Abstract
Type 2 diabetes mellitus is a heterogeneous inherited disorder characterized by chronic hyperglycemia resulting from pancreatic β-cell dysfunction and insulin resistance. Although the pathogenic mechanisms are not fully understood, manifestation of the disease most likely requires interaction between both environmental and genetic factors. In the search for such susceptibility genes, we have performed a genomewide scan in 58 multiplex families (comprising 440 individuals, 229 of whom were affected) from the Botnia region in Finland. Initially, linkage between chromosome 12q24 and impaired insulin secretion had been reported, by Mahtani et al., in a subsample of 26 families. In the present study, we extend the initial genomewide scan to include 32 additional families, update the affectation status, and fine map regions of interest, and we try to replicate the initial stratification analysis. In our analysis of all 58 families, we identified suggestive linkage to one region, chromosome 9p13-q21 (nonparametric linkage [NPL] score 3.9; P<.0002). Regions with nominal P values <.05 include chromosomes 2p11 (NPL score 2.0 [P<.03]), 3p24-p22 (NPL score 2.2 [P<.02]), 4q32-q33 (NPL score 2.5 [P<.01]), 12q24 (NPL score 2.1 [P<.03]), 16p12-11 (NPL score 1.7 [P<.05]), and 17p12-p11 (NPL score 1.9 [P<.03]). When chromosome 12q24 was analyzed in only the 32 additional families, a nominal P value <.04 was observed. Together with data from other published genomewide scans, these findings lend support to the hypothesis that regions on chromosome 9p13-q21 and 12q24 may harbor susceptibility genes for type 2 diabetes.
Type 2 diabetes mellitus (non–insulin-dependent diabetes mellitus [NIDDM]) is a multifactorial, heterogeneous disorder characterized by chronic hyperglycemia resulting from pancreatic β-cell dysfunction and insulin resistance. Manifestation of NIDDM is thought to require interaction between genetic and environmental factors, but the pathogenic mechanisms are not fully understood (Beck-Nielsen and Groop 1994; Groop and Tuomi 1997). Both segregation analysis and twin studies indicate that there is a genetic component of NIDDM, with an estimated recurrence risk of ∼3.5 (Rich 1990). Several genes predisposing to monogenic forms of diabetes, including maturity-onset diabetes of the young (MODY), have been identified in recent years (Froguel et al. 1993; Yamagata et al. 1996a; Yamagata et al. 1996b; Horikawa et al. 1997; Stoffers et al. 1997).
Dissection of the complex—and, most likely, polygenic—late-onset NIDDM has been more difficult, although some encouraging progress toward identification of NIDDM or diabetes-related quantitative susceptibility genes has been reported recently (Hanis et al. 1996; Mahtani et al. 1996; Hanson et al. 1998; Imperatore et al. 1998; Bektas et al. 1999; Duggirala et al. 1999; Elbein et al. 1999; Altshuler et al. 2000; Ehm et al. 2000; Ghosh et al. 2000; Mitchell et al. 2000; Vionnet et al. 2000; Luo et al. 2001; Parker et al. 2001; Permutt et al. 2001). Notable among these studies is the report of linkage between NIDDM in Mexican American sib pairs and the NIDDM1 (MIM 601283) locus on chromosome 2q37 (Hanis et al. 1996). This linkage was further strengthened when interaction with a locus on chromosome 15 was taken into account (Cox et al. 1999), and a subsequent linkage-disequilibrium search in this region identified association between NIDDM and variation in or around the CAPN10 (MIM 605286) gene (Horikawa et al. 2000).
Here we present results from a genomewide search for genes conferring increased susceptibility to late-onset NIDDM in 58 families, 26 of which have been described elsewhere (Mahtani et al. 1996). In the present study, we extend the family panel by including 223 individuals (109 of whom were affected) from 32 additional families (mean family size 7.0). To be included in the extended panel, a family had to have at least two affected siblings with an age at onset <70 years (which is less stringent than the age at onset <60–65 years that had been required in the previous study). The subjects who were unaffected at the time of the initial report were reinvestigated after 3 years, and five subjects were found to have developed overt NIDDM. Therefore, in total, 440 subjects from 58 families (229 affected; mean family size 7.6) were included in the present study (table 1). All nongenotyped individuals and individuals who were unavailable for phenotyping were considered to have an unknown affectation status. The families in this study are from the Botnia region on the western coast of Finland (Groop et al. 1996; Mahtani et al. 1996). The population history of the region is likely to restrict the number of distinct founder mutations and could therefore aid in genetic studies of complex diseases (de la Chapelle 1993; de la Chapelle and Wright 1998; Wright et al. 1999; Peltonen et al. 2000).
