Abstract
Mecp2 is a transcriptional repressor protein that is mutated in Rett syndrome, a neurodevelopmental disorder that is the second most common cause of mental retardation in women. It has been shown that the loss of the Mecp2 protein in Rett syndrome cells alters the transcriptional silencing of coding genes and microRNAs. Herein, we have studied the impact of Mecp2 impairment in a Rett syndrome mouse model on the global transcriptional patterns of long non-coding RNAs (lncRNAs). Using a microarray platform that assesses 41,232 unique lncRNA transcripts, we have identified the aberrant lncRNA transcriptome that is present in the brain of Rett syndrome mice. The study of the most relevant lncRNAs altered in the assay highlighted the upregulation of the AK081227 and AK087060 transcripts in Mecp2-null mice brains. Chromatin immunoprecipitation demonstrated the Mecp2 occupancy in the 5′-end genomic loci of the described lncRNAs and its absence in Rett syndrome mice. Most importantly, we were able to show that the overexpression of AK081227 mediated by the Mecp2 loss was associated with the downregulation of its host coding protein gene, the gamma-aminobutyric acid receptor subunit Rho 2 (Gabrr2). Overall, our findings indicate that the transcriptional dysregulation of lncRNAs upon Mecp2 loss contributes to the neurological phenotype of Rett syndrome and highlights the complex interaction between ncRNAs and coding-RNAs.
Keywords: non-coding RNA, Rett syndrome, Mecp2, mice, chromatin immunoprecipitation
Introduction
Epigenetic regulation, whose mechanisms involve chromatin remodeling by many different enzymes/complexes/molecular scaffolds that targets DNA methylation and histone code modifications,1 is also mediated by non-coding RNAs (ncRNAs).2-5 ncRNAs include the most studied member microRNAs, but also include long non-coding RNAs (lncRNAs). LncRNAs are transcripts of at least 200 nucleotides transcribed from all over the genome, including intergenic regions, antisense, overlapping or intronic to protein-coding genes.2-6 LncRNAs have a broad range of functions, such as enhancer-like activity,7 establishment of repressive chromatin in genomic regions8 or entire chromosomes,9 intronic antisense transcripts capable of binding to histone modifiers thereby regulating the transcriptional output of the host gene,6 alternative splicing and other post-transcriptional RNA modifications10 that determine the activity of our genome.
In this regard, the fine tuning of gene expression is critical in the human central nervous system (CNS). It is widely known that higher order cognitive and behavioral function are a CNS prerogative sustained by huge and intricate cell networks acting both locally and globally.11 One main aim of modern neurobiology is the ultimate understanding of the transcriptional programs that give rise to distinct neural networks, which are, in turn, formed by several neuronal and glial subtypes. Herein, alterations in the epigenetic modulation of gene expression could lead to several neurodevelopmental disorders, the most evident example represented by Rett syndrome (RTT).12,13 RTT (OMIM 312750) is an X-linked neurodevelopmental disorder that affects females at a frequency of 1:10,000 live births. The girls appear normal until 6–18 mo of age, when they lose their acquired skills and develop autistic features and mental retardation along with the typical stereotypic hand movement.14 Rett syndrome is the second leading cause of mental retardation in women after Down syndrome. Nearly 95% of typical RTT is due to mutations in the gene encoding the transcriptional regulator Methyl-CpG binding protein 2 (MECP2 in humans; Mecp2 in mice).15,16 Mecp2 is a basic nuclear protein that acts mainly as a transcriptional repressor, binding preferentially to methylated DNA sequences.17,18 In human cancer cells, the binding of MECP2 to the hypermethylated CpG islands of tumor suppressor genes19-21 and microRNAs22 is associated with transcriptional silencing. The loss of MECP2 in Rett syndrome patients and mice models is associated with a dysregulated pattern of coding-gene23-28 and microRNA expression.29,30
Herein, we investigate the role of lncRNAs in the physiopathology of Rett syndrome by comparing the transcriptome profiles of Mecp2-null mice brains31 vs. wild-type animals. In a similar pattern to Rett syndrome patients, Mecp2-null mice do not show the RTT-like phenotype just after birth, but 4–5 wk later,31 resembling the natural history of the disease in humans. The identified lncRNAs dysregulated in the Rett syndrome mice models provide important clues to understand the neurological phenotype of the disorder; furthermore, they illustrate the imbricate relationship between coding and ncRNA transcripts.
