Abstract
Prokaryotic transcripts constitute almost always uninterrupted intervals when mapped back to the genome. Split reads, i.e., RNA-seq reads consisting of parts that only map to discontiguous loci, are thus disregarded in most analysis pipelines. There are, however, some well-known exceptions, in particular, tRNA splicing and circularized small RNAs in Archaea as well as self-splicing introns. Here, we reanalyze a series of published RNA-seq data sets, screening them specifically for non-contiguously mapping reads. We recover most of the known cases together with several novel archaeal ncRNAs associated with circularized products. In Eubacteria, only a handful of interesting candidates were obtained beyond a few previously described group I and group II introns. Most of the atypically mapping reads do not appear to correspond to well-defined, specifically processed products. Whether this diffuse background is, at least in part, an incidental by-product of prokaryotic RNA processing or whether it consists entirely of technical artifacts of reverse transcription or amplification remains unknown.
Keywords: RNA-seq, self-splicing introns, split tRNAs, circular sRNAs
Introduction
Common wisdom has it that prokaryotic transcripts correspond to intervals on the genomic DNA. In archaea, several exceptions to this simple rule are well known. As in eukaryotes, some of their tRNAs have introns that are spliced out by dedicated splicing endonucleases.1,2 In contrast to Eukarya, enzymatically spliced introns can also be found in mRNAs3 and in rRNAs.4 In some archaeal species, furthermore, tRNAs are composed of pieces that are independently transcribed from different genomic locations.2,5-8
Archaeal non-coding RNAs often are processed to yield a circular form. Large ORF-containing introns derived from rRNAs form stable RNA species in Pyrobaculum organotrophum.9 Circular forms of both 23S and 16S rRNAs appear as processing intermediates during rRNA maturation.10 Circularized RNAs are produced from tRNA introns in Haloferax volcanii,5 and a circularized box C/D snoRNA from Pyrococcus furiosus11 turned out to be typical for box C/D snoRNAs, see also reference 8 for an example in Nanoarchaeum equitans. A recent study based on RNase R-treated RNA libraries systematically mapped circularized RNAs and showed that circularized RNAs are also abundant in Sulfolobus solfataricus and its relatives.12 In contrast to this rather complex situation in Archaea, eubacterial transcriptomes are not known to harbor spliced transcripts with the exception of the hosts of self-splicing group I and group II introns.13-15
It is well known, on the other hand, that reverse transcription can generate artifactual sequences that look like splicing products, i.e., by leaving out stable RNA secondary structure features.16-18 Most analyses of prokaryotic RNA-seq data thus completely neglect sequencing reads that do not map as a single, uninterrupted interval. Of course, this strategy also hides any true splicing or circularization products. The purpose of this contribution is to systematically explore the content of the “trash bin” of RNA-seq analysis, aiming at the identification of atypically processed RNAs.
In the much more complex transcriptomes of Eukarya, unexpected types of transcripts have recently received considerable attention. In particular, circularized RNAs have turned out to be not only abundant but also to convey important regulatory functions.19-22 Similar observations have been made for fusion transcripts.23,24 This begs the question whether prokaryotic transcriptomes might also harbor unexpected treasures.
Results
We re-evaluated published RNA-seq data from four Archaea and six Eubacteria. The basic statistics of the data sets, which were produced by different labs with different read lengths at different times, are compiled in the Methods section and in the Electronic Supplement. Our analysis focuses specifically on those reads that do not map as single, uninterrupted intervals to the respective reference genome.