Table 1.
Clinical Characteristics of the Individuals from the 58 Families Included in the Genomewide Scan
|
Mean ± SE |
||||||||
| Serum Level |
||||||||
| Age(years) |
Glucose(mmol/liter) |
Insulin(mU/liter) |
||||||
| Group | No. of Individuals (M/F) | At Time of Study | At Onset | BMI(kg/m2) | fB | 2hB | fB | 2hB |
| Nonaffected | 211 (103/108) | 52.8 ± 1.2 | … | 26.9 ± .3 | 5.8 ± .05 | 6.6 ± .1 | 9.8 ± .4 | 53.1 ± 3.2 |
| Affected | 229 (111/118) | 64.3 ± .8 | 57.5 ± .8 | 28.8 ± .3 | 9.6 ± .2 | 15.0 ± .5 | 15.9 ± 1.1 | 76.2 ± 4.6 |
All subjects have given their consent to be included in the study, which has been approved by the local ethics committee. Families with either type 1 diabetes or MODY were excluded. Type 1 diabetes was considered present if the patient (a) had either glutamic acid decarboxylase antibodies or fasting c-peptide concentrations <0.3 nmol/liter or (b) had required insulin treatment <3 mo after diagnosis (Mahtani et al. 1996). Diabetes was diagnosed on the basis of World Health Organization criteria (Alberti and Gries 1988): either (1) a previous diagnosis of NIDDM, with treatment with oral agents and/or insulin, or (2) either fasting blood (fB)-glucose >6.7 mmol/liter (preferred) or a modified 2–h blood (2hB) glucose level of >8.5 mmol/liter.
During a 3-year follow-up of our subjects with impaired glucose tolerance, 25% with a 2hB glucose >8.5 mmol/liter developed manifest NIDDM, compared with 3% of those with a 2hB glucose <8.5 mmol/liter (P<.0001). Furthermore, additional prospective studies also have shown that individuals with such 2hB-glucose levels have a very high risk of developing diabetes (Saad et al. 1988; Charles et al. 1991).
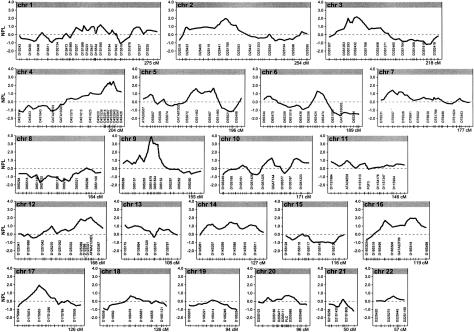
Genotypes were determined as described elsewhere (Mahtani et al. 1996), and all data were subjected to an extensive error-checking process. Data were checked for Mendelian segregation, by PEDMANAGER software (by M.P.R. and M.J.D.). The genotyping for the relevant locus was repeated for the entire family in question when Mendelian incompatibilities were found. We also checked the identity by descent (IBD) match between the observed and the expected values, in all possible sibships in the entire set. There were no deviations from the expected values, which indicates that, within our data set, there was a low incidence of genotyping errors, sample mixups, or incorrectly defined kinship. The initial scan included 387 polymorphic microsatellite markers distributed throughout the genome. In this study, we have added 65 markers in regions of potential interest from the first round of analysis. The total scan thus includes 452 polymorphic microsatellite markers (fig. 1). The mean sex-averaged distance between these markers is ∼7 cM (range 0.1–14.1 cM), and the average information content in the genomewide scan is 0.7 (estimated from data).
Figure 1.
Multipoint NPL analysis results for the NIDDM genomewide scan. Multipoint NPL scores were calculated by the GENEHUNTER version 2.0 software package (see the GENEHUNTER software-distribution web site). In each graph, the left vertical axis indicates the NPL score, represented by a thick line, the horizontal axis indicates the length of each chromosome, and the tick marks on the horizontal axis indicate the positions of the microsatellite markers; not all genotyped markers are represented. The shaded area indicates the recommended genomewide threshold (3.3 [Lander and Kruglyak 1995]) for suggestive linkage to a region.