Results
Identification of candidate lncRNAs misregulated in RTT
We utilized a well-established mouse model of RTT that mimics the human disease31 to perform lncRNA microarray expression analyses using pairs of wild-type (WT) and Mecp2-null (KO) 9-wk animals. RNA was extracted from total brain and hybridized to an lncRNA microarray platform. The Mouse long non-coding RNA array (Arraystar) contains more than 41,232 probes representing unique lncRNAs. The probes were designed according to NCBI RefSeq, UCSC, RNAdb2.0, NRED, Fantom3.0 and UCRs annotations. Two biological replicates were used for each sample and condition. Repeat sequences and ncRNAs shorter than 200 bp are not represented in the microarray. The lncRNA expression data obtained are freely available at the Gene Expression Omnibus database: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fvavvcewquegcfc&acc=GSE43689.
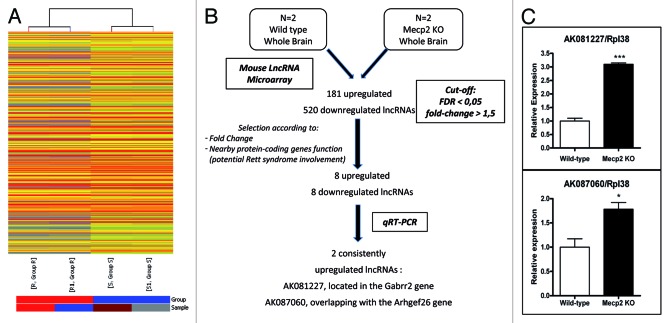
Unsupervised hierarchical clustering of the lncRNA microarray expression data indicates that the Mecp2-null mouse brains show a distinct pattern of lncRNA transcription in comparison to the wild-type animals (Fig. 1A). Among the 41,232 lncRNAs transcripts included in the microarray, we found 701 (1.7%) lncRNAs (Table S1) that had a different expression pattern in WT and Mecp2-null brain samples with a score of < 0.05 in the false discovery rate (FDR) test and a > 1.5-fold expression change (Fig. 1B). Among these significantly altered lncRNAs, overall downregulation of transcripts was the most common feature (520 of 701, 74%), while upregulation occurred in the remaining 26% (181 of 701) (Fig. 1B).
Figure 1. Dysregulation of the lncRNAs transcriptome in Mecp2-null mice brains. (A) Hierarchical clustering of the lncRNA microarray expression data shows distinguishable gene expression profiles between wild-type (S, S1) and Mecp2-null (R, R1) mice brains. (B) Flow-chart used to identify candidate misregulated lncRNAs in the RTT mouse model. (C) qRT-PCR of AK081227 and AK087060 normalized with RPL38. Error bars represent SE. Five biological replicates were used for each condition. * p < 0.05 *** p < 0.001.
Validation of lncRNA candidates in RTT
For practical purposes to reduce the lncRNAs to be further studied and to enrich those potentially involved in RTT, we selected candidate transcripts with a fold expression change > 2 that were associated with an annotated protein-coding gene where a biological function in neurons or glial cells have been proposed according to the scientific literature and through GO term enrichment on “negative regulation of neuron differentiation” (p = 0,008), “nerve development” (p = 0,007), “dendrite regeneration” (p = 0,004), “regulation of nervous system development” (p < 0,001), “regulation of excitatory postsynaptic membrane potential” (p < 0,001) and “positive regulation of synapse assembly” (p < 0,001). Using the above described criteria, we produced a short list of 16 lncRNAs: eight upregulated and eight downregulated in Mecp2-null mouse brains (Table S2). We then extracted brain RNA from five new pairs of wild-type and Mecp2-null 9-wk animals and analyzed the expression level of each one of the 16 candidate lncRNAs. The analyses were performed using quantitative PCR on reverse-transcribed RNA, three biological replicates per sample were developed and the statistical significance was assessed by two-tailed t-tests. Although for most cases, the expression analysis of each single candidate lncRNA matched the microarray data (data not shown), two lncRNAs exhibited a statistically significant difference: AK081227 (two-tailed t-test p < 0.0001) and AK087060 (two-tailed t-test p < 0.0248), both of them upregulated in Mecp2-null brain mouse (Fig. 1C).
