Abstract
It has been known for several years that heterozygous mutations of three members of the fibroblast growth-factor–receptor family of signal-transduction molecules—namely, FGFR1, FGFR2, and FGFR3—contribute significantly to disorders of bone patterning and growth. FGFR3 mutations, which predominantly cause short-limbed bone dysplasia, occur in all three major regions (i.e., extracellular, transmembrane, and intracellular) of the protein. By contrast, most mutations described in FGFR2 localize to just two exons (IIIa and IIIc), encoding the IgIII domain in the extracellular region, resulting in syndromic craniosynostosis including Apert, Crouzon, or Pfeiffer syndromes. Interpretation of this apparent clustering of mutations in FGFR2 has been hampered by the absence of any complete FGFR2-mutation screen. We have now undertaken such a screen in 259 patients with craniosynostosis in whom mutations in other genes (e.g., FGFR1, FGFR3, and TWIST) had been excluded; part of this screen was a cohort-based study, enabling unbiased estimates of the mutation distribution to be obtained. Although the majority (61/62 in the cohort sample) of FGFR2 mutations localized to the IIIa and IIIc exons, we identified mutations in seven additional exons—including six distinct mutations of the tyrosine kinase region and a single mutation of the IgII domain. The majority of patients with atypical mutations had diagnoses of Pfeiffer syndrome or Crouzon syndrome. Overall, FGFR2 mutations were present in 9.8% of patients with craniosynostosis who were included in a prospectively ascertained sample, but no mutations were found in association with isolated fusion of the metopic or sagittal sutures. We conclude that the spectrum of FGFR2 mutations causing craniosynostosis is wider than previously recognized but that, nevertheless, the IgIIIa/IIIc region represents a genuine mutation hotspot.
Introduction
Craniosynostosis, the premature fusion of one or more cranial sutures, occurs with a birth prevalence of 1/2,100–1/2,500 (Hunter and Rudd 1976; Lajeunie et al. 1995a). At present, causative mutations of single genes can be identified in ∼20% of cases. The major genes involved encode the transcription factor TWIST and three fibroblast growth-factor receptors (FGFRs)—FGFR1, FGFR2, and FGFR3 (reviewed by Muenke and Wilkie 2001). FGFRs are signal-transduction molecules that traverse the cell membrane; the binding of fibroblast growth factors (FGFs) to the extracellular region promotes FGFR dimerization, leading to trans-autophosphorylation of the intracellular tyrosine kinase region. FGFRs share a similar sequence structure, characterized by three extracellular immunoglobulin-like domains (IgI, IgII, and IgIII), a single-pass transmembrane segment, and a split tyrosine kinase (TK1/TK2) domain (Coulier et al. 1997; reviewed by Johnson and Williams 1993).
Heterozygous mutations of FGFR2, located at 10q26, were first described in the autosomal dominant disorder, Crouzon syndrome (CS [MIM 123500]) (Jabs et al. 1994; Reardon et al. 1994). Subsequently, FGFR2 mutations were also identified in Pfeiffer syndrome (PS [MIM 101600]) (Lajeunie et al. 1995b; Rutland et al. 1995; Schell et al. 1995), Apert syndrome (AS [MIM 101200]) (Wilkie et al. 1995), and Beare-Stevenson cutis gyrata syndrome [MIM 123790] (Przylepa et al. 1996); these phenotypes are distinguished from CS principally by specific limb and dermatological features (reviewed by Muenke and Wilkie 2001). Additional entities in which FGFR2 mutations have been described include Jackson-Weiss syndrome (Jabs et al. 1994) and some cases of Antley-Bixler syndrome (Chun et al. 1998; Reardon et al. 2000). Both these phenotypes overlap that of PS; the Jackson-Weiss designation is probably best reserved for the original family segregating the A344G mutation (Cohen 2001), whereas the value of assigning a diagnostic category of “Antley-Bixler syndrome caused by FGFR2 mutation” distinct from PS is debatable (Gripp et al. 1999).
The great majority of pathogenic FGFR mutations are missense, and all confer gain of function to the mutated protein; some mutations are highly recurrent. The gain-of-function mechanisms identified for FGFR2 mutations are (a) selectively enhanced FGF-binding affinity (Anderson et al. 1998), (b) illegitimate FGF-binding specificity (Yu et al. 2000), (c) FGF-independent covalent dimerization (Neilson and Friesel 1995; Robertson et al. 1998), and (d) ectopic spliceoform expression (Oldridge et al. 1999); these mechanisms account for the dominant inheritance of all the associated phenotypes. An exclusive paternal origin of de novo mutations, associated with advanced paternal age, has been described in AS, CS, and PS (Moloney et al. 1996; Glaser et al. 2000).
According to published reports, the mutational spectrum of FGFR2, the major forms of which are encoded by 19 exons, is very narrow. Of the distinct mutations identified, 94% (including all those causing AS and PS) occur either in one or the other two exons, IIIa (exon 8) and IIIc (exon 10), or in the intron sequence flanking exon IIIc (reviewed by Muenke and Wilkie 2001). Tissue-specific alternative splicing joins the constitutive IIIa exon to either the IIIb (exon 9) or IIIc exons, generating distinct isoforms of the FGFR2 molecule (i.e., FGFR2b/keratinocyte growth-factor receptor and FGFR2c, respectively) with different FGF-binding properties (Miki et al. 1992; Ornitz et al. 1996); hence, the exon IIIc mutations selectively affect only the FGFR2c isoform. The only mutations of FGFR2 described outside these regions have been a single mutation, Y105C, of the IgI domain in a child with atypical CS (Pulleyn et al. 1996); two closely spaced mutations, S372C and Y375C, in the extracellular juxta-membrane region causing Beare-Stevenson syndrome (Przylepa et al. 1996; Krepelová et al. 1998); and a single mutation, G384R, in the transmembrane region in a patient with nonsyndromic craniosynostosis (Pulleyn et al. 1996).
The known mutation spectrum of FGFR2 has remained essentially unchanged since the review by Wilkie (1997). Surprisingly, however, no complete genomic screen of FGFR2 in patients with craniosynostosis has been published. There are several possible reasons for this: the genomic structure of human FGFR2 has been determined only relatively recently (Zhang et al. 1999); the concentration of mutations in exons IIIa and IIIc may have discouraged investigators from looking elsewhere in the gene; and negative findings from small-scale genome screens may have remained unpublished. Three developments led us to reassess the situation. First, during the past 7 years we have collected a large sample of patients with craniosynostosis, in whom no mutation at the known hotspots could be identified. Second, reliable genomic sequence for FGFR2 is now available from the Human Genome Sequencing project; third, the development of denaturing high-performance liquid chromatography (DHPLC) technology provides a rapid, highly sensitive means of mutation detection (reviewed by Xiao and Oefner 2001). Our analysis provides the most complete picture of the spectrum of FGFR2 mutations to date and identifies new regions of the molecule (i.e., the IgII, TK1, and TK2 domains) in which mutations occur in craniosynostosis.
Patients and Methods
Patients and Samples
This study was approved by the Central Oxford Research Ethics Committee, and informed consent was given by patients or their parents before blood samples were obtained. DNA was obtained by standard proteinase K digestion and phenol-chloroform extraction.
Samples were obtained from three categories of patients (table 1). Category 1 (“Oxford”) comprised patients who had been attending the Oxford Craniofacial Unit since 1993, including nearly all those (n=112) who had undergone major craniofacial procedures and had been born during July 1995–October 1999 (inclusive). Complete clinical documentation and correction for ascertainment bias was possible for this sample. Category 2 (“non-Oxford”) comprised patients, from a diversity of sources, who had either proven or suspected craniosynostosis and who had been referred to the Oxford sample for molecular genetic diagnosis. Category 3 (“Pfeiffer”) comprised patients with a diagnosis of PS who had been referred to the National Institutes of Health–affiliated group and in whom no mutation of FGFR1, FGFR3, or exons IIIa and IIIc of FGFR2 had previously been found (Cornejo-Roldan et al. 1999). This sample included two families (designated “4” and “10” by Schell et al. [1995]) in which segregation of 10q markers was consistent with linkage to FGFR2. In this category, two patients who tested positive for an FGFR2 mutation had their diagnosis changed to CS after clinical review. Patients in the Oxford and non-Oxford samples had previously been screened by DHPLC, for (a) mutations in exon 7 of FGFR1, encoding the P252R mutation (Muenke et al. 1994), (b) mutations in exons 7 and 10 of FGFR3, encoding the P250R and A391E mutations, respectively (Meyers et al. 1995; Bellus et al. 1996; Moloney et al. 1997), and (c) coding regions of TWIST (Elanko et al. 2001), as well as for heterozygous deletions of TWIST (Johnson et al. 1998). Patients with mutations in these other genes, as well as those with craniofrontonasal syndrome, which maps to the X chromosome (Feldman et al. 1997), were excluded from the analysis. Owing to positive ascertainment bias, we also excluded patients with AS who in were the non-Oxford sample; we have reported elsewhere on the spectrum of FGFR2 mutations in AS (Slaney et al. 1996; Oldridge et al. 1997, 1999).
