Abstract
In Bacilli, there are three experimentally validated ribosomal-protein autogenous regulatory RNAs that are not shared with E. coli. Each of these RNAs forms a unique secondary structure that interacts with a ribosomal protein encoded by a downstream gene, namely S4, S15, and L20. Only one of these RNAs that interacts with L20 is currently found in the RNA Families Database. We created, or modified, existing structural alignments for these three RNAs and used them to perform homology searches. We have determined that each structure exhibits a narrow phylogenetic distribution, mostly relegated to the Firmicute class Bacilli. This work, in conjunction with other similar work, demonstrates that there are most likely many non-homologous RNA regulatory elements regulating ribosomal protein biosynthesis that still await discovery and characterization in other bacterial species.
Keywords: ribosomal protein, Rfam, gram-positive, Bacillus subtilis, Geobacillus stearothermophilus, Infernal, ribosomal leader sequence
Introduction
There are a wide variety of different bacterial regulatory RNAs ranging from riboswitches that require complex secondary- and tertiary- structure motifs for function1 to sRNAs that typically act predominantly through base-pairing interactions.2,3 Some of the first regulatory RNAs to be described are those that autogenously regulate ribosomal protein biosynthesis in Escherichia coli.4 These regulatory RNAs typically occur within 5′-untranslated or intergenic regions of transcripts encoding ribosomal proteins. When transcribed, these RNA sequences form secondary structures that interact with a specific ribosomal protein binding partner to regulate an entire ribosomal protein operon.5 The mechanism of gene regulation can be either transcriptional6 or translational,7-9 thus allowing these mRNA structures to act as a means of feedback inhibition. To date, over 10 such RNAs regulating more than half of ribosomal protein genes have been described in E. coli. However, recent work has shown that most of these RNAs are narrowly distributed to Gammaproteobacteria.10
Progress toward understanding the regulation of ribosomal protein biosynthesis in gram-positive bacteria, including model organisms such as Bacillus subtilis, has been much more limited. Ribosomal proteins are typically universally distributed and well conserved across bacterial phyla.11 In addition, the over 50 genes encoding ribosomal proteins occur in long multi-gene operons whose structure is largely, but not completely, conserved.12 Despite the universal nature of these proteins, their regulation does not appear to be conserved. Of the E. coli ribosomal protein regulatory RNAs, only two (interacting with ribosomal proteins L1 and L10)10,13 are widely present in Firmicutes (> 85% of all sequenced Firmicutes). A third RNA structure (interacting with ribosomal protein S2) has been identified in > 50% of sequenced Firmicutes.10,14 In addition to these widely distributed RNAs, RNA structures interacting with ribosomal proteins S4, S15 and L20 have been identified and experimentally validated in the Firmicute class Bacilli.15-17 While each of these ribosomal proteins is also a regulator in E. coli, the RNAs show little or no homology to the E. coli RNAs with the same function. Currently, Rfam alignments are only available for one of these RNAs (L20, RF00558).18 While several additional putative RNA structures associated with ribosomal proteins have been identified in comparative genomic studies,19 none have been experimentally validated.
We utilized the RNA homology search program Infernal,20 coupled with our high-capacity genomic context visualization tool,21 to identify homologs of the three experimentally validated ribosomal-protein autogenous regulatory RNAs found in Bacilli. The alignments produced in this work assess the phylogenetic distribution of these RNA structures, and integrate experimental data with comparative genomics to gain new insight into the structure and function for these RNAs.
Results
L20-interacting RNA
The RNA structure interacting with ribosomal protein L20 to regulate the infC-L30-L20 operon was discovered independently by two studies at approximately the same time. One study experimentally analyzed the RNA,17 while the other identified the RNA structure through a comparative genomics approach.19 In contrast to the L20-interacting RNA present in Gammaproteobacteria, the L20-interacting RNA in B. subtilis regulates transcription rather than translation, and does not directly precede the gene encoding its binding partner, rplT.7 Instead, the protein-binding site is located at the start of the operon, preceding and regulating the genes infC, rpmI and rplT.17 The current Rfam alignment for this RNA (Rfam: RF00558) (generated by comparative genomics in Yao et al.19), representing the L20-bound form, was used as the starting alignment for this study.