Mecp2 binding to the 5′-end genomic loci of the lncRNAs AK081227 and AK087060
One important function of Mecp2 is to act as a transcriptional repressor that binds to the promoters located in the 5′-end genomic region of its target coding genes19-21 and microRNAs.22,29,30 Thus, we speculated whether Mecp2 was directly involved in regulating AK087060 and AK081227 gene expression by analyzing its capability of binding to their promoters. To this end, we performed chromatin immunoprecipitation (ChIP) experiments with formaldehyde-cross-linked nuclear extracts from WT and Mecp2 KO brains followed by semi-quantitative PCR. We immunoprecipitated the cross-linked chromatin with a histone H3 antibody to serve as an internal control of the ChIP assay (Fig. 2A–C). We also ensured that the Mecp2 antibody was specific for the protein and reliable in the ChIP assay by checking Mecp2 occupancy on the promoter of Xist, a previously reported lncRNA involved in X-inactivation whose promoter is occupied by Mecp2 in male wild-type mice (where Xist is silenced in the only X chromosome).32 As expected, we found Mecp2 bound to the Xist promoter in wild-type, but not in Mecp2 KO mouse brain chromatin (Fig. 2A).
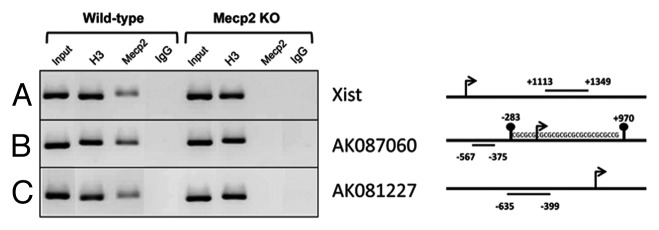
Figure 2. Mecp2 is bound to the 5′-end genomic loci of the lncRNAs AK081227 and AK087060 in wild-type mouse brain and lost in Mecp2-null mice. Semiquantitative chromatin immunoprecitation assay for a fraction of the total DNA (5%) (input), an antibody anti-H3 total (IP control), an antibody anti-Mecp2 and a negative control (mouse IgG) on the 5′-end genomic loci of the lncRNAs Xist (A), AK081227 (B) and AK087060 (C). The regions amplified by PCR were represented as straight lines.
Once these controls were established, we proceeded to address the Mecp2 occupancy for the AK081227 and AK087060 5′-end genomic loci. The PCR primers for the ChIP assay were designed in a region of 500 bp upstream to the transcription start site (TSS) within the corresponding lncRNAs proximal promoters. Importantly, we found that AK081227 and AK087060 5′-end loci were occupied by the Mecp2 protein in wild-type mouse brains, while the Mecp2 protein was absent from the 5′-end of the two lncRNAs in the Mecp2-null mouse brains that overexpress the AK087060 and AK081227 transcripts (Fig. 2B and C). DNA methylation differences in the two studied lncRNAs 5′-ends were studied by bisulfite genomic sequencing of multiple clones and were not detected in the brain of wild-type vs. Mecp2 KO samples (data not shown). These data support the role of Mecp2 as a transcriptional repressor of the transcripts located in the vicinity of its binding sites to DNA, herein regulating the expression of the lncRNAs AK081227 and AK087060.
Upregulation of the lncRNA AK081227 in RTT is associated with downregulation of its host gene gamma-aminobutyric acid receptor subunit rho 2
One of the main challenges in research with lncRNAs is the identification of a particular molecular or cellular function.6 One possibility is that lncRNAs act locally regulating the expression levels of neighboring RNA transcripts.6 Herein, we have studied the possible effect of AK081227 and AK087060 in the activity of their associated protein-coding genes. To address this issue, we have analyzed the expression levels of the two identified lncRNAs by qRT-PCR in comparison to their corresponding host genes in four functional relevant brain regions (frontal cortex, hypothalamus, thalamus and cerebellum) from new pairs of wild-type and Mecp2-null 9-wk animals.
