Abstract
Vitamin A (all-trans retinol) and its active metabolites, collectively called retinoids, exert potent effects on stem cell differentiation and thus, the formation of the entire organism, in part via the modulation of the epigenome. All-trans retinoic acid (RA), through binding to the retinoic acid receptors (RARs), alters interactions of the RARs with various protein components of the transcription complex at numerous genes in stem cells, and some of these protein components of the transcription complex then either place or remove epigenetic marks on histones or on DNA, altering chromatin structure and leading to an exit from the self-renewing, pluripotent stem cell state. Different epigenetic mechanisms, i.e. first, primarily H3K27me3 marks and then DNA methylation, may be employed by embryonic stem cells and other stem cells for control of early vs. late stages of cell differentiation. Creating these stable epigenetic changes requires the actions of many molecules, including tet1, polycomb protein complexes (PRCs), miRNAs, DNA methyltransferases (DNMTs), and telomerase reverse transcriptase. A more complete understanding of retinoid-dependent stem cell differentiation should reward us with new insights into the failure to maintain a differentiated state that is an essential part of neoplastic cell transformation and cancer.
Keywords: retinoic acid, differentiation, review, histone methylation, DNA methylation, Polycomb/PRC
1.1.1 Retinoids can act as Epigenetic Modifiers during Cell Differentiation
Cell differentiation, the process by which a less specialized cell, such as a stem cell, becomes more specialized in terms of its functions in a multi-cellular organism, can be viewed as a process in which epigenetic changes result in alterations in the genes expressed by the cell as it becomes more specialized, i.e. more differentiated. Thus, stem cell differentiation is a process that involves a series of epigenetic changes, defined as modifications to the genome that do not involve a change in the nucleotide sequence. Vitamin A (all-trans retinol) and its active metabolites, collectively called retinoids, exert potent effects on stem cell differentiation and thus, the formation of the entire organism, in part via the modulation of the epigenome. These epigenetic changes, many of which are induced in stem cells by an active biological metabolite of vitamin A, all-trans retinoic acid (RA), can be long-term, stable changes. Such changes include DNA methylation and histone modifications, including methylation and acetylation of specific amino acids in the various histones that surround DNA in the nucleus. The goal of this review is to address the question of how the RA signaling pathway interacts with the enzymes that generate epigenetic modifications in stem cells, and the focus is on more recent publications in this area.
1.1.2 Retinoids act as Ligand-activated Transcription Factors to Modulate Gene Expression
Two types of transcription factors, the retinoic acid receptors (multiple isoforms of RARα, β, and γ) and the retinoid X receptors (multiple isoforms of RXRα, β, and γ), members of the nuclear receptor family of proteins, mediate most of the actions of RA in stem cells [1]. In the nucleus, the agonist RA binds to RARα, β, and/or γ. When RA is present, this RXR/RAR heterodimer complex, bound to DNA, activates transcription of RA primary response genes, one of the first steps in the RA associated differentiation process [1]. Many “immediate early” genes, such as the transcription factor Hoxa1 [2], are “primary response” genes and possess enhancers containing an RARE (retinoic acid DNA response element) to which the RXR/RAR heterodimer can bind. In some cell types, RA addition also triggers persistent mitogen activated protein kinase signaling [3] to induce differentiation.
Additional proteins, including co-activators and co-repressors, bind to the RXR/RAR complex, modulating the sensitivity of cells to RA’s differentiation-inducing effects and integrating the effects of other signaling pathways. The RXR/RAR heterodimer binds co-activator proteins, such as Ncoa3 (GCID: GC20P046130, also known as pCip; p160; KAT13b; Actr; Aib1; Rac3; Src3; Tram1), and recruits these proteins to RA “primary target“ gene enhancers. Ncoa3 and some other co-activators possess histone acetyltransferase activity. Co-activator recruitment is dependent on the appropriate RAR binding at the RARE of the target gene [4-9]. The nuclear receptor interaction domains (NRID) of co-activator proteins contain three highly conserved motifs with a consensus amino acid sequence LXXLL; such LXXLL motifs mediate direct interaction with the AF-2 domain of RARs upon ligand binding [10]. Ncoa3 ubiquitination is also controlled by RA, and Ncoa3 degradation is involved both in the transcription of RA target genes and in the growth inhibitory actions of RA [11]. Binding of Ncoa3 then allows p300 (GC22P041487, KAT3B) and the related protein CBP (GC03P020081, KAT2B, P/CAF, GCN5, CREB Binding protein) to bind at the transcriptional activation complex. The p160 co-activators propagate the activating signal through at least two activation domains, AD1 and AD2, which recruit secondary co-activator proteins. AD1 binds CBP or p300 [12], whereas AD2 recruits the co-activator associated arginine methyltransferase 1 (CARM1) [13]. Both Ncoa3 and p300 and/or CBP are involved in RA transcriptional activation of target genes in stem cells [5]. Thus, retinoids, via their receptors and interacting proteins, initiate the transcription of primary response genes.