Table 1.
Sample Analyzed for FGFR2 Mutations
|
Proportion of Patients with FGFR2 Mutation |
|||
| Diagnosis | Oxford Sample | Non-Oxford Sample | Pfeiffer Sample |
| Syndromic: | |||
| AS | 29/29 | … | … |
| CS | 18/20 | 8/14 | |
| PS | 12/12 | 9/11 | |
| Other | 0/18 | 1/16 | … |
| Nonsyndromic: | |||
| Multiple | 0/5 | … | |
| Bicoronal | 2/12 | … | |
| Unicoronal | 1/24 | … | |
| Metopic | 0/17 | … | |
| Sagittal | 0/13 | … | |
| Lambdoid | 0/2 | … | |
| Subtotal | 3/73 | 0/28a | |
| Total | 62/152 | 18/69 | 5/38b |
Full documentation of sutures affected was not available.
Patients with identified mutations had diagnoses of either CS or PS.
Mutation Analysis
Exon numbering follows the nomenclature proposed by Ingersoll et al. (in press) (GenBank accession numbers AF410480 and AF360695); nucleotide numbering of the cDNA starts at the ATG initiation codon and is based on an FGFR2c spliceform (Genbank accession numbers NM_000141) that omits exons 7, 9, 20, and 21. Exons of FGFR2 were identified by comparing the cDNA sequences isolated by Dionne et al. (1990) and Miki et al. (1992) versus the genomic sequence of bacterial-artificial-chromosome clone RP11-7P17 (Genbank accession number AC009988). This method was preferred to the use of the sequences published by Zhang et al. (1999), owing to several discrepancies in the sizes of exons and the sequences of intron/exon boundaries. We did, however, include in the analysis three additional alternatively spliced exons (7, 20, and 21) tabulated by Zhang et al. (1999) and Ingersoll et al. (in press). Expression of these exons was originally detected in a stomach-cancer cell line (Katoh et al. 1992; Itoh et al. 1994), and their physiological significance is uncertain.
The primer sequences used for PCR amplification of FGFR2 are shown in table 2. All primers were newly designed, except for those amplifying exon IIIa (6/7AF-5/6R2), which have been described elsewhere (Slaney et al. 1996; Oldridge et al. 1999). The 6/7AF primer is not subject to allele-specific dropout as reported for a different forward primer that has been used to amplify this exon (Wong et al. 2001). Initially, we analyzed the mutation hotspots in exons IIIa and IIIc: 74 patients testing positive for a mutation in one of these two exons were excluded from the screen of the remainder of FGFR2. In the remaining 185 patients, the entire FGFR2 gene was screened by DHPLC. The single exception was the patient with the Y105C mutation, which previously had been detected by an unpublished RNA-based screening method (Johnson et al. 2000).
Table 2.
Primers and Conditions for Genomic Amplification of FGFR2
|
Primer(5′→3′) |
Temperature(s)(°C) |
||||
| Exon | Forward | Reverse | Fragment Size(bp) | Annealing | DHPLC Analysis |
| 2 | TCCCTGACTCGCCAATCTCTTTC | TGCCCCCAGACAAATCCCAAAAC | 341 | 55 | 57, 63 |
| 3 | CACTGACCTTTGTTGGACGTTC | GAGAAGAGAGAGCATAGTGCTGG | 380 | 64 | 61 or 62 |
| 4 | TGGAGAAGGTCTCAGTTGTAGAT | AGACAGGTGACAGGCAGAACT | 232 | 55 | 58, 62 |
| 5 | CAAAGCGAAATGATCTTACCTG | AGAAATGTGATGTTCTGAAAGC | 291 | 62 | 58, 63 |
| 6 | GCTAGGATTGTTAAATAACCGCC | AAACGAGTCAAGCAAGAATGGG | 226 | 62 | 59, 62 |
| 7 (5′) | TGAGTTTGCCTCTCCTCGTGTG | CCTTCTACAGTTGCCCTGTTGG | 390 | 62 | 58 |
| 7 (3′) | GATGTGCTGTAGCAGACCTTTGG | ATCATCACAGGCAAAACCTGGG | 360 | 62 | 58, 61 |
| 8 (IIIa) | GGTCTCTCATTCTCCCATCCC | CCAACAGGAAATCAAAGAACC | 325 | 62 | 61, 64 |
| 9 (IIIb) | AATGCTAAGACCTTCCTGGTTGG | CAGTCTCCCAAAGCACCAAGTC | 284 | 55 | 56, 60 |
| 10 (IIIc) | CCTCCACAATCATTCCTGTGTC | ATAGCAGTCAACCAAGAAAAGGG | 257 | 62 | 58, 60 |
| 11 | TGCGTCAGTCTGGTGTGCTAAC | AGGACAAGATCCACAAGCTGGC | 341 | 64 | 59, 62 |
| 12 | TGACTTCCAGCCTTCTCAGATG | AGTCTCCATCCTGGGACATGG | 252 | 64 | 60, 63 |
| 13 | CCCCATCACCAGATGCTATGTG | TTGATAAGACTCTCCACCCAGCC | 221 | 55 | 59, 63, 66 |
| 14 | TAGCTGCCCATGAGTTAGAGG | ATCTGGAAGCCCAGCCATTTC | 250 | 62 | 58, 60 |
| 15 | TGTTTTGCTGAATTGCCCAAG | TCCACCCAGCCAAGTAGAATG | 294 | 55 | 59, 61 |
| 16 | CTGGCGGTGTTTTGAAATTAG | CCTTTCTTCCTGGAACATTCTG | 242 | 60 | 57, 60 |
| 17 | AGCCCTATTGAGCCTGCTAAG | CCAGGAAAAAGCCAGAGAAAAG | 177 | 62 | 55, 59 |
| 18 | GGTTTTGGCAACGTGGATGGG | GGTATTACTGGTGTGGCAAGTCC | 250 | 60 | 62 |
| 19 | ACACCACGTCCCCATATTGCC | CTCACAAGACAACCAAGGACAAG | 243 | 60 | 58, 61 |
| 20 | TCTGCCAAAATTGTTGTTTCTAGT | GGTCTGGAACTCCTGACCTCA | 208 | 60 | 57, 60, 63 |
| 21 | TCCCACGTCCAATACCCACATC | TACTGTTCGAGAGGTTGGCTGAG | 196 | 62 | 58, 60 |
| 22 | CGTCCAATACCCACATCTCAAG | TTCCCAGTGCTGTCCTGTTTGG | 363 | 60 | 59 |
We amplified 60 ng of DNA in a 30-μl reaction that included 1× GeneAmp PCR Buffer (Applied Biosystems), 1.25–1.5 mM MgCl2 (except for exons 14 and 17, which employed 2.5 mM MgCl2), primers at 0.4 mM, and dNTPs at 120 mM (final concentrations), with 0.6 U of AmpliTaq Gold (Applied Biosystems), and 0.1 U of Pwo DNA polymerase (Roche). PCR was performed in a Dyad DNA Engine (MJ Research), under a heated lid, in Skirted Thermo-Fast 96 plates covered by Adhesive PCR Film (both from ABgene), with an initial incubation at 95°C for 10 min, followed by 30–32 cycles of denaturation at 94°C for 60 s, annealing (at temperatures given in table 2) for 45 s, and extension at 72°C for 30 s, and with a final extension for 10 min. Heteroduplex products were obtained by heating the mix to 95°C for 5 min, followed by cooling at 1.5°C/min. Five microliters of the product was analyzed by DHPLC, on the WAVE DNA Fragment Analysis System (Transgenomic), at the temperatures given in table 2. Fragments exhibiting an abnormal retention pattern were sequenced on the ABI PRISM 377 DNA sequencer, with BigDye terminator mix (Applied Biosystems). In the case of the 514_515GC→TT mutation, the PCR product was cloned into pGEM-T Easy (Promega), and individual clones were sequenced.