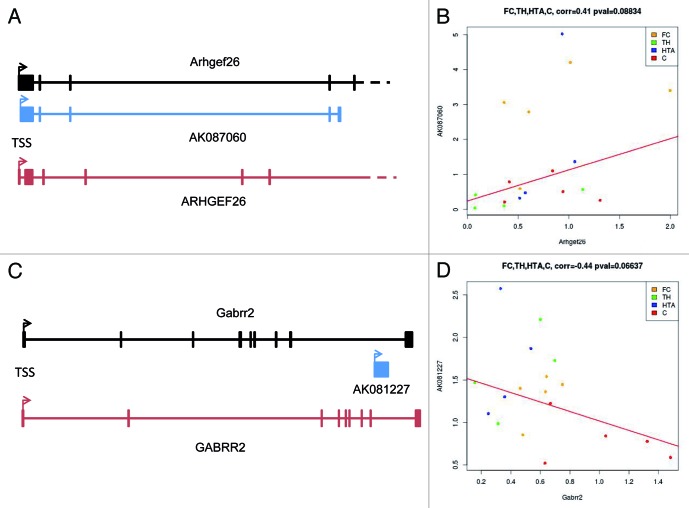
AK087060 is transcribed from 225 bp downstream of the first exon of the Arhgef26 gene (Fig. 3A). The Arhgef26 protein is a Rho guanine nucleotide exchange factor (GEF) that has a role in the actin-driven endocytic process known as macropinocytosis that contributes to repulsive turning in the axons of neurons33 and retinal cone growth.34 Thus, a role for this protein in a neurological disorder, such as Rett syndrome, could be invoked. In this regard, in humans, the equivalent RNA transcript is originated 357 bp downstream of the first exon of the ARHGEF26 promoter with a 79% nucleotide homology to AK087060. We found that the observed upregulation of AK087060 in Mecp2 KO mice had a statistical association with an increase in the expression levels of its host gene Arhgef26 in the four studied brain regions (Pearson's correlation test = 0.41, p = 0.08) (Fig. 3B). Since this particular lncRNA is transcribed from just 225 bps downstream of the coding gene, common promoter regulatory mechanism for AK087060 and Arhgef26 seems an unlikely scenario. In addition, AK087060 could be a promoter-associated lncRNA.6 These lncRNAs are transcribed around the TSS proximal region and are able to reclute other regulatory factors6 that impact on the expression of the associated-coding gene, in this case Arhgef26.
Figure 3. Genomic context and associated coding-genes for the identified lncRNAs. (A) AK087060 originates 200 bps downstream from the transcription start site of the mRNA for the Arhgef26 gene. Genomic organization of the human gene is also shown (ARHGEF26). (B) Upregulation of AK087060 in Mecp2 KO mice is associated with an increase in the expression levels of the host gene in the four studied brain regions (Pearson's correlation test = 0.41, p = 0.08). (C) AK081227 is transcribed from the last intron of the Gabrr2 gene. Genomic organization of the human gene is also shown (GABRR2). (D) Upregulation of AK087060 in Mecp2 KO mice is associated with a downregulation in the expression levels of the host gene in the four studied brain regions (Pearson's correlation test = 0.44, p = 0.06). Orange dots, frontal cortex; green dots, thalamus; blue dots, hipothalamus; red dots, cerebellum.
The lncRNA/mRNA equation is completely different for AK081227. This lncRNA is transcribed from an intronic region of the GABA receptor subunit rho-2 (Gabrr2) gene (Fig. 3C). Many autistic and neurodevelopmental disorders, including Rett syndrome, have been linked to dysfunction in particular aspects of GABAergic inhibitory neurotransmission in the brain.35,36 Most importantly, the expression of another GABA receptor subunit member (GABRB3) is reduced in Rett syndrome.24,37 Thus, the host gene of the identified lncRNA is a likely candidate to be altered in Rett syndrome. In this regard, in humans, the equivalent lncRNA has a 30% nucleotide homology to AK081227. Herein, we found that the upregulation of AK081227 in Mecp2 KO mice was statistically associated with a downregulation of the expression levels of its host gene Gabrr2 in the four studied brain regions (Pearson's correlation test = 0.44, p = 0.06) (Fig. 3D). Thus, the expression of the lncRNA AK081227 might act locally to interfere in the mRNA transcript levels of Gabrr2 and contribute to the neurological phenotype of Rett syndrome. In this regard, we have also recently identified another intronic lncRNA that finely regulates the expression of the host gene38 in what might become a common theme in the complex interaction between ncRNAs and coding-RNAs.