Because the extent of genetic homogeneity and the mode of inheritance of NIDDM are unknown, evidence of linkage was assessed by a nonparametric method. We have used the GENEHUNTER (Kruglyak et al. 1996) version 2 software package (see the GENEHUNTER software-distribution web site), which performs complete multipoint analysis of the statistical significance of IBD allele sharing, at each location in the genome, among all affected family members and which also estimates the information content (i.e., how much of the total genetic information in a segment has been extracted). Allele frequencies used in the analysis were those observed in the original 26 families with NIDDM and in 20 unrelated normoglycemic control subjects (i.e., spouses without family history of diabetes) from the Botnia region.
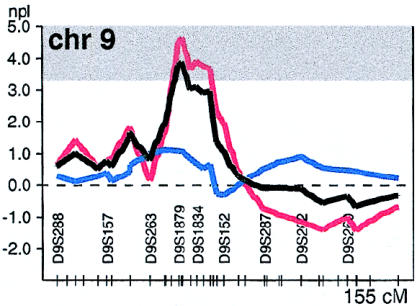
In our linkage analysis, the strongest linkage was between NIDDM and a region on chromosome 9q21. This region had shown nominal significance (P<.05) in the initial study. In the present, expanded study, the evidence is increased and shows suggestive evidence of linkage, with a nonparametric linkage (NPL) score of 3.9 (P<.0002), at markers D9S166/D9S301. The region under the 1-LOD support interval (D9S1874–D9S153) on chromosome 9p13-q21 spans ∼20 cM of the centromere (fig. 2). To determine the empirical genomewide significance of our particular data sets, simulations were performed by assignment of artificial genotype data to the families' structures (by GENSIM; M.J.D., unpublished data). These simulations matched our data sets, with regard to marker heterozygosity, individual affection status, individuals genotyped, and proportion of missing data. Genotypes for 100 replicates of the genomewide scan (2,200 chromosomes) were generated by a dense map of markers covering the entire genome (with marker density uniformly matching that of our fine-mapped regions). In only 8 of the 100 simulated genomewide scans was the observed NPL score of 3.9 exceeded (Pcorrected<.08), indicating that in <1/10 genomewide scans would one observe such a peak by chance.
Figure 2.
Multipoint NPL score on chromosome 9, in the entire family, represented as described in the legend to figure 1 but in greater detail. The results for the initial 26 families are represented by a red line (D9S166 [NPL score 4.61 {P<.0001}]), the results for the additional 32 families are represented by a blue line (D9S166 [NPL score 1.09 {P=.14}]), and the combined results for all 58 families are represented by a black line (D9S166 [NPL score 3.9 {P<.0002}]). The shaded area indicates the recommended genomewide threshold (3.3 [Lander and Kruglyak 1995]) for suggestive linkage to a region.
Overlapping results, with nominal statistical significance, can be found in Pima Indians, Mexican Americans, and Han Chinese (Hanis et al. 1996; Imperatore et al. 1998; Pratley et al. 1998; Luo et al. 2001). Hanis et al. (1996) reported some evidence (P<.01) of linkage between NIDDM and marker D9S175—4 cM from D9S166, the marker that showed the strongest linkage in our study—on chromosome 9q in their sample of 440 Mexican American sib pairs. Furthermore, Imperatore et al. (1998) obtained modest two-point LOD scores (1.28 and 1.48) for linkage between a region on chromosome 9q and NIDDM associated with retinopathy and nephropathy. Pratley et al. (1998) also reported modest evidence of linkage (LOD score 1.46), between chromosome 9q and a quantitative phenotype of 2-h insulin concentration during an oral glucose-tolerance test (OGTT), in 363 nondiabetic Pima Indians. All of these studies have 1-LOD–support intervals that overlap with those in the present study. Recently, Luo et al. (2001) have reported suggestive linkage to chromosome 9p13-q21 (NPL score 2.9 [P<.0005]) in 282 patients with NIDDM who are from 102 families of Han Chinese origin, a result that directly overlaps with our finding. However, several other studies found no evidence of linkage to this region (Norman et al. 1997; Elbein et al. 1999; Ghosh et al. 2000), emphasizing the difficulty in evaluation of linkage results.
In addition to the suggestive linkage to chromosome 9q21, six regions in our analysis displayed nominal P<.05, including chromosomes 2p11 (NPL score 2.0 [P<.03]), 3p24-p22 (NPL score 2.2 [P<.02]), 4q32-q33 (NPL score 2.5 [P<.01]), 12q24 (NPL score 2.1 [P<.03]), 16p12-11 (NPL score 1.7 [P<.05]), and 17p12-p11 (NPL score 1.9 [P<.03]) (table 2).