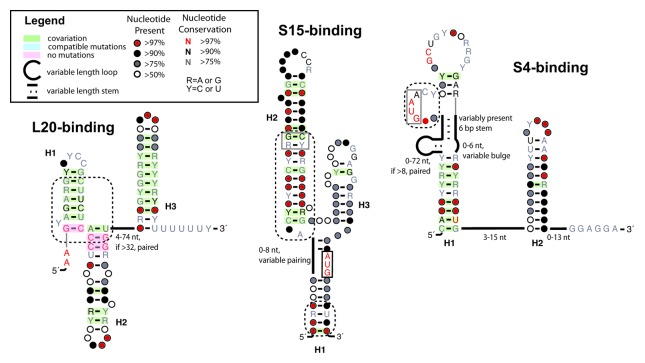
The L20-interacting RNA structure consists of three pairing elements (Fig. 1; Fig. S1), including a terminator stem that was experimentally confirmed using in vitro transcription termination assays.17 A fourth pairing element directly preceding the terminator was noted in previous studies.17 This fourth stem is present in > 75% of species in the alignment, but shows no conservation of individual nucleotides. In addition, nuclease footprinting assays indicate it is unlikely to be involved in L20 binding (Fig. 1).17
Figure 1. Consensus sequence and secondary structures of Bacilli ribosomal regulatory elements. Start codons (AUG) are depicted inside a black box when occurring within the RNA structure. Gray boxes indicate areas of high conservation and possible binding. Dotted boxes surround areas proposed to be important for binding. Co-varying base pairs are shaded red or green only when Watson-Crick base pairing is in > 95% of the aligned sequences. Helix numbering is consistent with previously published data for each RNA.
Toe-printing and RNase probing assays have determined that L20 binds specifically at the junction of Helices 1 and 2, stabilizing Helix 2 in the process.17 This region of the RNA bears striking resemblance to the L20 binding site on the 23S rRNA, and our studies show that this region is highly conserved, with few or no mutations. The terminator stem does not appear to be involved in L20 binding,17 and there is co-variation throughout the stem, indicating that the secondary structure, rather than the sequence, is important for function. There is also a rigorously conserved pair of adenosines before the start of the first helix, but their effect on binding is unknown.
Of the RNAs examined here, the L20-binding RNA has the greatest penetration in sequenced Firmicutes (Fig. 2). It is found in most Bacilli, and many Clostridia species. A few homologs of the L20-interacting RNA are also identified in Thermotogae and Actinobacteria. While Thermotogae are relatively closely related to Firmicutes,22 the presence in Actinobacteria species suggests possible horizontal transfer events.
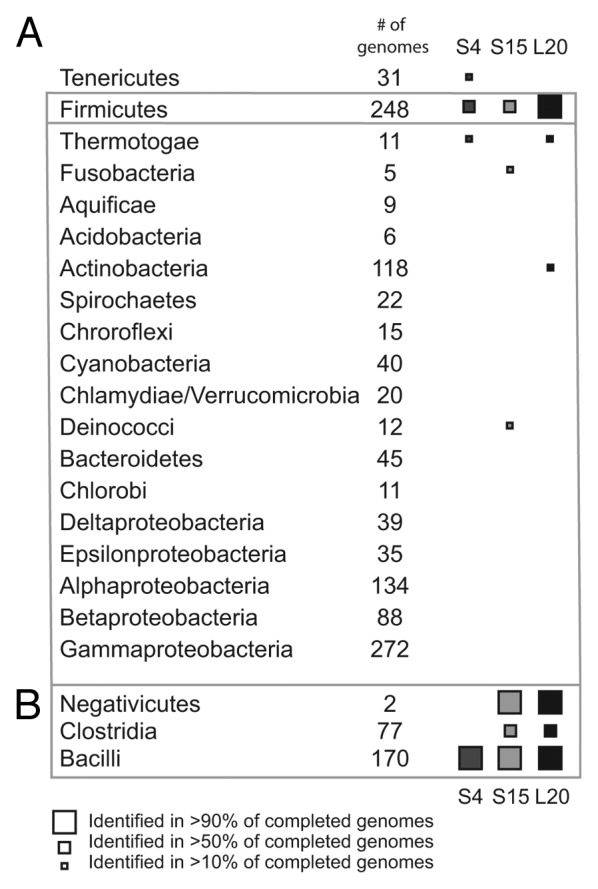
Figure 2. Phylogenetic distribution of Bacilli autogenous ribosomal regulators. (A) Distribution of autogenous regulators of ribosomal protein synthesis in eubacterial phyla. (B) Distribution of autogenous regulators of ribosomal protein synthesis for classes within the phylum Firmicutes.
S15-interacting RNA
The mRNA structure interacting with ribosomal protein S15 regulates only rpsO.23 Like its E. coli counterpart,24 the RNA structure identified in Bacilli overlaps with the beginning of the coding region for rpsO.23 The RNA was first identified in B. stearothermophilus (subsequently reclassified as Geobacillus stearothermophilus),25 and RNA-protein interaction was experimentally validated initially utilizing in vitro approaches. Regulatory activity of the RNA was subsequently demonstrated using E. coli as a surrogate organism.23 There is no Rfam alignment for this RNA, nor has it been identified in previous comparative genomic works.