Discussion
The current report represents the first lncRNA profiling in Rett syndrome, data that can be used to identify the transcripts from this class that are regulated by Mecp2 and that could explain the physiopathology of the disease. We observed a distinct lncRNA transcriptome between the brain of wild-type and Mecp2-null mice, where the expression of 701 lncRNAs was significantly different. Due to the limited information available about the function of each lncRNA, we selected only those whose higher fold expression change overlapped with protein-coding genes whose function was related to neurons or glia cells for further validation and study. In this last setting, the most relevant observation was that the release of the transcriptional silencing of the lncRNA AK081227 was associated with a downregulation of its host gene, Gabrr2. The importance of lncRNAs as cis-acting regulators is rapidly being recognized.6 LncRNAs can guide chromatin change in cis in a co-transcriptional manner (tethered by RNA polymerase) or as a complementary target for small regulatory RNAs. Regarding our case, there is increasing knowledge that lncRNAs overlapping with introns of protein-coding genes, whether originating from splicing or produced by independent transcriptional units, may recruit several classes of coactivator (e.g., trithorax group proteins)39 or corepressor (polycomb-group members like histone methyltransferase EZH2)38 complexes and, therefore, act as guides for the establishment of activating or repressive histone marks all over the host gene. In this regard, the exact mechanisms regulating the inverse expression levels of the non-coding (AK081227) and coding (Gabrr2) transcripts are currently under investigation and should be the focus of further developments in this area.
The observation that the intronic lncRNA AK081227 upregulation in the Rett syndrome brain is associated with the depletion of its host coding gene Gabrr2 can have a relevant impact in the understanding of the described neurological disorder. At the physiological level, post-mortem analysis of RTT brains showed altered levels of neurotransmitters such as glutamate and biogenic amines as well as changes in the abundance of some neurotransmitter receptors. In mouse models of RTT, analysis of spontaneous miniature excitatory and inhibitory postsynaptic currents indicated a shift in the excitatory/inhibitory balance, with increased excitatory and decreased inhibitory neurotransmission in the hippocampus and cortex.40 Consistently, studies of Mecp2-knockout mouse models revealed abnormalities in long-term potentiation (LTP) and impaired synaptic plasticity.41 Recent findings underline that Mecp2 deficiency in GABAergic neurons recapitulates most of the features displayed by Mecp2-null mice, including altered synaptic activity and plasticity.42 Since GABA is the major inhibitory neurotransmitter in the brain, most of the synaptic defects seen in RTT could be a direct consequence of the Mecp2 loss in GABA neurons. Additionally, genes necessary for GABAergic function, like Dlx5 and GABRB3, have already been associated with RTT.24,37 Gabrr2, as a member of GABA(C) receptor class, has been reported to be expressed in various brain regions.43,44 The fact that we found Gabrr2 downregulated in frontal cortex is consistent with the dysfunction of GABAergic signaling seen in the frontal cortex of RTT patients.45 Moreover, Gabrr2 was also downregulated in thalamus and hypothalamus, with the latter being one of the most affected region in RTT. As for the thalamus, a recent study showed that Mecp2 regulates GABAergic synapses differentially in excitatory and inhibitory neurons in the thalamus.46
Overall, our data provides the first hint that the lack of the transcriptional regulatory effect of Mecp2 in Rett syndrome leads to a dysregulation of lncRNA expression in the affected mice brain. Future data mining of the obtained lncRNA transcriptomes deposited in the public genomic databases could provide further clues about the impact of the altered lncRNAs on other transcripts; however, the neuropathological relevance of the inverse association between AK081227 (the intronic lncRNA) and Gabrr2 (the host coding-gene) already provides proof of principle for the existence of disrupted cis-regulated mechanisms in the disease.