All mutations were confirmed by an independent method (i.e., restriction-enzyme digestion, allele-specific oligonucleotide [ASO] hybridization, or combined primer-mismatch/restriction-enzyme digestion). The methods used for the newly described mutations were as follows (mismatches in oligonucleotides are underlined): 514_515GC→TT, loss of HaeIII site; 842A→G, gain of MwoI site; 1645A→C, ASO 5′-TATCATACATCTTCTTGG-3′; 1694A→G, mismatched forward primer 5′-TCAACAGGGCCTCTCTATGCCATAGTTG-3′ with BslI digestion; 1922A→G, mismatched forward primer 5′-ATGTTTTGGTAACAGAAAACAATGTGCTGA-3′ with DdeI digestion; 1977G→T, ASO 5′-ATTGGTGGTATTTTTGT-3′; 1988G→A, ASO 5′-TTTGCAGGAGCGGCTTCC-3′; and 2032A→G, ASO 5′-CTGTTTGATGGAGTATAC-3′. A high likelihood of correct paternity and sample identity was established for the de novo mutations, by demonstration of consistent segregation patterns for at least seven microsatellites, each of heterozygosity >0.67 and located on different chromosomes, in parent-child trios. Every mutation was shown to be absent from a panel of at least 128 control chromosomes. For variants considered to be nonpathogenic, allele frequencies were obtained for patients in the Oxford and non-Oxford samples only.
Results
In total, we detected 85 independent FGFR2 mutations in the 259 patients who met our selection criteria. Table 1 provides an overview of the number of patients studied and of the proportion who were positive for FGFR2 mutations; table 3 lists the individual mutations that were found. Our screening strategy for the Oxford and non-Oxford samples involved the initial examination of the two exons, IIIa and IIIc, that represent the known FGFR2-mutation hotspots. Excluding the 29 AS cases, we detected 45 mutations of 22 different types in these two exons (table 3). Many of these mutations involve patients described in earlier reports (Oldridge et al. 1995, 1997, 1999; Rutland et al. 1995; Przylepa et al. 1998; Glaser et al. 2000; Johnson et al. 2000). All exon IIIa and IIIc mutations were associated with AS, CS, or PS, except for the 943G→T (A315S) and 1032G→A (A344A) mutations, which, in one and two instances, respectively, occurred in nonsyndromic coronal synostosis (see Steinberger et al. 1996; Johnson et al. 2000). One unusual mutation is 842A→G (Y281C), which previously had been described only in abstract form (Tsai et al. 2000). In our family, this mutation presented with a very mild phenotype. The proband was an adult female who, as a child, had been diagnosed with CS and who had never required surgery; several of her relatives had a similar facial appearance, but none had come to medical attention, and they declined molecular investigation.
Table 3.
Pathogenic FGFR2 Mutations Identified in the Study
|
No. of Unrelated Patients with Phenotype (No. Corroborated by Molecular Analysis)a |
||||||||
| Oxford Sample |
Non-Oxford Sample |
Pfeiffer Sample |
||||||
| Nucleotide Change | Amino Acid Substitution | Domain | Familial | De Novo | Familial | De Novo | Familial | De Novo |
| Exon 3: | ||||||||
| 314A→G | Y105C | IgI | 1 CS (1) | |||||
| Exon 5: | ||||||||
| 514_515GC→TT | A172F | IgII | 1 PS (1) | |||||
| Exon 8: | ||||||||
| 755C→G | S252W | IgII-IgIII linker | 18 AS (11) | …b | …b | |||
| 755_757CGC→TCT | S252L, P253S | IgII-IgIII linker | 1 PS (1) | |||||
| 758C→G | P253R | IgII-IgIII linker | 1 AS (1) | 10 AS (10) | …b | …b | ||
| 799T→C | S267P | IgIIIa | 1 CS (1) | |||||
| 826T→G | F276V | IgIIIa | 1 CS | |||||
| 833G→T | C278F | IgIIIa | 5 CS, 1 PS (3) | 1 CS (1) | ||||
| 842A→G | Y281C | IgIIIa | 1 CS | |||||
| 866A→C | Q289P | IgIIIa | 1 CS (1) | |||||
| 870G→T | W290C | IgIIIa | 1 PS (1) | 1 PS (1) | ||||
| Exon 10: | ||||||||
| 940-2A→T | IgIIIc splice acceptor | 1 PS (1) | ||||||
| 940-2A→G | IgIIIc splice acceptor | 1 PS (1) | 1 PS (1) | |||||
| 943G→T | A315S | IgIIIc | 1 Ns (1) | |||||
| 1012G→C | G338R | IgIIIc | 1 CS | |||||
| 1018T→C | Y340H | IgIIIc | 1 CS | |||||
| 1019A→G | Y340C | IgIIIc | 1 PS | |||||
| 1021A→C | T341P | IgIIIc | 2 PS (1) | |||||
| 1024T→C | C342R | IgIIIc | 4 PS (2) | 1 PS | ||||
| 1025G→A | C342Y | IgIIIc | 1 CS | 4 CS (4) | 1 PS (1) | |||
| 1025G→C | C342S | IgIIIc | 1 PS (1) | 1 PS (1) | ||||
| 1025G→T | C342F | IgIIIc | 1 CS | |||||
| 1026C→G | C342W | IgIIIc | 1 CS (1) | |||||
| 1032G→A | A344A | IgIIIc (cryptic splice donor) | 2 CS, 2 Ns (1) | |||||
| 1040C→G | S347C | IgIIIc | 1 CS (1) | 1 CS | ||||
| 1061C→G | S354C | IgIIIc | 1 CS (1) | |||||
| Exon 11: | ||||||||
| 1124A→G | Y375C | Juxta-membrane | 1 PS | |||||
| Exon 14: | ||||||||
| 1645A→C | N549H | TK1 | 2 CS | |||||
| Exon 15: | ||||||||
| 1694A→G | E565G | TK1 | 1 PS (1) | |||||
| Exon 16: | ||||||||
| 1922A→G | K641R | TK2 | 1 PS (1) | 1 PS (1) | ||||
| 1977G→T | K659N | TK2 | 1 O (1) | |||||
| Exon 17: | ||||||||
| 1988G→A | G663E | TK2 | 1 PS | |||||
| 2032A→G | R678G | TK2 | 1 CS (1) | |||||
Each patient was classified as either a familial or a de novo case, on the basis of family history. The numbers in parentheses indicate the number of cases in which this was corroborated by molecular analysis. Ns = nonsyndromic; O = other, unclassified syndrome.
AS was excluded from the non-Oxford sample.
The remaining 185 patients in all three samples (except for 1 with a Y105C mutation that we had detected by an earlier, RNA-based analysis) were screened by DHPLC, for mutations in the entire remainder of the FGFR2 open reading frame. In 11 unrelated individuals, we detected nine distinct heterozygous missense substitutions that are likely to be pathogenic (table 3). In addition, we found 17 further sequence variations (table 4). Of these, 15 are either synonymous or occur in noncoding DNA, and they are unlikely to be pathogenic, whereas the nonsynonymous substitution M186T has already been identified as a polymorphism. At present, the clinical significance of the remaining nonsynonymous variant, S57L, is uncertain.
Table 4.
Additional FGFR2 Sequence Variants in 146 Patients Negative for Mutation in Exons IIIa and IIIc
| Location (No. of Variants) and Nucleotide Changea | Amino AcidSubstitution | Frequency ofRarer Alleleb | Reference |
| Exon 2 (2): | |||
| c−74G→A | … | 1/286 | |
| c−46G→A | … | 1/286 | |
| Exon 3 (4): | |||
| 110−54C→T | … | 2/286 | |
| 159G→A | A53A | 1/286 | rs1047102c |
| 170C→T | S57L | 1/286 | |
| 294G→A | T98T | .024 | rs1047101c |
| Exon 5 (1): | |||
| 557T→C | M186T | 2/286 | rs755793c |
| Exon 6 (1): | |||
| 696G→A | V232V | .243 | rs1047100c |
| Exon 7 [3′] (1) | |||
| 340G→Ad | .071 | rs1801043c | |
| Exon 11 (1): | |||
| 1085−41G→A | … | 1/286 | |
| Exon 12 (1): | |||
| 1288−23G→C | … | 2/286 | |
| Exon 15 (2): | |||
| 1673−47G→A | … | 2/288 | |
| 1673−10C→T | … | 1/288 | |
| Exon 16 (2): | |||
| 1864−17T→G | … | .014 | Ingersoll et al. (in press) |
| 1941C→T | L647L | 1/282 | |
| Exon 18 (1): | |||
| 2058−31A→G | … | .018 | Ingersoll et al. (in press) |
| Exon 19 (1): | |||
| 2301+15T→C | … | .420 | Ingersoll et al. (in press) |
Direction of nucleotide substitution is from the more common allele to the less common allele.