Archaea
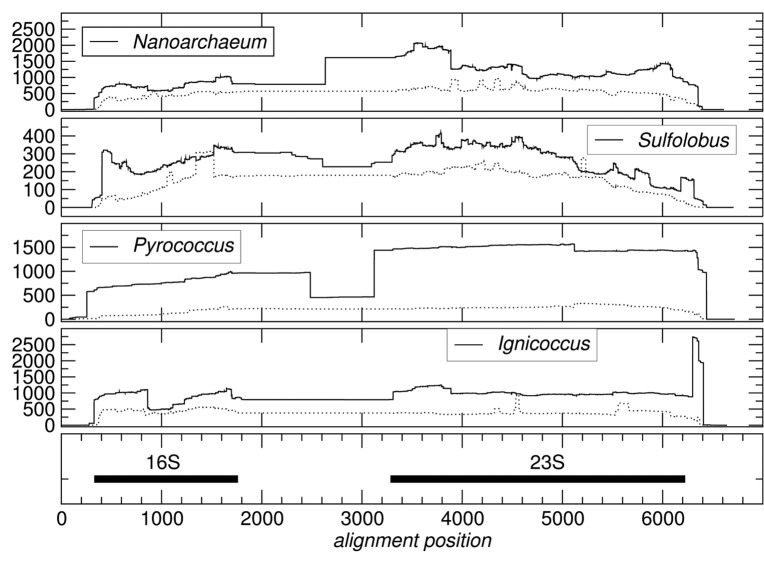
Several types of “atypical” RNAs are well known in Archaea. The most prominent forms among them are circularized RNAs. As expected, we observe circularized precursor forms for both the 16S and 23S RNAs, see e.g. reference 10. Large numbers of additional circularized products are observed, see Figure 1. The rRNA loci also feature substantial numbers of apparently spliced reads, see Figure S1.
Figure 1. Density of circularized (thick line) and “spliced” reads (thin line) at the ribosomal rRNA loci. Coordinates refer to a multiple sequence alignment of the four Archaea species. For Nanoarchaeum, two separate RNA genes are concatenated with 160 Ns as linker.
Most snoRNAs in Archaea also form circularized transcripts. Somewhat surprisingly, these are readily detectable from RNA-seq data even without prior treatment of the libraries to enrich circularized products as in the recent work of reference 12. The association of circularized products with small ncRNAs allows us to detect a number of novel ncRNA species in each of the four Archaea, Table 1. The number of new candidates depends strongly on the species, presumably in response to the quality of the available genome annotation.
Table 1. Novel ncRNAs in Archaea.
| Coordinates | Reads | Note | ||
|---|---|---|---|---|
|
Pyrococcus furiosus | ||||
| 128135 |
128190 |
? |
8 |
|
| 258945 |
259007 |
? |
915 |
|
| 505270 |
505323 |
? |
4 |
|
| 505760 |
505814 |
+ |
1 |
|
| 860511 |
860567 |
? |
3 |
|
|
Sulfolobus solfataricus | ||||
| 434665 |
434719 |
- |
2 |
|
| 1275505 |
1275576 |
? |
71 |
3 variants |
|
Nanoarchaeum equitans | ||||
| 432130 |
432227 |
+ |
159 |
5′ of 16S |
| 396865 |
396957 |
+ |
95 |
3′ of 23S |
| 339418 |
339570 |
? |
53 |
mult. |
| 248142 |
248285 |
? |
1 |
|
|
Ignicoccus hospitalis | ||||
| 28125 |
28202 |
? |
883 |
|
| 54013 |
54076 |
- |
9 |
|
| 62481 |
62544 |
? |
3 |
|
| 62543 |
62607 |
? |
7 |
~previous |
| 69658 |
69725 |
? |
411 |
|
| 74304 |
74365 |
? |
4 |
|
| 507227 |
507289 |
? |
112 |
|
| 576736 |
576811 |
? |
828 |
|
| 598273 |
598363 |
? |
41 |
|
| 599309 |
599358 |
? |
2 |
|
| 617433 |
617521 |
+ |
2017 |
~ Iho-sR86 |
| 720628 |
720706 |
? |
18 |
|
| 734264 |
734345 |
+ |
20 |
3′ of 23S |
| 824008 |
824070 |
+ |
8 |
~ Iho-sR109 |
| 1000660 |
1000778 |
+ |
6 |
~ Iho-sR131 |
| 1000717 |
1000778 |
+ |
3 |
~ Iho-sR131 |
| 1066825 |
1066891 |
? |
500 |
|
| 1266699 | 1266795 | ? | 461 | mult. |
Since the RNA-seq data are not strand specific, the reading direction remains undetermined in most cases (indicated by ?). Promoter or terminator elements annotated in the UCSC Archaea Browser identify a likely reading direction (indicated by +/-). Read support was added up for alternative junctions within a few nucleotides. In the Note column, ‘mult.’ designates the presence of multiple products, and ~ denotes loci adjacent to annotated ncRNAs.