Materials and Methods
Animal model
Four-week-old B6.129P2(C)-Mecp2tm.11Bird /J (stock number: 003890) heterozygous females (Mecp2+/−) were acquired from the Jackson Laboratory. In brief, the mutant strain was generated by replacing exons 3 and 4 of Mecp2 in embryonic stem cells with the same exons flanked by loxP sites. Homozygous Mecp2 lox/lox females were mated with mice with ubiquitous Cre expression to bring about gene disruption. The offspring from the crosses of Mecp2+/− females with C57BL/6J males were genotyped by PCR. Mice were kept under specific pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations. Lighting conditions (lights on from 08:00–20:00 h) and temperature (22°C) were kept constant. Animals were allowed ad libitum access to food and water and were inspected every day. Tissue samples were obtained from hemizygous Mecp2-null males (Mecp2-/y, KO) and their wild-type (WT) littermates after establishing RTT-like symptoms in the defective animals (at about 8–10 wk of age). Mice were euthanized in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research. Tissues were frozen on dry ice immediately after removal and stored at -80°C until use.
RNA extraction
To extract RNA, frozen tissues were ground into powder with mortar and pestle and resuspended in Trizol reagent (Life technologies). The RNA purification was performed on the RNA-containing aqueous phase with RNeasy mini kit (Qiagen). After elution with RNase-free water and treatment with turbo DNase (Ambion), the RNA is ready for all kinds of applications. Quantification and quality check were performed with Nanodrop and Agilent 2100 Bioanalyzer (Agilent Technologies), respectively.
Sample preparation and microarray analysis
Briefly, 1 ug of total RNA was labeled with Cy3 using Agilent Quick Amp Labeling Kit and microarray hybridization was performed at 65°C for 17 h in Agilent’s SureHyb Hybridization Chambers. After being washed in an ozone-free environment, the slides were scanned using the Agilent DNA microarray scanner (part number G2505B). Data was extracted using Agilent Feature Extraction Software (version 10.5.1.1) and normalization was performed using the Agilent FE one-color scenario (mainly median normalization). Finally, four samples were hybridized, two biological replicates for each condition (Mecp2 KO mice and their wild-type littermates). False discovery rate (FDR) test was performed on lncRNA that pass the cut-off (t test < 0.05, fold change > 1.5).
Quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR)
Two ug of RNA for each sample were retro-transcribed to cDNA using random hexamers from ThermoScript™ RT-PCR System for First-Strand cDNA Synthesis kit (Life Technologies). A negative sample (no ThermoScript enzyme) was performed to exclude DNA contamination. Primers for the reaction were designed according to Primer3 software and tested with a cDNA serial dilution to check amplification linearity and unique amplification product. Ppia and Rpl38 were used as house-keeping genes for normalization. 50 ng cDNA were used for each PCR reaction together with 5 ul 2X SYBR green PCR master mix (Life technologies), 250 nM of each primer and water up to 10 ul. Mouse primers sequences used were the following: Ppia Forward 5′-CAA ATG CTG GAC CAA ACA CAA-3′ Reverse 5′-GTT CAT GCC TTC TTT CAC CTT-3′; Rpl38 Forward 5′-AGG ATG CCA AGT CTG TCA AGA-3′ Reverse 5′-TCC TTG TCT GTG ATA ACC AGG G-3′; AK081227 Forward 5′-TCG GTC AGT GCA TTT GGG CTG T-3′ Reverse 5′-TCG GTC CAC TGT CTC AGG AGT GC-3′; Gabrr2 Forward 5′-CAA GGG GAA CGA CGT GCG GA-3′; AK087060, Forward 5′-GAA CGA CGT GCG GA-3′3’.TGT ATG GCG TCC ATC TCT TCG G-3′ and Reverse 5′-GTC CTC CTC TCT GCA ATT GCT TAG-3′; Arhgef26, Forward 5′-GGC CCT TGA TAT CGA CTC TGA TGA-3′ and 5′-CTT TTC ACC GCG GAG AGC TGG-3′. Quantitative PCR was performed for 40 cycles on an ABI 7900HT sequence detection system (Life Technologies) under the thermal cycling conditions recommended by the manufacturer.