Expressed as a decimal fraction if >.01; expressed as a proportion (no. of alleles identified/no. of alleles tested) if <.01.
See the Single Nucleotide Polymorphism web site.
Nucleotide number in exon 7.
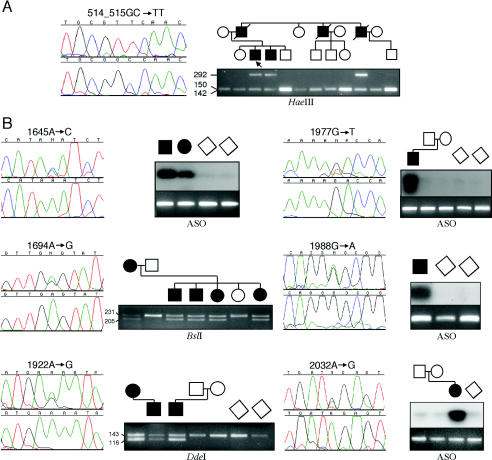
The pathogenic mutations of FGFR2 that were outside the exon IIIa/IIIc hotspot occurred in seven different exons, encoding one mutation in IgI, one in IgII, one in the juxta-membrane region, two (in a total of three patients) in TK1, and four (in a total of five patients) in TK2 (table 3). Two of the mutations have been published previously. The Y105C mutation, present in a boy with CS and marked facial asymmetry, has been reported, in a single case, by Pulleyn et al. (1996). The Y375C mutation, present in a girl who had severe PS including cloverleaf skull, bilateral choanal atresia, prominent labia majora, and a sacral appendage and who died on the 5th day of life, has been described in three cases of Beare-Stevenson syndrome (Przylepa et al. 1996; Krepelová et al. 1998). Interestingly, retrospective review of our case did not reveal the characteristic cutis gyrata described in previously published affected individuals, although the severe clinical course was typical. The remaining seven mutations, identified in nine unrelated cases, are newly described; figure 1 shows representative sequencing results and confirmatory tests for each mutation.
Figure 1.
Identification of novel FGFR2 mutations, in the IgII domain (A) and in the TK1 and TK2 domains (B), in patients with craniosynostosis. The left side of each panel shows the DNA-sequence electropherogram of an individual with the heterozygous mutation (above), compared with that of a normal control (below), except that individually cloned alleles were sequenced in the case of the double-nucleotide substitution 514_515GC→TT; the right side of each panel shows the independent confirmation, by restriction digestion or ASO blotting, in family and control samples.
One mutation, A172F, was found in the IgII domain in a family with PS. In 11 individuals in this family, a previous study had shown that the segregation of 10q markers was consistent with a mutation in FGFR2 (Schell et al. 1995). The mutation, which involves the substitution of two consecutive nucleotides, is the first described in the IgII domain in any FGFR and is of historic interest because the family was originally described by Pfeiffer (1964), giving the disorder its eponymous title. Paradoxically, the phenotype is somewhat atypical, since affected family members do not have the usual “crouzonoid” appearance (characterized by exorbitism, midface hypoplasia, and a prominent beaked nose) and have distinctive abnormalities of the hands and feet (short and broad first digits, with brachydactyly, symphalangism, and cutaneous syndactyly of other digits). We identified the mutation in three affected individuals, but in none of seven unaffected individuals at 50% prior risk (fig. 1A). To assess the pathogenic significance of the mutation, we aligned amino acid sequences of selected immunoglobulin-like domains, as shown in figure 2A. The mutated residue is located at the turn between β-sheet strands A′ and B, which make contacts with FGF ligand and heparin cofactor (Plotnikov et al. 1999; Pellegrini et al. 2000; Stauber et al. 2000). The peptide backbone is in a strained conformation at this position and is occupied by either glycine or alanine, in many telokin-type immunoglobulin folds (Harpaz and Chothia 1994; Bateman and Chothia 1995). In addition, according to one version of the structure of the quaternary FGF-FGFR binding complex, the two A172 side chains contact each other across the receptor:receptor interface (Plotnikov et al. 1999; Stauber et al. 2000).
Figure 2.
Amino acid sequence alignment around the sites of mutation, in the IgII domain (A) and in the TK1 and TK2 domains (B). In each case the FGFR2 sequence is shown at the top, with the substituted amino acids immediately above (red). Unfilled and hatched rectangles delimit regions of α-helix and β-sheet secondary structure, respectively, according to Plotnikov et al. (1999) (in the case of panel A) and Mohammadi et al. (1996) (in the case of panel B). Sequence identities of aligned proteins with FGFR2 are indicated by a dot. In panel A, Ig domains from human FGFRs, together with those from Drosophila melanogaster heartless (HTL) and Caenorhabditis elegans egg-laying defective 15 (EGL-15), are compared with MYLK, which is synonymous with telokin and provides a prototype for I-set Ig domains (Harpaz and Chothia 1994). In panel B, the positions of mutations in human FGFR3, D. melanogaster HTL, C. elegans EGL-15, and human KIT, RET, and MET are indicated by boxes, according to whether functional studies have shown the mutations to be activating (green) or inactivating (blue) or have not been undertaken (gray). Data are from DeVore et al. (1995); Gisselbrecht et al. (1996); Jeffers et al. (1997); Pelet et al. (1998); Iwashita et al. (1999); Longley et al. (1999); Stenberg et al. (1999; also see the KinMutBase protein-alignment web site); Bellus et al. (2000); Mortier et al. (2000); Iwashita et al. (2001); and the Human Gene Mutation Database.
The remaining six mutations were identified in the tyrosine kinase domains (TK1 and TK2) and are the first mutations described in the intracellular part of FGFR2. Three of these mutations—K641R, K659N, and R678G—were shown to have arisen de novo, providing strong evidence for pathogenicity, and the segregation of the E565G mutation, identified in the pedigree reported as family “10” by Schell et al. (1995), was concordant with the phenotype in six individuals. The N549H mutation, identified in two individuals with sporadic CS, is likely to be pathological, because it occurs at the residue equivalent to the mutation hotspot, in FGFR3, for hypochondroplasia (see below). No comparable supporting data are available for the pathogenicity of the G663E mutation; however, this is a nonconservative substitution that was not observed in 128 control chromosomes.
The sequence context of the six TK1/TK2 mutations is shown in figure 2B. All occurred at residues that are entirely conserved between the four human FGFRs but that are only variably conserved among invertebrate FGFR orthologues and other receptor tyrosine kinases. Significantly, the N549 and K659 residues in FGFR2 are exactly homologous, respectively, to the N540 and K650 residues of FGFR3, which are mutated in bone dysplasias. Mutations of the N540 residue to K, T, and S (but not H) and of the K650 residue to N and Q have been described in hypochondroplasia (Bellus et al. 2000; Mortier et al. 2000); biochemical studies indicate that these mutations are activating (see the Discussion section). No mutation at the residues equivalent to E565, K641, G663, or R678 has been reported in FGFR3. However, as illustrated in figure 2B, a patchwork of activating and inactivating mutations of other receptor tyrosine kinases, notably KIT, RET, and MET, has been described in similar regions of these proteins (see the Discussion section).
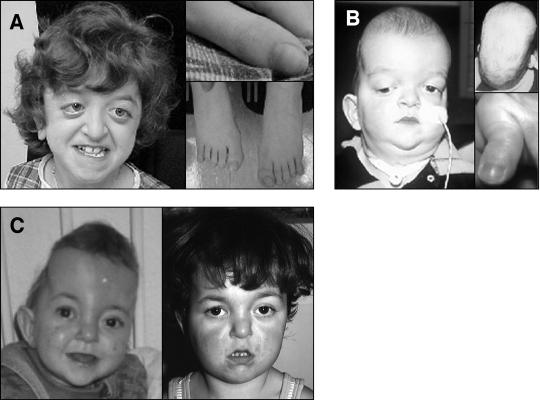
The clinical features associated with mutations of the TK1/TK2 region are summarized in table 5, and three individuals are illustrated in figure 3A–C. All patients were classified as having syndromic craniosynostosis and, to varying degrees, exhibited a crouzonoid facies that, in several cases, became more obvious with age (fig. 3C). In some cases, this was associated with broadening of the thumbs and halluces (compare figs. 3A and B). Hydrocephalus and developmental delay were overrepresented compared with their frequency in typical CS and PS (table 5). One patient heterozygous for the K641R mutation had marked scaphocephaly owing to predominant sagittal suture synostosis (fig. 3B). The phenotype associated with the familial E565G mutation was strikingly variable, ranging from a crouzonoid appearance with normal intelligence to cloverleaf skull with early death (table 5).