1.1.3 Retinoids also Act by Effecting Changes in Epigenetic Marks on Histones, Creating a Heritable Change in Chromatin Responsiveness
Some examples of the epigenetic changes that occur in response to RA and their relationship to cell differentiation are highlighted here. PHF8 (GC0XM053979, also called ZNF422, JHDM1F), a histone lysine demethylase that preferentially acts on histones in the monomethyl or dimethyl states, is involved in the response of stem cells to RA. PHF8, which requires Fe(2+) ion, 2-oxoglutarate, and oxygen for its catalytic activity, has been reported to act as an RARα co-activator, and PHF8 knockdown in P19 teratocarcinoma stem cells results in reduced RA-induced neural differentiation [14]. Additionally, in acute promyelocytic leukemia sensitivity to RA, a drug used widely in treatment of this cancer, depends on the enzymatic activity of PHF8; forced expression of PHF8 can resensitize RA resistant acute promyelocytic leukemia cells to RA, while reduced expression of PHF8 can result in RA resistance [15].
Conversely, SETD6 (GCID:GC16P058549), a lysine methyltransferase, places monomethyl groups on substrates in ES cells. RA treatment rapidly removes these monomethyl marks, and similarly, a genetic reduction in SETD6 expression in ES cells results in differentiation and loss of self-renewal ability [16].
CARM1 (GCID: GC19P010982; also known as PRMT4) is a protein arginine N-methyltransferase that catalyzes the transfer of a methyl group from S-adenosyl-L-methionine to the side chain nitrogens of arginine residues to form methylated arginine derivatives and S-adenosyl-L-homocysteine. Depletion of CARM1 in embryonic stem (ES) cells results in loss of pluripotency and induction of differentiation [17]. CARM1 acts as a “secondary” co-activator and enhances transcription, but only in the presence of Ncoa3 or its co-activator family members [13]. Importantly, knockdown of Ncoa3 (see above) not only compromises the expression of pluripotency markers but also impairs the differentiation of ES cells. Ncoa3 binds to the Nanog (a stem cell marker) promoter and recruits the histone arginine methyltransferase CARM1 to activate Nanog expression [18]. Importantly, the ability of Ncoa3 to interact with p300 and CBP depends on the methylation state of CARM1-dependent methylation sites located in a glutamine-rich region within the carboxy terminus of Ncoa3 [19].
ASH2L (GC08P03763) is a core protein in the mixed lineage leukemia (MLL) methyltransferase complex; the MLL complex is not able to trimethylate histone H3K4 without ASH2L. Trimethylation of H3K4 is generally associated with open chromatin. Knockdown of ASH2L results in reduced H3K4 methylation; high H3K9 methylation, an epigenetic mark associated with silenced chromatin; and loss of ES cell pluripotency [20]. We found that ASH2L was recruited to the Hoxa1 and Hoxb1 genes and enhancer regions upon RA treatment of ES cells [6].