Chromatin immunoprecipitation in brain tissues
Frozen wild-type and Mecp2 KO male brains were reduced to powder with mortar and pestle. The pulverized brain tissues were cross-linked with 1% formaldehyde for 8 min and the reaction was blocked by adding glycine to a final concentration of 0.125 M. After washing two times with ice-cold PBS, cell pellets were resuspended in cell lysis buffer (HEPES 5mM, KCl 85 mM, NP40 0.5% pH 8.0) supplemented with protease inhibitor cocktail (Complete EDTA-free, Roche) and the lysate was homogenized with a douncer to facilitate cell membrane break. The nuclear pellet was then resuspended in Nuclei lysis buffer (TRIS-HCl 50 mM, EDTA 10 mM, SDS 1% pH 8.1) and subsequently sonicated with Bioruptor (Diagenode) for 30 min (30 sec ON, 30 sec OFF cycles). The chromatin size of the fragments obtained was 150–400 bp. Samples were diluted with Dilution buffer (SDS 0.01%, Triton X-100 1.1%, EDTA 1.2 mM, NaCl 165 mM, TRIS-HCl 16.7 mM pH 8.1). Magnetic beads were used for the pre-clearing of diluted chromatin (over-night at 4°C) and for incubation with anti-total H3 (ab1791, abcam) and anti-Mecp2 (m9317, sigma). Non-related mouse IgG antibody (12–371, Millipore) was used as a negative control. The Beads-Antibody complexes were then incubated with pre-cleared chromatin for 2 h at 4°C in agitation. The immune-complexes were washed: twice with low salt Buffer (TRIS-HCl 50 mM pH 8.0, NaCl 150 mM, SDS 0.1%, NP-40 1%, EDTA 1 mM, Deoxicolate Na 0.5%), twice with high Salt Buffer (TRIS-HCl 50 mM pH 8.0, NaCl 500 mM, SDS 0.1%, NP-40 1%, EDTA 1 mM, Deoxicolate Na 0.5%), twice with LiCl Buffer (TRIS-HCl 50 mM pH 8.0, LiCl 250 mM, SDS 0.1%, NP-40 1%, EDTA 1 mM, Deoxicolate Na 0.5%) and twice with TE Buffer (TRIS-HCl 10 mM pH 8.0, EDTA 0.25 mM). Cross-linked chromatin was then eluted from the magnetic beads by adding elution Buffer (NaHCO3 100 mM, SDS 1%). Samples were de-crosslinked overnight at 65°C and incubated with Proteinase K at 50 ug/ml final concentration for 1 h. Finally, DNA was purified with PCR purification kit (Qiagen). The primers used for ChIP analysis were the following: AK081227 promoter, Forward 5′-TTG TCC CCA CTA AGAG ACA G-3′ and Reverse 5′-CCT GTA CTC TGC TAT GCT TAC TC-3′; AK087060, Forward 5′-CTG TGT GAC TTT CAA ACA TAC AG-3′ and Reverse 5′-CTT CAC TGG GCC ACT TGT G-3′; Xist, Forward 5′-CCT GTA CGA CCT AAA TGT CC-3′ and Reverse 5′-GTA TTA GTG TGC GGT GTT GC-3′.
Supplementary Material
Acknowledgments
Funding was provided by the European Community's Seven Framework Programme (FP7/2007-2013) under grant agreement Nº PITN-GA-2009-238242 DISCHROM project and ERC grant agreement nº. 268626 EPINORC project, E-RARE EuroRETT network (Carlos III Health Institute Project nº. PI071327), the Fondation Lejeune, MINECO Project nº SAF2011-22803, Cellex Foundation, Botin Foundation, the Catalan Association of Rett Syndrome and the Health and Science Departments of the Catalan Government (Generalitat de Catalunya). M.E. is an ICREA Research Professor.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24286
References
- 1.MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88:170–83. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068–75. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 7.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wutz A. RNA-mediated silencing mechanisms in mammalian cells. Prog Mol Biol Transl Sci. 2011;101:351–76. doi: 10.1016/B978-0-12-387685-0.00011-1. [DOI] [PubMed] [Google Scholar]
- 10.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gräff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–72. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 13.Smeets EE, Pelc K, Dan B. Rett Syndrome. Mol Syndromol. 2012;2:113–27. doi: 10.1159/000337637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagberg B, Aicardi J, Dias K, Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann Neurol. 1983;14:471–9. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 15.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 16.Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, et al. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–21. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–14. doi: 10.1016/0092-8674(92)90610-O. [DOI] [PubMed] [Google Scholar]