Enzymatic splicing in tRNAs is a well-known phenomenon in Archaea. It is typically invisible in RNA-seq data, however, because tRNAs are normally multi-copy genes and tRNAs with introns often have nearly identical paralogs without an intron. In this situation, mature tRNAs are mapped to the intron-less locus even if the molecule in reality was produced by splicing from the locus with intron. In 21 cases, the intron is visible as a circularized by-product of splicing, see Table S4.
In Sulfolobus, an enzymatically spliced intron interrupts the coding sequence of the cbf5 gene.25 This case is readily detectable in the form of multiple splitreads of the “normal” type. A second well-supported candidate is located close to the annotated translation start site of the putative protein SSO1586. With a length of 144 nt it preserves the reading frame. Since the entire sequence of the putative protein is conserved it might encode a functional isoform.
An interesting case of trans-splicing are the split tRNAs reported in Nanoarchaeum.6,8,26 Some of them are not directly observable as split reads, however. This is the case e.g., for tRNA-Lys and tRNA-Gln, which have nearly identical paralogs that attract the mature tRNA reads to the unspliced loci irrespective of their true origin. tRNA-Met and tRNA-Glu are visible at least with a few reads, while tRNA-His is invisible. This is explained by the high conservation of tRNA genes and the fact that the RNA-seq data used here comprise a mixture of N. equitans and I. hospitalis. The tRNA-His sequence in I. hospitalis thus captures the trans-spliced tRNA-His reads from N. equitans. No other split tRNAs were observed in the data sets analyzed here.
Given the high expression levels of rRNAs, it is not surprising that a large fraction (ranging from 25% in Ignicoccus to 75% in Nanoarchaeum) of split reads maps to the rRNA loci, see Table 2. The number of spliced reads nevertheless is systematically smaller than the number of reads crossing a circularization point, see Figure 1.
Table 2. Splice junction overlaps with CRISPR and rRNA.
| Species | all | CRISPR | rRNA |
|---|---|---|---|
| Eubacteria | |||
|
Bacillus cereus |
11,808 |
0 |
7,955 |
|
Escherichia coli |
68,704 |
6 |
37,616 |
|
Salmonella enterica |
7,445 |
0 |
6,050 |
|
Pseudomonas PA14 |
33,349 |
528 |
16,819 |
|
Helicobacter pylori 26695 |
148,734 |
0 |
114,388 |
|
Synechocystis PCC6803 |
40,371 |
7 |
786 |
| Archaea | |||
|
Nanoarchaeum equitans |
22,185 |
0 |
16,607 |
|
Ignicoccus hospitalis |
49,904 |
0 |
12,615 |
|
Pyrococcus furiosus |
83,426 |
23,544 |
23,502 |
| Sulfolobus solfataricus | 22,197 | 150 | 13,781 |
Surprisingly, about a quarter of the split read data for Pyrococcus maps to the CISPR loci. It is tempting to speculate that inclusion of an organism’s own sequences in CRISPRs is akin to an autoimmune reaction. Without further validation, however, we cannot rule out that artifacts in reverse transcription or amplification are responsible for these “trans-spliced” reads.