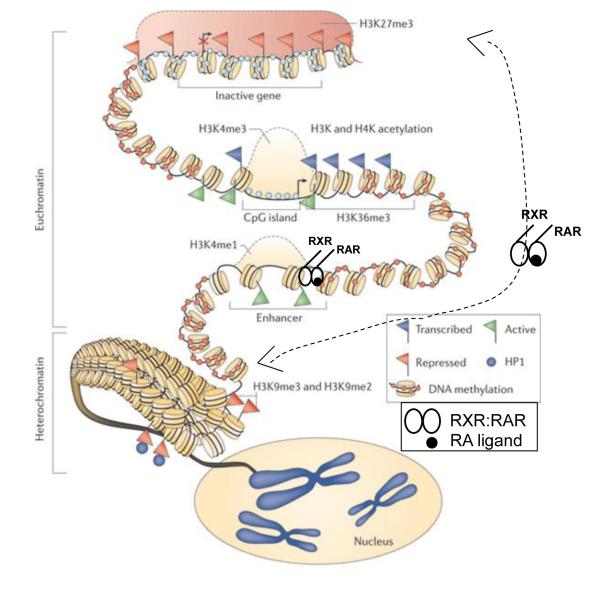
Thus, numerous enzymes that alter the modifications on histones are involved in transcriptional activation of specific genes in stem cells, and many of these enzymes are modulated by RA treatment of stem cells. These data suggest that RA, through binding to the RARs, alters interactions of the RARs with various protein components of the transcription complex at numerous genes in stem cells, and some of these protein components of the transcription complex then either place or remove epigenetic marks on histones, altering chromatin structure and leading to an exit from the self-renewing, pluripotent stem cell state (Fig. 1) (for rev. Gudas and Wagner [1]).
Figure 1.
Interaction of RA Signaling System with Various Epigenetic Modifications. (modified from Baylin et al [44]).
The exit from the self-renewing state of ES cells is also accompanied by an induction of the silencing-associated histone H3 Lys 9 dimethylation and trimethylation (H3K9Me2/Me3) marks. The protein Heterochromatin Protein 1 (HP1) (CBX5, HP1α GC12M054624, KMT1A) can exert long term gene silencing in concert with the enzyme Suv39H1; methylation of the histone H3 tail by Suv39H1 was found to repress transcription in an HP1-dependent manner [21, 22]. Conversely, the H3K9me2 and H3K9me3 demethylase enzymes JMJD1a (KDM3a, GC02P086667) and JMJD2c (KDM4c, GC0P006720) are positively regulated in ES cells and knockdown of these enzymes results in ES cell differentiation [23]. JMJD2c prevents HP1 from binding to the Nanog promoter. Thus, an increase in H3K9me3 marks and HP1 binding is strongly associated with the RA-associated transition to the differentiated state from the stem cell state (Fig. 1).
1.2 Polycomb Complexes are Important Mediators of the Epigenetic Changes Induced by RA
Polycomb group proteins control the transition from stem cells to differentiated cells [24, 25]. Polycomb group-repressive complexes (PRC1 and PRC2) mark many genes for silencing in embryonic stem (ES) cells. Moreover, the protein Ezh2 is a component of the PRC2 complex that places the H3K27me3 epigenetic mark recognized by PRC1 factors, which in turn bring Ring1 to mono-ubiquitinylate histone H2A lysine 119 (H2Aub) [26]. The PRC2 complex somehow senses the chromatin environment and local chromatin compaction occurs before the H3K27me3 mark is added [27]. However, Ezh2 can, when phosphorylated, also act as a transcriptional activator [28]. We have shown that, although complete erasure of the polycomb repressive mark H3K27me3 is not necessary to initiate Hoxa1 transcription in ES cells, RA/RARγ signaling is necessary to erase H3K27me3 from activated Hox genes during embryonic stem cell differentiation [6, 7]. Additionally, RARγ is not required to establish the bivalent character (DNA regions decorated with both active (H3K4me3) and repressive (H3K27me3) marks) of Hox clusters [6, 7]. The enzyme tet1, which carries out DNA demethylation, is necessary for chromatin binding of the PRC2 complex; greater than 95% of PRC2 targets in ES cells are tet1 targets, and tet1 knockdown leads to a loss of PRC2 recruitment [29].
We have also shown that both the Nr2F1 (Coup-TF1) and the Hoxa5 gene are transcriptionally activated by RA in WT ES cells and display increased levels of the permissive H3K9/K14ac and tri-methylated histone H3 lysine 4 epigenetic marks in response to RA. However, while in response to RA the PRC2 proteins and H3K27me3 marks are greatly decreased at the Hoxa1 and Hoxa5 promoters, these marks are initially increased at the Nr2F1 promoter. Functional depletion of the essential PRC2 protein Suz12 enhanced the RA-associated transcription of Nr2F1, Nr2F2, Meis1, Sox9, and BMP2, but had no effect on the Hoxa5, Hoxa1, Cyp26a1, Cyp26b1 and RARβ2 transcript levels in wild type ES cells. Our data thus indicate that upon RA addition to stem cells, PRC2 recruitment attenuates the RA-associated transcriptional activation of a subset of genes, while at other genes the PRC2 complex is removed after RA addition, resulting in maximal RA-associated transcriptional activation. Such a mechanism would permit the fine-tuning of transcriptional networks during the differentiation of ES cells [30].