- 18.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–78. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–45. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66:8342–6. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Serra L, Ballestar E, Ropero S, Setien F, Billard LM, Fraga MF, et al. Unmasking of epigenetically silenced candidate tumor suppressor genes by removal of methyl-CpG-binding domain proteins. Oncogene. 2008;27:3556–66. doi: 10.1038/sj.onc.1211022. [DOI] [PubMed] [Google Scholar]
- 22.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 23.Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, et al. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet. 2005;116:91–104. doi: 10.1007/s00439-004-1200-0. [DOI] [PubMed] [Google Scholar]
- 24.Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005;14:483–92. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:2247–56. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- 26.Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, et al. FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum Mol Genet. 2007;16:640–50. doi: 10.1093/hmg/ddm007. [DOI] [PubMed] [Google Scholar]
- 27.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–9. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urdinguio RG, Lopez-Serra L, Lopez-Nieva P, Alaminos M, Diaz-Uriarte R, Fernandez AF, et al. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS One. 2008;3:e3669. doi: 10.1371/journal.pone.0003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urdinguio RG, Fernandez AF, Lopez-Nieva P, Rossi S, Huertas D, Kulis M, et al. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of Rett syndrome. Epigenetics. 2010;5:656–63. doi: 10.4161/epi.5.7.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, et al. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2010;107:18161–6. doi: 10.1073/pnas.1005595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–6. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 32.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–68. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolpak AL, Jiang J, Guo D, Standley C, Bellve K, Fogarty K, et al. Negative guidance factor-induced macropinocytosis in the growth cone plays a critical role in repulsive axon turning. J Neurosci. 2009;29:10488–98. doi: 10.1523/JNEUROSCI.2355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo D, Standley C, Bellve K, Fogarty K, Bao ZZ. Protein kinase Cα and integrin-linked kinase mediate the negative axon guidance effects of Sonic hedgehog. Mol Cell Neurosci. 2012;50:82–92. doi: 10.1016/j.mcn.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medrihan L, Tantalaki E, Aramuni G, Sargsyan V, Dudanova I, Missler M, et al. Early defects of GABAergic synapses in the brain stem of a MeCP2 mouse model of Rett syndrome. J Neurophysiol. 2008;99:112–21. doi: 10.1152/jn.00826.2007. [DOI] [PubMed] [Google Scholar]
- 36.Coghlan S, Horder J, Inkster B, Mendez MA, Murphy DG, Nutt DJ. GABA system dysfunction in autism and related disorders: from synapse to symptoms. Neurosci Biobehav Rev. 2012;36:2044–55. doi: 10.1016/j.neubiorev.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–70. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 39.Iglesias-Platas I, Martin-Trujillo A, Cirillo D, Court F, Guillaumet-Adkins A, Camprubi C, et al. Characterization of novel paternal ncRNAs at the Plagl1 locus, including Hymai, predicted to interact with regulators of active chromatin. PLoS One. 2012;7:e38907. doi: 10.1371/journal.pone.0038907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21:217–27. doi: 10.1016/j.nbd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 42.Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–9. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grabert J, Jost B, Patz S, Wahle P, Schmidt M. GABA(C) receptors are expressed in GABAergic and non-GABAergic neurons of the rat superior colliculus and visual cortex. Exp Brain Res. 2009;199:245–52. doi: 10.1007/s00221-009-1918-y. [DOI] [PubMed] [Google Scholar]
- 44.Frazao R, Nogueira MI, Wässle H. Colocalization of synaptic GABA(C)-receptors with GABA (A)-receptors and glycine-receptors in the rodent central nervous system. Cell Tissue Res. 2007;330:1–15. doi: 10.1007/s00441-007-0446-y. [DOI] [PubMed] [Google Scholar]
- 45.Blue ME, Naidu S, Johnston MV. Altered development of glutamate and GABA receptors in the basal ganglia of girls with Rett syndrome. Exp Neurol. 1999;156:345–52. doi: 10.1006/exnr.1999.7030. [DOI] [PubMed] [Google Scholar]
- 46.Zhang ZW, Zak JD, Liu H. MeCP2 is required for normal development of GABAergic circuits in the thalamus. J Neurophysiol. 2010;103:2470–81. doi: 10.1152/jn.00601.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.