Table 2.
Regions Displaying Nominal P Values <.05, in the Analysis of the 58 Families Included in the Genomewide Scan
| Chromosome | Markers Included | Best Marker(s) | NPL Score (P) |
| 2p11 | D2S286–D2S1790 | D2S1777 | 2.0 (<.03) |
| 3p24-p22 | D3S3038–D3S2409 | D3S1561/D3S1768 | 2.2 (<.02) |
| 4q32-q33 | D4S1595–D4S3047 | D4S3015/D4S2951 | 2.5 (<.01) |
| 9q21 | D9S1874–D9S153 | D9S166/D9S301 | 3.9 (<.002) |
| 12q24 | D12S2070–D12S324 | D12S304– D12S1614 | 2.1 (<03) |
| 16p12-11 | D16S420 | D16S420 | 1.7 (<.05) |
| 17p12-p11 | D17S122–D17S953 | D17S953 | 1.9 (<.03) |
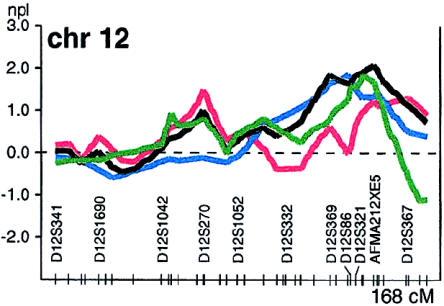
In the analysis of chromosome 12, which used diabetes as the phenotype in the 32 additional families only, the observed NPL score was 1.8 (P<.04) for marker D12S366 on chromosome 12q24 (fig. 3). Together with our previous evidence for linkage to this region (Mahtani et al. 1996), this strengthens the case for the presence of a susceptibility gene/factor in this region.
Figure 3.
Multipoint NPL score on chromosome 12 in two NIDDM genomewide scans, represented as described in the legend to figure 1. The results for the initial 26 families are represented by a red line (D12S304 [NPL score 1.2 {P<.2}]), the results for the additional 32 families are represented by a blue line (D12S366 [NPL score 1.8 {P<.04}]), the combined results for all 58 families are represented by a black line (D12S304–D12S1614 [NPL score 2.1 {P<.03}]), and the results of the new analysis of the 338 families described by Parker et al. (2001) are represented by a green line (D12S378 [NPL score 1.8 {P<.03}]).
We then investigated whether our initial report of linkage with chromosome 12q24 in families with NIDDM and with impaired insulin secretion could be replicated in the 32 additional families. These families were divided into quartiles of family means of log-transformed insulin levels 30 min after OGTT (30-min OGTT) in the affected individuals, after exclusion of outliers (see the Human Genetics Group web site) (Mahtani et al. 1996). Five families were excluded because of insufficient data—that is, fewer than one-third of the affected subjects had detectable levels of insulin 30-min OGTT. Therefore, 27 families were ranked according to the family means and were divided into quartiles. For the stratified analysis, we used the seven families from the lowest 30-min-OGTT insulin quartile (average, 2.7 affected individuals). In the analysis of chromosome 12q24 in these seven families, the observed NPL score was 0.7. We calculated the empirical P value for this NPL score of 0.7 by resampling 7 families from the 27 families, 10,000 times. In 74% of these runs, we observed NPL scores higher than the observed 0.7. These new data are not entirely incompatible with those previously reported (Mahtani et al. 1996), but they suggest that, if this region contains a diabetes-susceptibility gene, it is not restricted to an insulin-deficient phenotype.
In a separate study (Parker et al. 2001), our group reported a LOD score of 1.85 on chromosome 12q24 in an affected-sib-pair analysis of a subgroup of 117 sib pairs with high body-mass index (BMI) who were from Sweden and Finland. To further investigate this locus, we performed a nonparametric affected-pedigree-member analysis of the 338 families from that study (Parker et al. 2001), using the same NIDDM-affection criteria that have been described above. The families from that study (Parker et al. 2001) were, in general, smaller and consisted mainly of sibships, compared with the extended families in the present study. The patients in that other study resembled the patients from the current study, in terms of BMI (mean ± standard error [SE] = 29±0.2, compared to 28.8±0.3 in the present study), although the age at onset was slightly lower (mean ± SE = 52.1±0.4, compared to 57.5±0.8 in the present study) (Parker et al. 2001). Modest but overlapping evidence for linkage to NIDDM was observed to the region on chromosome 12q24, with an NPL score of 1.8 (fig. 3). Linkage at this locus is further supported by results from a recent meta-analysis including our own data (as described above) and two additional large European genomewide scans for NIDDM (Vionnet et al. 2000; Parker et al. 2001; Wiltshire et al. 2001), yielding a nominal P value of <.05, (F. Demenais and T. Kanninen, personal communication).