Recent, high-throughput, genome-wide measurements show that in ES cells promoters active in early developmental stages are generally CG rich and utilize H3K27me3 marks, generated by the PRCs, to silence genes in nonexpressing lineages during differentiation. In fact, there is a change in the protein composition of the PRC1 complex when ES cells leave the self-renewing state and start to differentiate [31]. This composition change allows PRC1 to target different genes in stem vs. differentiating cells. In contrast, promoters of genes generally expressed at later stages of differentiation/development utilize increased DNA methylation to establish and maintain gene silencing [32]. Thus, different epigenetic mechanisms, i.e. first, primarily H3k27me3 marks and later, DNA methylation, may be employed by ES and other stem cells for control of early vs. late stages of cell differentiation (Fig. 1).
The type of DNA methylation is also an important determinant of gene expression during ES cell differentiation. For example, upon RA treatment of NT2 human embryonyl carcinoma stem cells the active, anterior part of the Hoxa gene cluster becomes enriched in 5-hydroxymethylcytosine (5hmC), following closely the colinear activation of the gene cluster. This increase is paralleled by a reduction in 5-methylcytosine (5mC) marks. Depletion of the 5hmC generating dioxygenase enzyme Tet2 impairs the maintenance of Hoxa transcriptional activation and partially restores 5mC levels [33].
1.3 Telomerase Activity and DNA Methylation Play an Essential Role in Stabilizing Stem Cell Differentiation
The question of how the differentiation state is stably maintained is also a critical one, as loss of the differentiated phenotype can result in carcinogenesis and the generation of cancer stem cells or cancer initiating cells. ES cells that lack the catalytic subunit of the telomerase protein, telomerase reverse transcriptase (TERT, GC05M001253), have short telomeres and express very high levels of the homeobox transcription factor Nanog (GC12P007940) [34]; this high Nanog transcript expression is associated with a lower level of H3K27me3 epigenetic marks at the Nanog promoter and global genomic hypomethylation of DNA at CpGs. Addition of RA to the Tert-/- ES cells caused a reduction in Nanog transcript levels, but a few days later Nanog levels increased again in the Tert-/- but not in the wild type (WT) ES cells [34]. However, the RA-induced differentiation state could be stabilized in the Tert-/- cells by reintroduction of exogenous Tert into the cells, by shRNA knockdown of Nanog, or by forced expression of DNMT3b, a DNA methyltransferase [34]. Similarly, ES cells that lack all three DNMTs, DNMT1, DNMT3a, and DNMT3b, can differentiate, but when replated under pluripotency promoting culture conditions, the ES cells that lack the three DNMTs quickly revert back to an undifferentiated state while the WT do not [35]. Thus, DNA methylation appears to be critical for stabilizing the differentiation state, though it is not required for initial activation of stem cell differentiation and exit from the pluripotent, self-renewing state. How short telomeres result in global DNA hypomethylation is not yet understood.
Treatment of cells with DNA damaging agents and cellular oxidative stress can also recruit DNA methyltransferase 1 (DNMT1) to damaged chromatin. DNMT1 and DNMT3b then interact with members of Polycomb Repressive Complex 4; these protein complexes migrate from non-GC-rich to GC-rich areas, sometimes leading to heritable silencing of CpG island-containing promoters via increased DNA methylation [36, 37]. Such actions of DNMTs thus link alterations in DNA methylation state with aberrant differentiation and cancer development.