Eubacteria
In contrast to Archaea, split reads are expected to be very rare in Eubacteria. In fact, the only well-understood sources are self-splicing introns. In the six genomes considered here, eight group I and nine group II introns could be tentatively annotated computationally.
Not all of them are visible in the RNA-seq data in the form of split-reads. Only a group I intron in the initiator tRNA of Synechocystis27 and a group I intron in the recA gene of B. cereus28,29 are well represented in our data. All of the detectable group II introns are located in B. cereus.29,30 Only the B.c.I3 intron located within the DNA polymerase III subunit α is supported by many split reads. The two plasmid-borne introns designated B.c.I4 and B.c.I5 are visible only as a single split read each. More details on the self-splicing introns can be found in Tables S2 and S3.
Surprisingly, our mapping data also show a large number of split and circularized reads that cannot be explained by known splicing mechanisms. As in Archaea, a large fraction of the split reads again maps to the rRNA operons, see Table 2; Figure S1. With the exception of Synechocystis, rRNA accounts for the dominating part of the unusual RNAs. We have not been able to isolate candidates for well-defined stable processing products, however.
Beyond the self-splicing introns and the rRNA loci only a moderate number of “splice sites” is supported by multiple, non-identical reads. Among the most peculiar examples are tmRNAs with missing subsequences, Figure 2, which appear in several species. Although the excisions appear to be concentrated in the highly structured, pseudoknotted regions, only some of them are easily explained as “RTfact” resulting from the RT reading through the base of a stem and, thus, omitting the entire structural domain enclosed by the stem.

Figure 2. Mapping of “introns” observed in multiple reads from E. coli (colored sequence), H. pylori (•) and P. aeruginosa (*) to the tmRNA structure (E. coli tmRNA model from ref. 31) shows that the excisions are concentrated in the pseudoknotted regions. Counts in brackets indicate the number of split reads.
Cleavage of tRNAs as a response to stress, first discovered as response to phage infection in E. coli,32,33 is a general phenomenon in all domains of life, see e.g. references 34‒36. At least in some cases, tRNA cleavage seems to have evolved into an internal regulation mechanism.37 Fragments of tRNAs, furthermore, may act as regulatory ncRNAs in both Eukarya38,39 and Archaea.40 Healing of the cleaved tRNAs is likewise a frequently observed phenomenon, see e.g. references 41 and 42. The ligases involved in tRNA splicing in Eukarya43 and Archaea44 utilize the 2’,3′-cyclic phosphates generated by endonucleolytic cleavage. Members of the same protein family have also been found in Eubacteria, see reference 45 for a recent review of RNA ligases. The E. coli ligase RtcB, a component of the RNA repair operon, reseals tRNAs cleaved in the anticodon loop.42 It has been shown to be capable to catalyze tRNA splicing in yeast.46 It is not unreasonable to assume, therefore, that unexpected tRNA-derived RNAs, including “trans-splicing” products, appear as by-products of the tRNA cleavage/repair pathways and, hence, are present in the cell. In Helicobacter, for example, we find a transcript that looks like a spliced common precursor of two adjacent tRNAs, see Figure 3. In Salmonella, the matured tRNA-Gln-CTG is associated with circularized reads.
Figure 3. Unusual eubacterial reads associated with tRNAs. Above: Spliced fusion of two adjacent tRNAs in H. pylori. Redmarks indicate the 5′-side of the acceptor stem and D-stem, resp. The apparent intron extends roughly from the end of the acceptor stem of tRNA-Ile to the beginning of the D-stem of tRNA-Ala. The coverage suggests that the two adjacent tRNAs are produced from a single primary transcript. Below: A circularized tRNA in Salmonella.
Discussion
The preparation of RNA-seq libraries contains a reverse transcription step that may account for many of the observed non-canonical splicing events. Such RT artifacts have been investigated in detail, e.g., in reference 16‒18. While we cannot rule out in most cases that the observed reads are such “RTfacts,” there are plausible alternative mechanisms that could produce atypical transcript structures.