A number of previously published linkage studies have reported suggestive or tentative linkage between chromosome 12q and NIDDM (Bowden et al. 1997; Shaw et al. 1998; Ehm et al. 2000). In a large study involving four different populations, Ehm et al. have recently reported modest linkage (LOD score 1.4) to this region, on 12q24, in both a sample of white American subjects and a sample of African American subjects. Furthermore, this region was the only region, in their study, that showed overlapping linkage in two of the ethnic groups, with a linkage of P⩽.01 (Ehm et al. 2000). Bowden et al. reported a maximum multipoint LOD score of 1.45 for this region, in a sample consisting of white sib pairs with NIDDM and nephropathy (Bowden et al. 1997). Linkage to the NIDDM2 (MIM 601407) region, on chromosome 12q24, has also been reported, in an extended pedigree with late-onset NIDDM (LOD score 3.65 at recombination fraction .0008, telomeric to marker D12S321) (Shaw et al. 1998). Other studies, however, have been unable to replicate this finding (Duggirala et al. 1999; Elbein et al. 1999; Ghosh et al. 2000). Therefore, several published genomewide scans support the hypothesis that chromosome 12q24 might harbor a gene increasing susceptibility to NIDDM. In addition, some modest support for linkage to NIDDM has been found for a few other loci, on chromosomes 3p, 4q, and 17q, which also have been implicated in other genomewide scans.
The chromosome 3p21.2-p14.2 region where we find modest evidence of linkage has been linked to NIDDM or related traits, in several other genomewide scans (Hanis et al. 1996; Pratley et al. 1998; Duggirala et al. 1999; Ehm et al. 2000; Mitchell et al. 2000), and is an interesting candidate region for follow-up studies. We also found some modest evidence for linkage between NIDDM and chromosome 4q32-q33. Mitchell et al. (1995) also found evidence for linkage between a nearby locus on chromosome 4q28-31 and 2hB insulin during an OGTT in 382 Mexican American nondiabetic individuals. Duggirala et al. (1999) also found suggestive evidence for linkage between NIDDM and a close by region on chromosome 4q. The region on chromosome 17p12-q12 recently has been reported to be linked to plasma leptin levels, as part of a genomewide screen of 507 white nuclear families (Kissebah et al. 2000). Linkage of chromosome 17 to total cholesterol and HDL-cholesterol (HDL-C) has also been found in a genomewide scan of 232 multigenerational pedigrees randomly selected from the population (Klos et al. 2001). Furthermore, modest evidence of linkage to this region has recently been described in two large genomewide scans for NIDDM in white families (Vionnet et al. 2000; Wiltshire et al. 2001).
The collection of published genomewide scans of NIDDM also underscores the difficulty in the interpretation and replication of linkage findings. An unrealistic number of sib pairs might be needed in order for weak effects to be detected. A Pro12Ala variant in the PPARγ gene (MIM 601487) provides an example of such a situation; the variant was significantly associated with NIDDM, in a meta-analysis using a transmission/disequilibrium test (Altshuler et al. 2000). Despite a modest individual risk reduction, of ∼15%, associated with the rare Ala allele, the population-attributable risk was large, 20%–25%. By simulation, we estimated that 3 million sib pairs would have been needed in order to detect this effect in a linkage study. Furthermore, in a recent study, simulations have shown that one would not always expect a locus to be replicated over independent studies, even if it were present (Hirschhorn et al. 2001). Potential solutions must involve (a) the use of very large data sets, such as those studied by the ongoing International Type 2 Diabetes Linkage Analysis Consortium; (b) association studies designed to detect modest effects of common polymorphisms (Altshuler et al. 2000); and (c) the use of isolated populations (Peltonen et al. 2000).
In conclusion, this extension of our previously reported genomewide scan provides some support for linkage between NIDDM and regions on chromosome 9p13-q21 and 12q24. Further analysis of these regions will likely require comprehensive association or linkage-disequilibrium analysis. Fortunately, such extensive analyses are becoming increasingly possible with the availability of dense genetic maps (Sachidanandam et al. 2001).