1.4 MicroRNAs and DNA Methylation Play Key Roles in Controlling Retinoid-dependent Stem Cell Differentiation
MicroRNAs (miRNAs), small non-coding RNAs that regulate gene expression post-transcriptionally, are involved in the RA-associated differentiation of various types of cells and in cancer development. For example, two miRNAs upregulated during the differentiation of neuroblastoma cells--miR-9 and miR-103-- directly target the coding sequence and 3′ untranslated region of the transcription factor ID2 mRNA, respectively. The two miRNAs show an inverse correlation with ID2 expression during neuroblastoma cell differentiation induced by retinoic acid [38]. In addition, microRNAs miR-29a and miR-142-3p promote the phorbol 12-myristate 13-acetate-induced monocytic and RA-induced granulocytic differentiation of HL-60, THP-1, or NB4 cells. A decrease in miR-29a and miR-142-3p levels and their target protein levels was also observed in blasts from acute myeloid leukemia, suggesting that these microRNAs play a role in the regulation of the RA-associated differentiation of stem cells along the myeloid lineage [39]. Importantly, in human ES cells, CBP/p300 acetylates the protein p53 at lysine 373 in response to RA; this stabilizes p53. p53 then activates expression of miR-34a and miR-145, which repress the stem cell factors OCT4, KLF4, LIN28A, and SOX2 and reinforce the differentiation initiated by RA [40].
A recent report has shown that toll-like receptor TLR3 stimulation upregulates four miRNAs, miR-2b, -29c, -148b, and -152, in prostate and breast cancer lines. miR-148b and miR-152 target DNA methyltransferase 1 [41], and miR-29b and -29c target DNA methyltransferases 3A and 3B and silence these genes [42]. This results in the partial demethylation and re-expression of the RARβ2 gene in these tumor cells and the authors also present tumor xenograft data that the TLR3 agonist poly(I:C) and RA together inhibit prostate tumor growth [43]. These data thus link RA actions, DNA methylation, and expression of specific microRNAs.
Conclusions
Recent research has demonstrated that retinoids control gene expression by several inter-related mechanisms. Beyond regulating transcription by a classic ligand-dependent transcriptional activation mechanism, retinoids promote epigenetic changes both by influencing histone modifications and by influencing the methylation state of the DNA. These epigenetic changes can create heritable changes in gene expression and influence the ability of stem cells to respond to both retinoids and to other different signaling pathways. Creating these epigenetic changes requires the actions of many molecules, including miRNAs, DNMTs, and telomerase reverse transcriptase. A more complete understanding of retinoid-dependent cell differentiation will require us to dissect the complex interactions of all of these proteins; this effort should reward us with greater knowledge of differentiation and with new insights into the failure to maintain a differentiated state that is an essential part of neoplastic cell transformation and cancer.
Highlights.
Vitamin A and its metabolites (retinoids) exert effects on stem cell differentiation
RARs interact with transcription in stem cells, altering chromatin structure
Histone marks and then DNA methylation control stages of cell differentiation
Acknowledgments
Dr. Gudas was supported by NIH grants R01-CA043796, R01-DE010389, and R21-AA021484 while preparing this review. I thank Dr. John Wagner for critically reading this review, and Tamara Weissman for editorial assistance.
Abbreviations
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- CARM
co-activator associated arginine methyltransferase 1
- DNMT
DNA methyltransferase
- ES
embryonic stem
- miRNA
micro RNA
- MLL
mixed lineage leukemia
- PRC
Polycomb group-repressive complex
- RA
all-trans retinoic acid
- RAR
retinoic acid receptor
- RARE
retinoic acid DNA response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226:322–30. doi: 10.1002/jcp.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].LaRosa GJ, Gudas LJ. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol Cell Biol. 1988;8:3906–17. doi: 10.1128/mcb.8.9.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Congleton J, MacDonald R, Yen A. Src inhibitors, PP2 and dasatinib, increase retinoic acid-induced association of Lyn and c-Raf (S259) and enhance MAPK-dependent differentiation of myeloid leukemia cells. Leukemia. 2012;26:1180–8. doi: 10.1038/leu.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gillespie RF, Gudas LJ. Retinoic acid receptor isotype specificity in F9 teratocarcinoma stem cells results from the differential recruitment of coregulators to retinoic response elements. J Biol Chem. 2007;282:33421–34. doi: 10.1074/jbc.M704845200. [DOI] [PubMed] [Google Scholar]