On the other hand, the data contain a large number of true positive examples for both Archaea and Eubacteria in which splicing or circularization has been demonstrated in independent experiments. Hence, clearly not all of the observed split reads are technical artifacts. In some cases, the molecular mechanisms that lead to the “spliced” RNAs are well known. This is the case for the self-splicing introns and for the processing of tRNAs47 and rRNAs10 in Archaea. The splicing endonuclease in Archaea has a broad range of targets and is known to be involved also in trans-splicing of tRNAs from independently encoded fragments as well as in the splicing of mRNAs. Homologous enzymes are present also in diverse eubacterial species, where they form a tRNA cleavage/repair pathway (briefly reviewed in the previous section). Thus, there appears to be an ancient RNA repair system present in all domains of life, which could account for many or even most of the spliced and circularized RNAs observed here.
In E. coli, the stress-induced toxin MazF cleaves certain single-stranded mRNAs at or closely upstream of the start codon and removes a 43 nt fragment that comprises the anti-Shine-Dalgarno sequence from the 3′ terminus of the 16S rRNA.48 Ribosomes with the truncated 16S rRNA specifically translate leaderless mRNAs, presumably as a stress response.49 The abundance of leaderless transcripts, also in other proteobacteria,50,51 might imply that similar mechanisms are more widespread. In conjunction with a variety of RNA ligases,45 they might account for at least a part of the atypical sequences observed here.
Apparent splice junctions that are supported by multiple read counts, thus, are at least good candidates for atypically processed RNAs that deserve further attention. In Archaea, the combination of atypical reads and a local, (nearly) isolated peak of coverage provide at least a very strong indication for processed ncRNAs. In all four Archaea considered here, additional candidates (Table 1) could be identified.
On the other hand, several well-described cases of atypical transcripts, such as the trans-spliced tRNAs in Nanoarchaeum, were observed only in a very small number of reads. This can be explained only in part by the presence of unspliced paralogs that attract the processed reads to the contiguous locus in the mapping procedure because an unspliced alignment is always preferred over a spliced one. Low expression, or support by only a small number of splice junctions, thus does not necessarily imply that an atypical transcript is a technical artifact or, even if present in the cell, devoid of biological function.
Materials and Methods
Sequence data
Publicly available RNA-seq data were downloaded from the short read archive for four Archaea and six Eubacteria, see Table S5 for details. All these RNA-seq data were produced with non-strand-specific protocols. With the exception of the read data for Escherichia coli and Salmonella enterica, all reads are single ended with lengths between 30‒100 nts. The data sets also vary considerably in size and read qualities, see Table S5 and Figure S2. According to requirements, the raw reads were quality trimmed with FASTX-Toolkit and adaptor clipped with Cutadapt.52
Annotation
Annotation sources are the GFF files for each analyzed species downloaded from the NCBI(ftp://ftp.ncbi.nih.gov/genomes/Bacteria) and the Rfam (ftp://ftp.sanger.ac.uk/pub/database/Rfam/11.0/genome.gff3.tar.gz) ftp servers, respectively. From the NCBI files, all genes are extracted and the corresponding annotated elements, i.e., CDS, tRNA and rRNA, are used. All genes that did not code for one of these elements are grouped into the separate class, “other.” Since NCBI annotation files often miss non-coding RNAs (ncRNAs) and regulatory elements such as riboswitches, these were instead adopted from the Rfam GFF files. The sources are listed in Table S5, all annotation items are provided in the Electronic Supplement.
Since the Rfam annotation did not feature well-known group I introns, we reasoned that either the Rfam seed alignment (RF00028) does not cover the diversity of bacterial group I introns, or the presence of open reading frames in these introns hampers the infernal search. We therefore split the Rfam seed alignment as well as 14 alignments of group I subtypes (www.rna.whu.edu.cn/gissd/53) into 27 overlapping blocks along the 5′→3′ direction of the intron, constructed individual CMs, scanned the genomes for these sub-CMs and reconstructed potential group I introns by chaining adjacent hits in the correct order (non-overlapping 5′ and 3′ sub-CMs, and ≤ 5 kb distance between sub-CMs). The resulting eight candidates, of which all but two have been described in literature,27,29,30 are listed in Table S2.