Table 5.
Clinical Features of Patients with Tyrosine Kinase Domain Mutations in FGFR2
|
Status (No.)b |
|||||||||
| Patient | Amino AcidSubstitution | ClinicalDiagnosisa | No. and Sex of Affected | CRS | BroadThumbs | BroadHalluces | DD | HC | Additional Features (No.) |
| BL2403 | N549H | CS | 1 M | − | − | − | + | + | Macrocephaly |
| BL2622 | N549H | CS | 1 F | + | − | − | + | + | Arnold-Chiari malformation, choanal stenosis |
| BL864 | E565G | PS | 2 M, 3 F (plus 1 M and 1 Fc) | + (7) | + (2), − (2) | + (5) | + (3), − (4) | + (1) | Cloverleaf skull (1), congenital heart disease (1) |
| OX2066 | K641R | PS | 1 Md | + | + | + | − | + | Scaphocephaly, proximal symphalangism of index finger |
| BL814 | K641R | PS | 1 F, 1 M | + (2) | + (2) | + (2) | + (2) | + (1), − (1) | Imperforate anus (1) |
| OX1732 | K659N | O | 1 Fd | + | − | − | (+) | − | |
| OX1263 | G663E | PS | 1 M | + | + | + | + | − | |
| OX1278 | R678G | CS | 1 Fd | (+) | − | (+) | − | + | |
O = other, unclassified syndrome.
CRS = craniosynostosis; DD = developmental delay; HC = hydrocephalus; + = present; (+) = mildly affected; − = absent.
These two additional individuals were not subject to molecular testing.
Documented de novo mutation.
Figure 3.
Phenotype of patients heterozygous for FGFR2 mutations in the TK1 domain (A) and in the TK2 domain (B and C). A, Patient BL2622, with the N549H mutation, age 11 years. Note the crouzonoid appearance (left) and that thumbs and halluces are not significantly broadened (right). B, Patient OX2066, with the K641R mutation, age 8 mo. The facial appearance is not characteristic (left); note the tracheostomy. Viewed from above, the head has a markedly scaphocephalic contour; the thumb is broad with radial angulation (right). C, Patient OX1732, with the K659N mutation, age <1 year (left) and 3.4 years (right). The initial diagnosis was unclassified syndromic craniosynostosis. Note the marked turricephaly at the earlier age; the crouzonoid appearance is more evident with age.
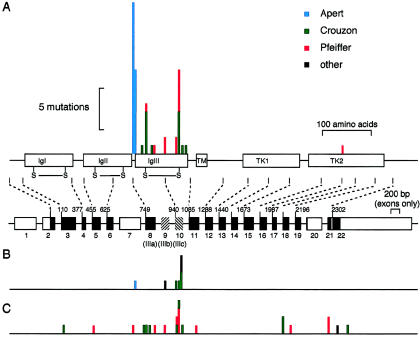
The prospective ascertainment and uniform clinical workup practiced at the Oxford Craniofacial Unit enables a fairly unbiased assessment of the spectrum of germline FGFR2 mutations that cause craniosynostosis. This can be examined in two ways. First, the distribution of de novo FGFR2 mutations (based on a sample size of 54), shown in figure 4A, is highly nonrandom: mutations of four amino acids—S252, P253, C278, and C342—account for 81% of these mutations. Only one of the TK1/TK2 mutations originated from the Oxford sample, attesting to their relative rarity. Second, the distribution of transmitted mutations (fig. 4B), comprising 13% of the mutations in the Oxford sample, is also highly nonrandom. Four of eight mutations involved the synonymous substitution 1032G→A, which, characteristically, is associated with a mild and variable phenotype (Steinberger et al. 1996). Overall, 61 of 62 mutations in the Oxford sample localized to either the IIIa exon or the IIIc exon. The spectrum of FGFR2 mutations in patients in the non-Oxford and Pfeiffer samples (fig. 4C) was noticeably different (only 13 of 23 had mutations in exon IIIa or exon IIIc). This reflects the bias of these samples toward atypical phenotypes and toward cases in which exon IIIa and IIIc mutations had already been excluded.
Figure 4.
Distribution of mutations identified in the complete screen of FGFR2. A, Distribution of de novo mutations in the Oxford sample (n=54), shown above a cartoon of the FGFR2 domain structure (drawn to scale) and a summary of the genomic organization (only exons are drawn to scale). Exons are shown as boxes, with coding regions in black, except for the alternatively spliced exons IIIb and IIIc, which are hatched. B, Distribution of inherited mutations in the Oxford sample (n=8). C, Distribution of all other mutations (n=23) described in the present report. Phenotypes shown are AS (blue), CS (green), PS (red), and other (syndromic or nonsyndromic) (black).
Discussion
In this study, we have undertaken the most complete screen for FGFR2 mutations in craniosynostosis to date, examining ∼0.9 Mb of DNA sequence and revealing unexpected diversity in the spectrum of mutations. As in any screening method, the value of our findings depends on the sensitivity of the detection method. Before embarking on this study, we evaluated the sensitivity of DHPLC by assessing its ability to detect 30 known single-nucleotide substitutions in eight different PCR fragments from the FGFR1, FGFR2, FGFR3, and TWIST genes; all were detected (i.e., sensitivity was 100%). We assessed the positive predictive value during this study by comparing the number of distinct positive signals in DHPLC screening (30) with the number of distinct sequence changes identified (26), which yields a positive predictive value of 87%. We conclude that DHPLC analysis using the WAVE system is a sensitive and specific method to screen for FGFR2 mutations in patients with craniosynostosis.
The identification of FGFR2 mutations in seven different exons outside the IIIa/IIIc hotspot highlights potential deficiencies in the mutation screens currently undertaken in diagnostic laboratories, which tend to concentrate on these two exons. On the basis of the phenotypes of patients testing positive for atypical mutations, the greatest pick-up rate is anticipated in patients with suspected diagnoses of either CS or PS. On the other hand, no FGFR2 mutation was found in any case of nonsyndromic sagittal or metopic synostosis (table 1), and the three FGFR2 mutations identified in patients with nonsyndromic coronal synostosis were all located in the IIIc exon (table 3). These observations suggest strategies for maximizing diagnostic pick-up while maintaining the cost-effectiveness of FGFR2 mutation screening. In this regard, we note that the efficiency of mutation detection varied markedly with the population studied; for example, in the Oxford sample, a molecular diagnosis was obtained in 90% and 100%, respectively, of patients with CS and patients with PS, but these figures fell to 57% and 82%, respectively, in the non-Oxford sample. This probably represents differences in the stringency of clinical diagnosis: we emphasize the importance of careful clinical assessment, particularly the recognition of the crouzonoid facies, in achieving maximum diagnostic yield. The overall contribution of mutations in FGFR2 to the burden of craniosynostosis, estimated from data for the 4.3-year period of complete ascertainment at the Oxford Craniofacial Unit, was 9.8% (11/112). This may slightly overestimate the true figure, owing to a bias toward referral of the more complex cases for surgical evaluation.
The newly identified FGFR2 mutations map to two regions of the protein, the IgII and TK1/TK2 domains. The most important consequence of the A172F mutation appears to be local unfolding of the IgII domain, potentially exposing the side chain of one of the paired cysteines, which lies seven residues downstream of the mutated alanine (fig. 2A); this unfolding may promote constitutive activation by covalent intermolecular dimerization of mutant receptor monomers. Experimental testing of this possibility will be important, given that a previously published investigation of the consequences of mutation of cysteine residues in IgII did not reveal any activation in a cellular-transformation assay (Robertson et al. 1998). The apparent paucity of IgII mutations, combined with the distinct limb and craniofacial phenotype in this family, raises the possibility of a more specific pathological mechanism; for example, it is conceivable that the proposed interaction between the two A172 residues across the receptor:receptor interface (Plotnikov et al. 1999; Stauber et al. 2000) could be strengthened by stacking of the substituted phenylalanine side chains in the mutant homodimer.