- [5].Gillespie RF, Gudas LJ. Retinoid regulated association of transcriptional co-regulators and the polycomb group protein SUZ12 with the retinoic acid response elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 embryonal carcinoma cells. J Mol Biol. 2007;372:298–316. doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kashyap V, Gudas LJ, Brenet F, Funk P, Viale A, Scandura JM. Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J Biol Chem. 2011;286:3250–60. doi: 10.1074/jbc.M110.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kashyap V, Laursen KB, Brenet F, Viale AJ, Scandura JM, Gudas LJ. RARγ is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells. J Cell Sci. 2013;126:999–1008. doi: 10.1242/jcs.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hedblom A, Laursen KB, Miftakhova R, Sarwar M, Anagnostaki L, Bredberg A, et al. CDK1 interacts with RARγ and plays an important role in treatment response of acute myeloid leukemia. Cell Cycle. 2013;12:1251–66. doi: 10.4161/cc.24313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Urvalek AM, Laursen KL, Gudas LJ. The Roles of Retinoic Acid and Retinoic Acid Receptors in Inducing Epigenetic Changes. 2013 doi: 10.1007/978-94-017-9050-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–68. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ferry C, Gaouar S, Fischer B, Boeglin M, Paul N, Samarut E, et al. Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proc Natl Acad Sci U S A. 2011;108:20603–8. doi: 10.1073/pnas.1102572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–14. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- [13].Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- [14].Qiu J, Shi G, Jia Y, Li J, Wu M, Dong S, et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–18. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- [15].Arteaga MF, Mikesch JH, Qiu J, Christensen J, Helin K, Kogan SC, et al. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013;23:376–89. doi: 10.1016/j.ccr.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Binda O, Sevilla A, LeRoy G, Lemischka IR, Garcia BA, Richard S. SETD6 monomethylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics. 2013;8:177–83. doi: 10.4161/epi.23416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu Q, Bruce AW, Jedrusik A, Ellis PD, Andrews RM, Langford CF, et al. CARM1 is Required in Embryonic Stem Cells to Maintain Pluripotency and Resist Differentiation. Stem Cells. 2009;27:2637–45. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wu Z, Yang M, Liu H, Guo H, Wang Y, Cheng H, et al. Role of nuclear receptor coactivator 3 (Ncoa3) in pluripotency maintenance. J Biol Chem. 2012;287:38295–304. doi: 10.1074/jbc.M112.373092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–34. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W, et al. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem. 2013;288:5039–48. doi: 10.1074/jbc.M112.424515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Loyola A, LeRoy G, Wang YH, Reinberg D. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 2001;15:2837–51. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nestorov P, Tardat M, Peters AH. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr Top Dev Biol. 2013;104:243–91. doi: 10.1016/B978-0-12-416027-9.00008-5. [DOI] [PubMed] [Google Scholar]
- [23].Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- [25].Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–7. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- [27].Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–5. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- [28].Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–93. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Laursen KB, Mongan NP, Zhuang Y, Ng MM, Benoit YD, Gudas LJ. Polycomb recruitment attenuates retinoic acid-induced transcription of the bivalent NR2F1 gene. Nucleic Acids Res. 2013;41:6430–43. doi: 10.1093/nar/gkt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- [32].Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–48. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bocker MT, Tuorto F, Raddatz G, Musch T, Yang FC, Xu M, et al. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nat Commun. 2012;3:818. doi: 10.1038/ncomms1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pucci F, Gardano L, Harrington L. Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell. 2013;12:479–86. doi: 10.1016/j.stem.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schmidt CS, Bultmann S, Meilinger D, Zacher B, Tresch A, Maier KC, et al. Global DNA hypomethylation prevents consolidation of differentiation programs and allows reversion to the embryonic stem cell state. PLoS One. 2012;7:e52629. doi: 10.1371/journal.pone.0052629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–19. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Annibali D, Gioia U, Savino M, Laneve P, Caffarelli E, Nasi S. A new module in neural differentiation control: two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2. PLoS One. 2012;7:e40269. doi: 10.1371/journal.pone.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119:4992–5004. doi: 10.1182/blood-2011-10-385716. [DOI] [PubMed] [Google Scholar]
- [40].Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, et al. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10:e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–90. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Galli R, Paone A, Fabbri M, Zanesi N, Calore F, Cascione L, et al. Toll-like receptor 3 (TLR3) activation induces microRNA-dependent reexpression of functional RARβ and tumor regression. Proc Natl Acad Sci U S A. 2013;110:9812–7. doi: 10.1073/pnas.1304610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]