For group II introns, we downloaded 35 intron sequences listed in the group II intron database (webapps2.ucalgary.ca/~groupii/ on Feb 4th 201354) for different strains of the species considered here. Of these, nine could be located in our reference genomes by blastn, Table S3.
Read mapping
All reads were mapped with segemehl, version 0.1.455,56 with the split read option -S. Depending on the read length of RNA-seq data, the minimum fragment length Z and the minimum fragment score U were set to combinations from -Z 20 -U 18 to -Z 14 -U 12. These small values are motivated by the need to emphasize split reads. For all other parameters, the default values were used. Reads that remained unmapped in the first pass were remapped with Remapper, a component of the segemehl suite.
Reads that were split-mapped were assigned to one of three categories: “normal,” same strand, same chromosome and insert between 15 nt and 200 kb and matched fragments co-linear with the genomic DNA; “circular,” same strand, same chromosome and junction distance less than 200 kb with fragment order inverted relative to genomic DNA; “(strand)switched,” same chromosome, junction distance less than 200 kb and fragments located on opposite strands. Splice sites determined by the read mapping were clustered with haarz, a component of the segemehl suite, to determine median split positions. The results of the mapping procedure are summarized in Table 3.
Table 3. Summary of mapped reads.
| Species | input | mappable | unsplit | split | normal | Split class circular | switch |
|---|---|---|---|---|---|---|---|
| Eubacteria | |||||||
|
Bacillus cereus |
15,498,220 |
15,264,233 |
15,250,993 |
13,240 |
3,853 |
1,631 |
6,324 |
|
Escherichia coli |
52,515,346 |
44,429,568 |
44,115,280 |
314,288 |
8,573 |
20,544 |
39,587 |
|
Salmonella enterica |
31,924,568 |
27,752,771 |
27,737,761 |
15,010 |
543 |
2,481 |
4,421 |
|
Pseudomonas PA14 |
78,141,620 |
65,573,260 |
65,300,316 |
272,944 |
12,271 |
8,706 |
12,372 |
|
Helicobacter pylori 26695 |
82,847,902 |
40,152,294 |
39,146,732 |
1,005,562 |
17,930 |
53,709 |
77,095 |
|
Synechocystis PCC6803 |
31,985,927 |
15,080,656 |
15,031,302 |
49,354 |
39,956 |
165 |
250 |
| Archaea | |||||||
|
Nanoarchaeum equitans‡ |
17,253,447 |
11,173,688 |
11,096,897 |
35,034 |
7,393 |
12,860 |
1,932 |
|
Ignicoccus hospitalis‡ |
17,253,447 |
5,302,517 |
5,181,769 |
76,039 |
6,254 |
39,994 |
3,656 |
|
Pyrococcus furiosus |
16,449,461 |
8,691,213 |
8,474,477 |
216,736 |
11,536 |
54,795 |
17,095 |
| Sulfolobus solfataricus | 17,356,356 | 11,965,214 | 11,921,178 | 44,036 | 3,681 | 6,893 | 11,623 |
Nanoarchaeum and Ignicoccus was mapped from an RNA library containing material from both species. *Additional information can be found in the Supplemental Material.
In order to estimate the false positive rate for split transcripts, we constructed artificial data sets of approximately 1.3 million reads of length 50 and 100 randomly selected as contiguous sequences from the E. coli genome sequence. An Illumina-specific error model was used to generate a realistic data set. We observed only eight and four reads that were mapped with a split. The number of split reads observed in our mapping data, Table 3, thus exceeds the expected number of false positives by several orders of magnitude.
