32. doi: 10.1017/S1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–9. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 59.Ayala YM, De Conti L, Avendaño-Vázquez SE, Dhir A, Romano M, D’Ambrogio A, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–88. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avendaño-Vázquez SE, Dhir A, Bembich S, Buratti E, Proudfoot N, Baralle FE. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 2012;26:1679–84. doi: 10.1101/gad.194829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol. 2010;17:306–12. doi: 10.1038/nsmb.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ali PS, Ghoshdastider U, Hoffmann J, Brutschy B, Filipek S. Recognition of the let-7g miRNA precursor by human Lin28B. FEBS Lett. 2012;586:3986–90. doi: 10.1016/j.febslet.2012.09.034. [DOI] [PubMed] [Google Scholar]
- 63.Jin J, Jing W, Lei XX, Feng C, Peng S, Boris-Lawrie K, et al. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–34. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramaniam S, Unsicker K. ERK and cell death: ERK1/2 in neuronal death. FEBS J. 2010;277:22–9. doi: 10.1111/j.1742-4658.2009.07367.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, et al. DIAGRAM Consortium. MAGIC Investigators The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 70.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.