For this study, we manually constructed a starting alignment consisting of the 3-helix junction necessary for binding and regulation16,23using BLAST to identify the initial homologs. The starting alignment contained sequences from several Geobacillus species, as well as a hand-aligned portion of the genomic region preceding rpsO in B. subtilis and Caldicellulosiruptor bescii DSM 6725 (a member of Clostridia). Utilizing this initial alignment we were able to identify the S15-binding RNA in most Bacilli species, and in both sequenced Negativiticutes species (Fig. 2). However, its incidence in Clostridia is considerably lower, resulting in a lower overall frequency in Firmicutes. We also identified sequences in Deinococci and Fusobacteria (two sequences in each phylum), suggesting potential horizontal transfer. However, it is difficult to make a definitive conclusion in this matter due to the small number of putative homologs and lack of any experimental data to verify them.
Although alternative structures may exist, our final secondary structure is presumably stabilized by interactions with S15 (Fig. 1; Fig. S2).16 Consistent with deletion studies suggesting the length of Helix 1 may vary,23 the sequence and length of Helix 1 in naturally occurring examples can range from nine to 17 base pairs. The putative “AUG” start codon within the loop of that helix (Fig. 1, black box) is highly conserved, appearing in > 97% of all sequences. Similarly to Helix 1, Helix 2 shows nucleotide sequence variability, but base pairing throughout the stem is largely maintained. While deletion studies have shown that the H2 helix may be reduced to 29 nucleotides and still retain functionality,16 our alignment shows that the full-length stem is maintained in > 90% of all sequences. A consecutive set of “G-C” and “G·U” pairs in H2, appearing as “G-C” and “R·Y” base pairs in Figure 1 due to sequence variability, were both expected to be highly conserved, as both base pairs are reported to be important for binding.16 However, only the “G-C” pair showed > 90% conservation, while the “G·U” pair (“R·Y” in Fig. 1) exhibited much greater sequence variability, though base pairing was maintained. Helix 3 includes a conserved “GGAGG” that based on its location relative to the conserved “AUG,” is likely part of the Shine-Delgarno sequence.
S4-interacting RNA
The B. subtilis S4-interacting RNA regulates only the gene encoding S4, rpsD. In E. coli, the operon containing S4 also contains four additional ribosomal genes, and the S4 protein regulates the synthesis of all of them.26 In B. subtilis, this gene cluster does not include rpsD.27,28 Rather rpsD is at a different location in the genome and is likely to be the only gene regulated by this RNA. B. subtilis S4 represses its own synthesis post-transcription initiation, but the mechanism of action remains unknown. The S4 protein is known to interact with 16S rRNA, and parts of the B. subtilis 5′-UTR (5′-untranslated region) have sequence and structural similarity to 16S.29 The S4-interacting RNA is found in only Firmicutes, Tenericutes and Thermotogae, although within Firmicutes, the RNA does not appear in Clostridia or in the two Negativiticute genomes analyzed (Fig. 2).
The starting alignment used here originated from the supplementary material of a comparative genomic screen of Firmicutes.19 However, the structure derived from comparative genomics did not match the experimental structure proposed by Grundy et. al.29 In particular, the proposed structure incorporates sequence demonstrated to have no regulatory activity. For the work presented here, the structure was manually edited to match the structural prediction based on experimental data.29
This structure contains two hairpins with variable bulges and stems (Fig. 1; Fig. S3). We found the hairpin branching from the first helix to be especially variable, both in sequence and in presence. The “GUAA” bulge (Fig. 1, gray box) remains conserved and is proposed to interact with the S4 protein,15,29 as it has sequence identity to a similar bulge on the 16S rRNA.30,31 Also, in most sequences aligned, the Shine-Dalgarno sequence follows relatively closely downstream of the second variable helix of the RNA (Fig. 1). We evaluated the presence of two pseudoknots proposed in the original description of this RNA structure.15 Mutations to these pseudoknots did not affect regulation,29 and our alignment shows no support for them.