Overlaps between mapped reads and annotation data were computed with the help of BEDTools.57
Analysis of rRNA loci
To compare rRNA split read patterns across species the following steps were performed: (1) For each species, operon structures have been defined based on the rRNA gene annotation. For Eubacteria, 16S-23S-5S rRNA operons are used and 16S-23S rRNA operon in Archaea. (2) The sequences of the rRNA operons including 300 nt flanking sequence have been extracted from the corresponding genomes. In species with multiple copies of rRNA operons, a clustalw58 alignment has been calculated and the consensus sequence extracted. Either the consensus sequence or the sequence of a unique encoded operon has been used as reference operon. The only exceptions are N. equitans and H. pylori. In N. equitans, the 16S and 23S rRNA are transcribed from separate loci, in H. pylori, the 16S rRNA and an operon comprising the 23S and 5S rRNAs are separated. The separate parts were concatenated with an intervening stretch of 160 Ns as reference sequence. (3) For each species, all rRNA gene overlapping reads have been remapped onto the reference operon. Hence, all rRNA reads are projected to a single locus for each species. (4) To compare split-read patterns between species, the species-specific reference operons were aligned using clustalw and the mapped read coordinates transferred onto the alignment.
Supplementary Material
Acknowledgments
This work was supported in part by the German Research Foundation (STA 850/7-2, under the auspicies of SPP-1258 “Sensory and Regulatory RNAs in Prokaryotes”). LIFE - Leipzig Research Center for Civilization Diseases, Universitӓt Leipzig is funded in part by means of the European Social Fund and the Free State of Saxony.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24972
References
- 1.Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9:1516–31. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugahara J, Yachie N, Arakawa K, Tomita M. In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge-helix-bulge splicing motifs. RNA. 2007;13:671–81. doi: 10.1261/rna.309507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshinari S, Itoh T, Hallam SJ, DeLong EF, Yokobori S, Yamagishi A, et al. Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem Biophys Res Commun. 2006;346:1024–32. doi: 10.1016/j.bbrc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Evolution of introns in the archaeal world. Proc Natl Acad Sci USA. 2011;108:4782–7. doi: 10.1073/pnas.1100862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salgia SR, Singh SK, Gurha P, Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA. 2003;9:319–30. doi: 10.1261/rna.2118203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randau L, Söll D. Transfer RNA genes in pieces. EMBO Rep. 2008;9:623–8. doi: 10.1038/embor.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujishima K, Sugahara J, Kikuta K, Hirano R, Sato A, Tomita M, et al. Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc Natl Acad Sci USA. 2009;106:2683–7. doi: 10.1073/pnas.0808246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012;13:R63. doi: 10.1186/gb-2012-13-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgaard JZ, Garrett RA. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene. 1992;121:103–10. doi: 10.1016/0378-1119(92)90167-N. [DOI] [PubMed] [Google Scholar]
- 10.Tang TH, Rozhdestvensky TS, d’Orval BC, Bortolin ML, Huber H, Charpentier B, et al. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002;30:921–30. doi: 10.1093/nar/30.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starostina NG, Marshburn S, Johnson LS, Eddy SR, Terns RM, Terns MP. Circular box C/D RNAs in Pyrococcus furiosus. Proc Natl Acad Sci USA. 2004;101:14097–101. doi: 10.1073/pnas.0403520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–42. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–68. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen H, Johansen SD. Group I introns: Moving in new directions. RNA Biol. 2009;6:375–83. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- 15.Edgell DR, Chalamcharla VR, Belfort M. Learning to live together: mutualism between self-splicing introns and their hosts. BMC Biol. 2011;9:22. doi: 10.1186/1741-7007-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–31. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Roy SW, Irimia M. When good transcripts go bad: artifactual RT-PCR ‘splicing’ and genome analysis. Bioessays. 2008;30:601–5. doi: 10.1002/bies.20749. [DOI] [PubMed] [Google Scholar]