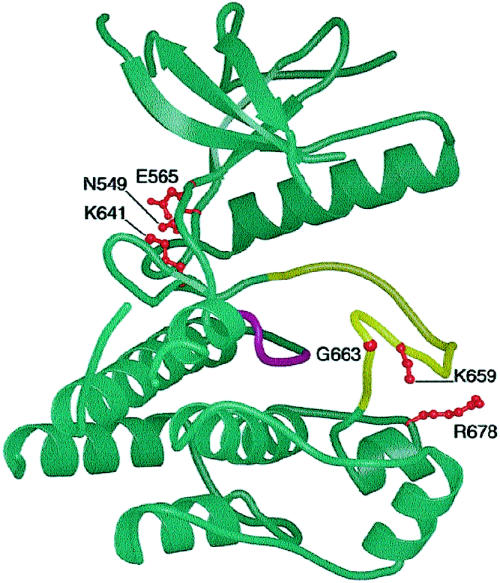
The position of the six mutations of the TK1 and TK2 domains in the FGFR1 structure published by Mohammadi et al. (1996) is shown in figure 5. All are located in the cleft between the TK1 and TK2 domains, which contains structures critical for tyrosine kinase activation, including the ATP and substrate peptide-binding regions, the catalytic loop, and the activation loop. Activation is accompanied by a change in the relative orientation of the TK1 and TK2 domains, the architecture of which is specific to each receptor tyrosine kinase (reviewed by Hubbard and Till 2000). Mutations of residues bordering the TK1/TK2 cleft may either activate or inhibit tyrosine kinase activity, and activation may occur by different mechanisms (reviewed by Robertson et al. 2000; Miller et al. 2001; see fig. 3B and additional references cited in the figure legend). As noted above, two of the FGFR2 mutations are located at conserved residues exactly equivalent to those of residues of FGFR3, at which mutations have been identified, and their activating nature has been experimentally verified. The most direct precedent is provided by the K659N mutation, for which the homologous K650N mutation in FGFR3, associated with hypochondroplasia, had been shown to exhibit weak autophosphorylation activity in transfected NIH 3T3 cells (Bellus et al. 2000). The N549H mutation occurs at the position equivalent to that of the N540K mutation, in FGFR3, which is also associated with hypochondroplasia; weak ligand-independent autophosphorylation of an immature receptor protein containing the N540K mutation was observed by Raffioni et al. (1998). No precedent exists to predict the consequences of mutations at the remaining four positions (i.e., E565, K641, G663, and R678); the only specific function ascribed to any of these residues is hydrogen bonding to the adenine ring of ATP, by E565 (Mohammadi et al. 1996). Although it is tempting to speculate (on the basis of the similarity of the consequent phenotypes) that these mutations are also activating, this will require experimental confirmation.
Figure 5.
Position of residues mutated in the TK1 (upper lobe) and TK2 (lower lobe) domains of FGFR2, superimposed on the structure of the tyrosine kinase domain of FGFR1, as determined by Mohammadi et al. (1996), drawn by means of BobScript (Esnouf 1997), based on Raster3D software (Merritt and Murphy 1994). The catalytic (magenta) and activation (yellow) loops, as well as the side chains at which mutations occur (red), are shown.
Comparison of their biological roles suggests that FGFR2 is more critical than FGFR3 for function during embryogenesis. Mice homozygous for null mutations of Fgfr2 die in utero, whereas mice homozygous for null mutations of Fgfr3 are viable (Colvin et al. 1996; Deng et al. 1996; Arman et al. 1998; Xu et al. 1998). It may be significant that the FGFR2 mutations that we have observed to date are associated, when mutated at the equivalent positions in FGFR3, with a mild phenotype (i.e., hypochondroplasia). More severe phenotypes (e.g., thanatophoric dysplasia type 2 and SADDAN syndrome) are observed with the FGFR3 mutations K650E and K650M, respectively (Tavormina et al. 1995, 1999), the equivalents of which have not been observed in FGFR2. The homologous FGFR2 mutations may cause severe—perhaps embryonic lethal—phenotypes.
The reasons for the high mutation rate in FGFR2 remain mysterious. Our study demonstrates that mutations causing craniosynostosis are widely distributed across the protein, yet the majority localize to four amino acids (i.e., S252 and P253, which are in the IgII-IgIII linker, and C278 and C342, which form the disulfide bridge of the IgIIIa/IIIc domain) at key structural points of the FGFR2 protein (fig. 4A). The gain of function conferred by these mutations, combined with both the exclusive paternal origin of mutations and their association with increased paternal age, leads us to speculate that the elevated germline rate of FGFR2 mutations arises as a consequence of positive selection of mutated spermatogonial stem cells (Oldridge et al. 1997). However, FGFR2 mutations may cause different phenotypes and may arise in distinct contexts; for example, the double mutation 755C→T;943G→T (S252L;A315S) leads to syndactyly but not to craniosynostosis (Wilkie et al., in press), whereas somatic mutations of FGFR2 have been identified in atypical acne (Munro and Wilkie 1998) and gastric carcinoma (Jang et al. 2001). Comparison of the FGFR2 mutational spectra in different contexts, including that of sperm, may yield further clues regarding the molecular basis of the elevated germline mutation rate.
Acknowledgments
We are grateful to the families who participated in this work; to the many clinicians—especially K. Becker, C. ffrench-Constant, C. Hill, H. Hughes, D. Pilz, J. Punt, E. Spalding, D. Trump, and N. Waterhouse—who contributed cases to the study; and to D. Moloney, M. Oldridge, and S. Walsh, for their help in earlier phases of the project. E. Y. Jones and R. Esnouf are thanked for their help with figure 5, S. Robertson for commenting on the manuscript. This work was funded by the Ministry of Education in Taiwan (support to S.-h.K.), the Overseas Research Students Awards Scheme (support to S.-h.K.), and the Wellcome Trust (support to A.O.M.W.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for FGFR2 [accession numbers AC009988, AF410480, AF360695, and NM_000141])
- Human Gene Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd/search.html (for mutations in KIT, RET, and MET)
- KinMutBase protein-alignment web site, http://protein.uta.fi/KinMutBase/ProtAlign.html (for mutations in kinase domains)
- Online Mendelian Inheritance in Man (OMIM), http://www3.ncbi.nlm.nih.gov/Omim/searchomim.html (for AS [MIM 101200], CS [MIM 123500], PS [MIM 101600], Beare-Stevenson cutis gyrata syndrome [MIM 123790], and FGFR2 [MIM 176943])
- Single Nucleotide Polymorphism web site, http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=2263 (for single-nucleotide polymorphisms in FGFR2)
References
- Anderson J, Burns HD, Enriquez-Harris P, Wilkie AOM, Heath JK (1998) Apert syndrome mutations in fibroblast growth factor receptor 2 exhibit increased affinity for FGF ligand. Hum Mol Genet 7:1475–1483 [DOI] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P (1998) Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA 95:5082–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Chothia C (1995) Outline structures for the extracellular domains of the fibroblast growth factor receptors. Nat Struct Biol 2:1068–1074 [DOI] [PubMed] [Google Scholar]
- Bellus GA, Gaudenz K, Zackai EH, Clarke LA, Szabo J, Francomano CA, Muenke M (1996) Identical mutations in three different fibroblast growth factor receptor genes in autosomal dominant craniosynostosis syndromes. Nat Genet 14:174–176 [DOI] [PubMed] [Google Scholar]
- Bellus GA, Spector EB, Speiser PW, Weaver CA, Garber AT, Bryke CR, Israel J, Rosengren SS, Webster MK, Donoghue DJ, Francomano CA (2000) Distinct missense mutations of the FGFR3 Lys650 codon modulate receptor kinase activation and the severity of the skeletal dysplasia phenotype. Am J Hum Genet 67:1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun K, Siegel-Bartelt J, Chitayat D, Phillips J, Ray PN (1998) FGFR2 mutation associated with clinical manifestations consistent with Antley-Bixler syndrome. Am J Med Genet 77:219–224 [DOI] [PubMed] [Google Scholar]
- Cohen MM Jr (2001) Jackson-Weiss syndrome. Am J Med Genet 100:325–329 [DOI] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM (1996) Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet 12:390–397 [DOI] [PubMed] [Google Scholar]
- Cornejo-Roldan LR, Roessler E, Muenke M (1999) Analysis of the mutational spectrum of the FGFR2 gene in Pfeiffer syndrome. Hum Genet 104:425–431 [DOI] [PubMed] [Google Scholar]
- Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D (1997) Of worms and men: an evolutionary perspective on the fibroblast growth factor (FGF) and FGF receptor families. J Mol Evol 44:43–56 [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P (1996) Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84:911–921 [DOI] [PubMed] [Google Scholar]
- DeVore DL, Horvitz HR, Stern MJ (1995) An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell 83:611–620 [DOI] [PubMed] [Google Scholar]
- Dionne CA, Crumley G, Bellot F, Kaplow JM, Searfoss G, Ruta M, Burgess WH, Jaye M, Schlessinger J (1990) Cloning and expression of two distinct high-affinity receptors cross-reacting with acidic and basic fibroblast growth factors. EMBO J 9:2685–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elanko N, Sibbring JS, Metcalfe KA, Clayton-Smith J, Donnai D, Temple IK, Wall SA, Wilkie AO (2001) A survey of TWIST for mutations in craniosynostosis reveals a variable length polyglycine tract in asymptomatic individuals. Hum Mutat 18:535–541 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model 15:132–134 [DOI] [PubMed] [Google Scholar]
- Feldman GJ, Ward DE, Lajeunie-Renier E, Saavedra D, Robin NH, Proud V, Robb LJ, Der Kaloustian V, Carey JC, Cohen MM Jr, Cormier V, Munnich A, Zackai EH, Wilkie AOM, Price RA, Muenke M (1997) A novel phenotypic pattern in X-linked inheritance: craniofrontonasal syndrome maps to Xp22. Hum Mol Genet 6:1937–1941 [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM (1996) heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev 10:3003–3017 [DOI] [PubMed] [Google Scholar]
- Glaser RL, Jiang W, Boyadjiev SA, Tran AK, Zachary AA, Johnson D, Walsh S, Oldridge M, Wall SA, Wilkie AOM, Jabs EW (2000) Paternal origin of FGFR2 mutations in sporadic cases of Crouzon and Pfeiffer syndromes. Am J Hum Genet 66:768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Zackai EH, Cohen MM Jr (1999) Not Antley-Bixler syndrome. Am J Med Genet 83:65–66 [PubMed] [Google Scholar]
- Harpaz Y, Chothia C (1994) Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol 238:528–539 [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Till JH (2000) Protein tyrosine kinase structure and function. Annu Rev Biochem 69:373–398 [DOI] [PubMed] [Google Scholar]
- Hunter AGW, Rudd NL (1976) Craniosynostosis. I. Sagittal synostosis: its genetics and associated clinical findings in 214 patients who lacked involvement of the coronal suture(s). Teratology 14:185–194 [DOI] [PubMed] [Google Scholar]
- Ingersoll RG, Paznekas WA, Tran AK, Scott AF, Jiang G, Jabs EW. Fibroblast growth factor receptor 2 (FGFR2): genomic sequence and variations. Cytogenet Cell Genet (in press) [DOI] [PubMed] [Google Scholar]
- Itoh H, Hattori Y, Sakamoto H, Ishii H, Kishi T, Sasaki H, Yoshida T, Koono M, Sugimura T, Terada M (1994) Preferential alternative splicing in cancer generates a K-sam messenger RNA with higher transforming activity. Cancer Res 54:3237–3241 [PubMed] [Google Scholar]
- Iwashita T, Kato M, Murakami H, Asai N, Ishiguro Y, Ito S, Iwata Y, Kawai K, Asai M, Kurokawa K, Kajita H, Takahashi M (1999) Biological and biochemical properties of Ret with kinase domain mutations identified in multiple endocrine neoplasia type 2B and familial medullary thyroid carcinoma. Oncogene 18:3919–3922 [DOI] [PubMed] [Google Scholar]
- Iwashita T, Kurokawa K, Qiao S, Murakami H, Asai N, Kawai K, Hashimoto M, Watanabe T, Ichihara M, Takahashi M (2001) Functional analysis of RET with Hirschsprung mutations affecting its kinase domain. Gastroenterology 121:24–33 [DOI] [PubMed] [Google Scholar]
- Jabs EW, Li X, Scott AF, Meyers G, Chen W, Eccles M, Mao JI, Charnas LR, Jackson CE, Jaye M (1994) Jackson-Weiss and Crouzon syndromes are allelic with mutations in fibroblast growth factor receptor 2. Nat Genet 8:275–279 [DOI] [PubMed] [Google Scholar]
- Jang JH, Shin KH, Park JG (2001) Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res 61:3541–3543 [PubMed] [Google Scholar]
- Jeffers M, Schmidt L, Nakaigawa N, Webb CP, Weirich G, Kishida T, Zbar B, Vande Woude GF (1997) Activating mutations for the Met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci USA 94:11445–11450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Horsley SW, Moloney DM, Oldridge M, Twigg SRF, Walsh S, Barrow M, Njølstad PR, Kunz J, Ashworth GJ, Wall SA, Kearney L, Wilkie AOM (1998) A comprehensive screen for TWIST mutations in patients with craniosynostosis identifies a new microdeletion syndrome of chromosome band 7p21.1. Am J Hum Genet 63:1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Wall SA, Mann S, Wilkie AOM (2000) A novel mutation, Ala315Ser, in FGFR2: gene-environment interaction leading to craniosynostosis? Eur J Hum Genet 8:571–577 [DOI] [PubMed] [Google Scholar]
- Johnson DE, Williams LT (1993) Structural and functional diversity in the FGF receptor multigene family. In: Vande Woude GF, Klein G (eds) Advances in cancer research. Vol 60. Academic Press, San Diego, pp 1–41 [DOI] [PubMed] [Google Scholar]
- Katoh M, Hattori Y, Sasaki H, Tanaka M, Sugano K, Yazaki Y, Sugimura T, Terada M (1992) K-sam gene encodes secreted as well as transmembrane receptor tyrosine kinase. Proc Natl Acad Sci USA 89:2960–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepelová A, Baxová A, Calda P, Plavka R, Kapras J (1998) FGFR2 gene mutation (Tyr375Cys) in a new case of Beare-Stevenson syndrome. Am J Med Genet 76:362–364 [PubMed] [Google Scholar]
- Lajeunie E, Le Merrer M, Bonaïti-Pellie C, Marchac D, Renier D (1995a) Genetic study of nonsyndromic coronal craniosynostosis. Am J Med Genet 55:500–504 [DOI] [PubMed] [Google Scholar]
- Lajeunie E, Ma HW, Bonaventure J, Munnich A, Le Merrer M, Renier D (1995b) FGFR2 mutations in Pfeiffer syndrome. Nat Genet 9:108 [DOI] [PubMed] [Google Scholar]
- Longley BJ Jr, Metcalfe DD, Tharp M, Wang X, Tyrrell L, Lu S-Z, Heitjan D, Ma Y (1999) Activating and dominant inactivating c-KIT catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc Natl Acad Sci USA 96:1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Murphy MEP (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr D50:869–873 [DOI] [PubMed] [Google Scholar]
- Meyers GA, Orlow SJ, Munro IR, Przylepa KA, Jabs EW (1995) Fibroblast growth factor receptor 3 (FGFR3) transmembrane mutation in Crouzon syndrome with acanthosis nigricans. Nat Genet 11:462–464 [DOI] [PubMed] [Google Scholar]
- Miki T, Bottaro DP, Fleming TP, Smith CL, Burgess WH, Chan AM-L, Aaronson SA (1992) Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci USA 89:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Ginalski K, Lesyng B, Nakaigawa N, Schmidt L, Zbar B (2001) Structural basis of oncogenic activation caused by point mutations in the kinase domain of the MET proto-oncogene: modeling studies. Proteins 44:32–43 [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Schlessinger J, Hubbard SR (1996) Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell 86:577–587 [DOI] [PubMed] [Google Scholar]
- Moloney DM, Slaney SF, Oldridge M, Wall SA, Sahlin P, Stenman G, Wilkie AOM (1996) Exclusive paternal origin of new mutations in Apert syndrome. Nat Genet 13:48–53 [DOI] [PubMed] [Google Scholar]
- Moloney DM, Wall SA, Ashworth GJ, Oldridge M, Glass IA, Francomano CA, Muenke M, Wilkie AOM (1997) Prevalence of Pro250Arg mutation of fibroblast growth factor receptor 3 in coronal craniosynostosis. Lancet 349:1059–1062 [DOI] [PubMed] [Google Scholar]
- Mortier G, Nuytinck L, Craen M, Renard J-P, Leroy JG, De Paepe A (2000) Clinical and radiographic features of a family with hypochondroplasia owing to a novel Asn540Ser mutation in the fibroblast growth factor receptor 3 gene. J Med Genet 37:220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S, Winter RM (1994) A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat Genet 8:269–274 [DOI] [PubMed] [Google Scholar]