We also performed homology searches with the original alignment derived from comparative genomics by Yao and coworkers,19 as this structure has the potential to be an alternative non-interacting conformation for the RNA. The final alignments of the two possible structures share significant taxonomic overlap, 172 out of 177 species, indicating that both structures are possible in most species. The majority of the alternative structure is quite similar to the S4-binding structure. H1 is largely intact in the alternative structure, with a conserved “GUAA” bulge near the top of the stem, as well as similarly conserved loops. There are some minor changes to the base pairing of H1, causing the protein binding site (“GUAA”) and the top loop to be smaller than their correlates in the S4-binding structure. More dramatically, the alternative structure lacks the H2 stem, and instead has an elongated H1 stem that partially overlaps the existing H2. In order to elongate the H1 stem, the alternative structure includes a 5′ extension of 25–30 nucleotides. Although this 5′ extension corresponds exactly with the transcriptional start in B. subtilis,27 deletion of this region was shown to have no impact on S4 protein binding.29 An alignment and secondary structure diagram for this alternative structure are included in the supplementary data (Figs. S4 and S5).
Discussion
Scientists have been aware of autogenous regulators of ribosomal protein synthesis since 1980.4 While there is a good understanding of the RNA structures that regulate ribosomal protein regulation in E. coli, our knowledge of these elements outside of E. coli is sorely lacking. While three of the E. coli RNA regulators are widely distributed, the majority are not. In Bacilli, experimentally validated RNA structures interacting with S4, S15 and L20 are known to regulate ribosomal proteins. These structures show no homology to RNAs interacting with homologous proteins from E. coli.
This study created alignments for two of the three RNAs unique to Bacilli and assessed the homologs in the context of previous experimental results. We have found that the three regulatory elements examined—interacting with S4, S15 and L20—have a narrow evolutionary distribution, even so narrow as to exclude the Firmicute class Clostridia from the S4 distribution. Based on various experimental analyses it is apparent that there are multiple evolutionarily distinct regulatory RNAs responding to the same ribosomal protein in different bacterial phyla.7,16,17,26,29,32 Furthermore, most of the characterized RNA structures responsible for regulating ribosomal protein biosynthesis appear to be narrowly distributed,10 and comparative genomic studies have discovered a number of putative RNA structures associated with ribosomal proteins in Firmicutes and other bacterial species that have yet to be verified.19,33 The combination of these observations suggests strongly that there are many distinct RNA structures responsible for ribosomal protein regulation that have yet to be identified and experimentally characterized. In the future, identifying and validating non-homologous regulatory RNAs that likely have the same function in non-model organisms will lead to a more comprehensive understanding of each RNA-protein interaction and to elucidation of the evolutionary trajectories for these regulatory RNAs.
Materials and Methods
Initial multiple sequence alignments were obtained as described below for each RNA. The seed alignment for the L20-binding RNA was downloaded from the Rfam database (Rfam families: RF00558), the S4-binding RNA was obtained via Yao et al. (2007) supplementary material19 and the S15-binding RNA was created manually via BLAST (completed genomes only) matches to the G. stearothermophilus sequence of interest and hand alignments of a few selected sequences as noted in the main text. The alignments were all manually edited to remove sequences not compatible with published experimental data and to adjust base pairing. Any changes to base paring are discussed in the main text.
Covariance models for each RNA were constructed and calibrated using Infernal 1.0 (cmbuild, cmcalibrate), and homologs identified for each RNA (cmsearch).20 Cmsearch was performed against a custom sequence database as described in Fu et al.10 using a lenient e-value cut-off. Potential homologs were assessed on the basis of genomic context, using a custom visualization tool (GenomeChart),21 and for fit to the existing alignment. Alignments were manually adjusted as necessary when sequences with variable-length helices and/or loops were added.