- 18.Houseley J, Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 23.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–11. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenkel-Morgenstern MFM, Lacroix V, Ezkurdia I, Levin Y, Gabashvili A, Prilusky J, et al. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012;22:1231–42. doi: 10.1101/gr.130062.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokobori S, Itoh T, Yoshinari S, Nomura N, Sako Y, Yamagishi A, et al. Gain and loss of an intron in a protein-coding gene in Archaea: the case of an archaeal RNA pseudouridine synthase gene. BMC Evol Biol. 2009;9:198. doi: 10.1186/1471-2148-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randau L, Münch R, Hohn MJ, Jahn D, Söll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–41. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 27.Biniszkiewicz D, Cesnaviciene E, Shub DA. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–35. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko M, Choi H, Park C. Group I self-splicing intron in the recA gene of Bacillus anthracis. J Bacteriol. 2002;184:3917–22. doi: 10.1128/JB.184.14.3917-3922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tourasse NJ, Kolstø AB. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: insights into their spread and evolution. Nucleic Acids Res. 2008;36:4529–48. doi: 10.1093/nar/gkn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tourasse NJ, Stabell FB, Reiter L, Kolstø AB. Unusual group II introns in bacteria of the Bacillus cereus group. J Bacteriol. 2005;187:5437–51. doi: 10.1128/JB.187.15.5437-5451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwieb C, Gorodkin J, Knudsen B, Burks J, Wower J. tmRDB (tmRNA database) Nucleic Acids Res. 2003;31:446–7. doi: 10.1093/nar/gkg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David M, Borasio GD, Kaufmann G. Bacteriophage T4-induced anticodon-loop nuclease detected in a host strain restrictive to RNA ligase mutants. Proc Natl Acad Sci USA. 1982;79:7097–101. doi: 10.1073/pnas.79.23.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–25. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–9. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Jöchl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, et al. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–89. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, et al. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–55. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keppetipola N, Nandakumar J, Shuman S. Reprogramming the tRNA-splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res. 2007;35:3624–30. doi: 10.1093/nar/gkm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem. 2011;286:7727–31. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konarska M, Filipowicz W, Gross HJ. RNA ligation via 2′-phosphomonoester, 3’5′-phosphodiester linkage: requirement of 2′,3′-cyclic phosphate termini and involvement of a 5′-hydroxyl polynucleotide kinase. Proc Natl Acad Sci USA. 1982;79:1474–8. doi: 10.1073/pnas.79.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Englert M, Sheppard K, Aslanian A, Yates JR, 3rd, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA. 2011;108:1290–5. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popow J, Schleiffer A, Martinez J. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci. 2012;69:2657–70. doi: 10.1007/s00018-012-0944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286:30253–7. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinemann IU, Söll D, Randau L. Transfer RNA processing in archaea: unusual pathways and enzymes. FEBS Lett. 2010;584:303–9. doi: 10.1016/j.febslet.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, et al. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–57. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moll I, Engelberg-Kulka H. Selective translation during stress in Escherichia coli. Trends Biochem Sci. 2012;37:493–8. doi: 10.1016/j.tibs.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 51.Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, et al. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012;40:2020–31. doi: 10.1093/nar/gkr904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17 [Google Scholar]
- 53.Zhou Y, Lu C, Wu QJ, Wang Y, Sun ZT, Deng JC, et al. GISSD: Group I intron sequence and structure database. Nucleic Acids Res. 2008;36(Database issue):D31–7. doi: 10.1093/nar/gkm766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Candales MA, Duong A, Hood KS, Li T, Neufeld RA, Sun R, et al. Database for bacterial group II introns. Nucleic Acids Res. 2012;40(Database issue):D187–90. doi: 10.1093/nar/gkr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann S, et al. A multi-split mapping algorithm for splicing, trans-splicing, and fusion detection in single-end reads. 2012 doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.