- Muenke M, Wilkie AOM (2001) Craniosynostosis syndromes. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 6117–6146 [Google Scholar]
- Munro CS, Wilkie AOM (1998) Epidermal mosaicism producing localised acne: somatic mutation in FGFR2. Lancet 352:704–705 [DOI] [PubMed] [Google Scholar]
- Neilson KM, Friesel RE (1995) Constitutive activation of fibroblast growth factor receptor-2 by a point mutation associated with Crouzon syndrome. J Biol Chem 270:26037–26040 [DOI] [PubMed] [Google Scholar]
- Oldridge M, Lunt PW, Zackai EH, McDonald-McGinn DM, Muenke M, Moloney DM, Twigg SRF, Heath JK, Howard TD, Hoganson G, Gagnon DM, Jabs EW, Wilkie AOM (1997) Genotype-phenotype correlation for nucleotide substitutions in the IgII-IgIII linker of FGFR2. Hum Mol Genet 6:137–143 [DOI] [PubMed] [Google Scholar]
- Oldridge M, Wilkie AOM, Slaney SF, Poole MD, Pulleyn LJ, Rutland P, Hockley AD, Wake MJC, Goldin JH, Winter RM, Reardon W, Malcolm S (1995) Mutations in the third immunoglobulin domain of the fibroblast growth factor receptor-2 gene in Crouzon syndrome. Hum Mol Genet 4:1077–1082 [DOI] [PubMed] [Google Scholar]
- Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SRF, Johnson D, Wall SA, Jiang W, Theda C, Jabs EW, Wilkie AOM (1999) De novo Alu element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. Am J Hum Genet 64:446–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M (1996) Receptor specificity of the fibroblast growth factor family. J Biol Chem 271:15292–15297 [DOI] [PubMed] [Google Scholar]
- Pelet A, Geneste O, Edery P, Pasini A, Chappuis S, Attié T, Munnich A, Lenoir G, Lyonnet S, Billaud M (1998) Various mechanisms cause RET-mediated signaling defects in Hirschsprung's disease. J Clin Invest 101:1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL (2000) Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407:1029–1034 [DOI] [PubMed] [Google Scholar]
- Pfeiffer RA (1964) Dominant erbliche Akrocephalosyndaktylie. Z Kinderheilkd 90:301–320 [PubMed] [Google Scholar]
- Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M (1999) Structural basis for FGF receptor dimerization and activation. Cell 98:641–650 [DOI] [PubMed] [Google Scholar]
- Przylepa KA, Moloney DM, Wall SA, Gagnon D, Hoganson G, Yin M, Wilkie AOM, Jabs EW (1998) A FGFR2 mutation causing type 2 Pfeiffer syndrome. J Craniofac Genet Dev Biol 18:6–7 [Google Scholar]
- Przylepa KA, Paznekas W, Zhang M, Golabi M, Bias W, Bamshad MJ, Carey JC, Hall BD, Stevenson R, Orlow SJ, Cohen MM Jr, Jabs EW (1996) Fibroblast growth factor receptor 2 mutations in Beare-Stevenson cutis gyrata syndrome. Nat Genet 13:492–494 [DOI] [PubMed] [Google Scholar]
- Pulleyn LJ, Reardon W, Wilkes D, Rutland P, Jones BM, Hayward R, Hall CM, Brueton L, Chun N, Lammer E, Malcolm S, Winter RM (1996) Spectrum of craniosynostosis phenotypes associated with novel mutations at the fibroblast growth factor receptor 2 locus. Eur J Hum Genet 4:283–291 [DOI] [PubMed] [Google Scholar]
- Raffioni S, Zhu Y-Z, Bradshaw RA, Thompson LM (1998) Effect of transmembrane and kinase domain mutations on fibroblast growth factor receptor 3 chimera signaling in PC12 cells. J Biol Chem 273:35250–35259 [DOI] [PubMed] [Google Scholar]
- Reardon W, Smith A, Honour JW, Hindmarsh P, Das D, Rumsby G, Nelson I, Malcolm S, Ades L, Sillence D, Kumar D, DeLozier-Blanchet C, McKee S, Kelly T, McKeehan WL, Baraitser M, Winter RM (2000) Evidence for digenic inheritance in some cases of Antley-Bixler syndrome? J Med Genet 37:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Winter RM, Rutland P, Pulleyn LJ, Jones BM, Malcolm S (1994) Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat Genet 8:98–103 [DOI] [PubMed] [Google Scholar]
- Robertson SC, Meyer AN, Hart KC, Galvin BD, Webster MK, Donoghue DJ (1998) Activating mutations in the extracellular domain of the fibroblast growth factor receptor 2 function by disruption of the disulfide bond in the third immunoglobulin-like domain. Proc Natl Acad Sci USA 95:4567–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SC, Tynan JA, Donoghue DJ (2000) RTK mutations and human syndromes: when good receptors turn bad. Trends Genet 16:265–271 [DOI] [PubMed] [Google Scholar]
- Rutland P, Pulleyn LJ, Reardon W, Baraitser M, Hayward R, Jones B, Malcolm S, Winter RM, Oldridge M, Slaney SF, Poole MD, Wilkie AOM (1995) Identical mutations in the FGFR2 gene cause both Pfeiffer and Crouzon syndrome phenotypes. Nat Genet 9:173–176 [DOI] [PubMed] [Google Scholar]
- Schell U, Hehr A, Feldman GJ, Robin NH, Zackai EH, de Die-Smulders C, Viskochil DH, Stewart JM, Wolff G, Ohashi H, Price RA, Cohen MM Jr, Muenke M (1995) Mutations in FGFR1 and FGFR2 cause familial and sporadic Pfeiffer syndrome. Hum Mol Genet 4:323–328 [DOI] [PubMed] [Google Scholar]
- Slaney SF, Oldridge M, Hurst JA, Morriss-Kay GM, Hall CM, Poole MD, Wilkie AOM (1996) Differential effects of FGFR2 mutations on syndactyly and cleft palate in Apert syndrome. Am J Hum Genet 58:923–932 [PMC free article] [PubMed] [Google Scholar]
- Stauber DJ, DiGabriele AD, Hendrickson WA (2000) Structural interactions of fibroblast growth factor receptor with its ligands. Proc Natl Acad Sci USA 97:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger D, Reinhartz T, Unsöld R, Müller U (1996) FGFR2 mutation in clinically nonclassifiable autosomal dominant craniosynostosis with pronounced phenotypic variation. Am J Med Genet 66:81–86 [DOI] [PubMed] [Google Scholar]
- Stenberg KAE, Riikonen PT, Vihinen M (1999) KinMutBase, a database of human disease-causing protein kinase mutations. Nucleic Acids Res 27:362–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Bellus GA, Webster MK, Bamshad MJ, Fraley AE, McIntosh I, Szabo J, Jiang W, Jabs EW, Wilcox WR, Wasmuth JJ, Donoghue DJ, Thompson LM, Francomano CA (1999) A novel skeletal dysplasia with developmental delay and acanthosis nigricans is caused by a Lys650Met mutation in the fibroblast growth factor receptor 3 gene. Am J Hum Genet 64:722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Shiang R, Thompson LM, Zhu Y-Z, Wilkin DJ, Lachman RS, Wilcox WR, Rimoin DL, Cohn DH, Wasmuth JJ (1995) Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet 9:321–328 [DOI] [PubMed] [Google Scholar]
- Tsai F-J, Yang CF, Wu JY, Tsai CH (2000) Mutation analysis of Crouzon syndrome. Eur J Hum Genet 8 Suppl 1:120 [Google Scholar]
- Wilkie AOM (1997) Craniosynostosis: genes and mechanisms. Hum Mol Genet 6:1647–1656 [DOI] [PubMed] [Google Scholar]
- Wilkie AOM, Patey SJ, Kan S-h, van den Ouweland AMW, Hamel BCJ. FGFs, their receptors, and human limb malformations: clinical and molecular correlations. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Wilkie AOM, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, Hayward RD, David DJ, Pulleyn LJ, Rutland P, Malcolm S, Winter RM, Reardon W (1995) Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet 9:165–172 [DOI] [PubMed] [Google Scholar]
- Wong L-JC, Chen T-J, Dai P, Bird L, Muenke M (2001) Novel SNP at the common primer site of exon IIIa of FGFR2 gene causes error in molecular diagnosis of craniosynostosis syndrome. Am J Med Genet 102:282–285 [DOI] [PubMed] [Google Scholar]
- Xiao W, Oefner PJ (2001) Denaturing high-performance liquid chromatography: a review. Hum Mutat 17:439–474 [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C (1998) Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125:753–765 [DOI] [PubMed] [Google Scholar]
- Yu K, Herr AB, Waksman G, Ornitz DM (2000) Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci USA 97:14536–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gorry MC, Post JC, Ehrlich GD (1999) Genomic organization of the human fibroblast growth factor receptor 2 (FGFR2) gene and comparative analysis of the human FGFR gene family. Gene 230:69–79 [DOI] [PubMed] [Google Scholar]