The search process was repeated three to four times per multiple sequence alignment, to expand sequence diversity. The counts for Figure 2 were calculated from the number of completed genomes within refseq46 based on the final alignments utilizing queries to our custom database. Consensus secondary structure diagrams were created from the alignments using GSC-weighting in R2R.34
Supplementary Material
Acknowledgments
This work is supported by a PhRMA Foundation Research Starter Grant in Informatics, the Alfred P. Sloan Foundation and Boston College startup funds.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24151
References
- 1.Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol. 2012;22:279–86. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desnoyers G, Bouchard MP, Massé E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2013;29:92–8. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein mRNA. Proc Natl Acad Sci USA. 1980;77:7084–8. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1994;47:331–70. doi: 10.1016/S0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 6.Zengel JM, Lindahl L. Ribosomal protein L4 stimulates in vitro termination of transcription at a NusA-dependent terminator in the S10 operon leader. Proc Natl Acad Sci USA. 1990;87:2675–9. doi: 10.1073/pnas.87.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillier M, Allemand F, Raibaud S, Dardel F, Springer M, Chiaruttini C. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA-messenger RNA molecular mimicry. RNA. 2002;8:878–89. doi: 10.1017/S1355838202029084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, et al. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–31. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Schlax PJ, Xavier KA, Gluick TC, Draper DE. Translational repression of the Escherichia coli alpha operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J Biol Chem. 2001;276:38494–501. doi: 10.1074/jbc.M106934200. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y, Deiorio-Haggar K, Anthony J, Meyer MM. Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucleic Acids Res. 2013;41:3491–503. doi: 10.1093/nar/gkt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yutin N, Puigbò P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenye T, Vandamme P. Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS Microbiol Lett. 2005;242:117–26. doi: 10.1016/j.femsle.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 13.Iben JR, Draper DE. Specific interactions of the L10(L12)4 ribosomal protein complex with mRNA, rRNA, and L11. Biochemistry. 2008;47:2721–31. doi: 10.1021/bi701838y. [DOI] [PubMed] [Google Scholar]
- 14.Meyer MM, Ames TD, Smith DP, Weinberg Z, Schwalbach MS, Giovannoni SJ, et al. Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique’. BMC Genomics. 2009;10:268. doi: 10.1186/1471-2164-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundy FJ, Henkin TM. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991;173:4595–602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott LG, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with its 5′-translational operator mRNA. J Mol Biol. 2001;314:413–22. doi: 10.1006/jmbi.2001.5165. [DOI] [PubMed] [Google Scholar]
- 17.Choonee N, Even S, Zig L, Putzer H. Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007;35:1578–88. doi: 10.1093/nar/gkm011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(Database issue):D226–32. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Z, Barrick J, Weinberg Z, Neph S, Breaker R, Tompa M, et al. A computational pipeline for high- throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller CA, Anthony J, Meyer MM, Marth G. Scribl: an HTML5 Canvas-based graphics library for visualizing genomic data over the web. Bioinformatics. 2013;29:381–3. doi: 10.1093/bioinformatics/bts677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf M, Müller T, Dandekar T, Pollack JD. Phylogeny of Firmicutes with special reference to Mycoplasma (Mollicutes) as inferred from phosphoglycerate kinase amino acid sequence data. Int J Syst Evol Microbiol. 2004;54:871–5. doi: 10.1099/ijs.0.02868-0. [DOI] [PubMed] [Google Scholar]
- 23.Scott LG, Williamson JR. The binding interface between Bacillus stearothermophilus ribosomal protein S15 and its 5′-translational operator mRNA. J Mol Biol. 2005;351:280–90. doi: 10.1016/j.jmb.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 24.Philippe C, Portier C, Mougel M, Grunberg-Manago M, Ebel JP, Ehresmann B, et al. Target site of Escherichia coli ribosomal protein S15 on its messenger RNA. Conformation and interaction with the protein. J Mol Biol. 1990;211:415–26. doi: 10.1016/0022-2836(90)90362-P. [DOI] [PubMed] [Google Scholar]
- 25.Nazina TN, Tourova TP, Poltaraus AB, Novikova EV, Grigoryan AA, Ivanova AE, et al. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int J Syst Evol Microbiol. 2001;51:433–46. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- 26.Jinks-Robertson S, Nomura M. Ribosomal protein S4 acts in trans as a translational repressor to regulate expression of the alpha operon in Escherichia coli. J Bacteriol. 1982;151:193–202. doi: 10.1128/jb.151.1.193-202.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundy FJ, Henkin TM. Cloning and analysis of the Bacillus subtilis rpsD gene, encoding ribosomal protein S4. J Bacteriol. 1990;172:6372–9. doi: 10.1128/jb.172.11.6372-6379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Lindahl L, Sha Y, Zengel JM. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-alpha cluster. J Bacteriol. 1997;179:7046–54. doi: 10.1128/jb.179.22.7046-7054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundy FJ, Henkin TM. Characterization of the Bacillus subtilis rpsD regulatory target site. J Bacteriol. 1992;174:6763–70. doi: 10.1128/jb.174.21.6763-6770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986;192:101–10. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 31.Vartikar JV, Draper DE. S4-16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J Mol Biol. 1989;209:221–34. doi: 10.1016/0022-2836(89)90274-X. [DOI] [PubMed] [Google Scholar]
- 32.Serganov A, Polonskaia A, Ehresmann B, Ehresmann C, Patel DJ. Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 2003;22:1898–908. doi: 10.1093/emboj/cdg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naville M, Gautheret D. Premature terminator analysis sheds light on a hidden world of bacterial transcriptional attenuation. Genome Biol. 2010;11:R97. doi: 10.1186/gb-2010-11-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinberg Z, Breaker RR. R2R--software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.