Acknowledgments
We thank the Botnian families for their support and willingness to participate and for making this project possible. We would also like to thank all people, at Whitehead and in Malmö, who have been involved in this project during the past years, for their devoted and skillful work. This work was supported by grants from the Sigrid Juselius Foundation, the JDF Wallenberg Foundation, the Ingabritt and Arne Lundbergs Foundation, the Finnish Diabetes Research Foundation, The Swedish Medical Research Council, the Novo-Nordisk Foundation, and a European Commission grant to Genome Integrated Force in Type 2 Diabetes (GIFT). M.M.M. was supported by a Centennial Fellowship from the Medical Research Council of Canada. C.M.L. is supported by the Foundation for Strategic Research, via the National Network for Cardiovascular Research, and by the Royal Physiographic Society, the Anna-Lisa and Sven Lundgrens Foundation, and the Dir. Albert Påhlssons Foundation. E.W. was supported by the Finnish Medical Society. M.I.M. was supported by a Medical Research Council (U.K.) Travelling Fellowship. L.K. was supported by a Special Emphasis Research Career Award from the National Human Genome Research Institute.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GENEHUNTER software-distribution web site, http://www.genome.wi.mit.edu/ftp/distribution/software/gh2/
- Human Genetics Group web site, http://www-genome.wi.mit.edu/humgen/ (for raw data)
- International Type 2 Diabetes Linkage Analysis Consortium, http://www.sfbr.org/external/diabetes/ [DOI] [PMC free article] [PubMed]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CAPN10 [MIM 605286], NIDDM1 [MIM 601283], and NIDDM2 [MIM 601407], and PPARγ [MIM 601487])
References
- Alberti KG, Gries FA (1988) Management of non-insulin-dependent diabetes mellitus in Europe: a consensus view. Diabet Med 5:275–281 [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, ES L (2000) The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Beck-Nielsen H, Groop L (1994) Metabolic and genetic characterization of prediabetic states: sequence of events leading to non-insulin-dependent diabetes mellitus. J Clin Invest 94:1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektas A, Suprenant ME, Wogan LT, Plengvidhya N, Rich SS, Warram JH, Krolewski AS, Doria A (1999) Evidence of a novel type 2 diabetes locus 50 cM centromeric to NIDDM2 on chromosome 12q. Diabetes 48:2246–2251 [DOI] [PubMed] [Google Scholar]
- Bowden DW, Sale M, Howard TD, Qadri A, Spray BJ, Rothschild CB, Akots G, Rich SS, Freedman BI (1997) Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes 46:882–886 [DOI] [PubMed] [Google Scholar]
- Charles MA, Fontbonne A, Thibult N, Warnet JM, Rosselin GE, Eschwege E (1991) Risk factors for NIDDM in white population: Paris prospective study. Diabetes 40:796–799 [DOI] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hanis CL, Bell GI, Kong A (1999) Loci on chromosomes 2 (NIDDM1) and 15 interact to increase susceptibility to diabetes in Mexican Americans. Nat Genet 21:213–215 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A (1993) Disease gene mapping in isolated human populations: the example of Finland. J Med Genet 30:857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Blangero J, Almasy L, Dyer T, Williams K, Leach R, O'Connell P, Stern M (1999) Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehm MG, Karnoub MC, Sakul H, Gottschalk K, Holt DC, Weber JL, Vaske D, Briley D, Briley L, Kopf J, McMillen P, Nguyen Q, Reisman M, Lai EH, Joslyn G, Shepherd NS, Bell C, Wagner MJ, Burns DK (2000) Genomewide search for type 2 diabetes susceptibility genes in four American populations. Am J Hum Genet 66:1871–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein S, Hoffman M, Teng K, Leppert M, Hasstedt S (1999) A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes 48:1175–1182 [DOI] [PubMed] [Google Scholar]
- Froguel P, Zouali H, Vionnet N, Velho G, Vaxillaire M, Sun F, Lesage S, Stoffel M, Takeda J, Passa P, Permutt MA, Beckmaan JS, Bell GI, Cohen D (1993) Familial hyperglycemia due to mutations in glucokinase: definition of a subtype of diabetes mellitus. N Engl J Med 328:697–702 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, et al (2000) The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 [PMC free article] [PubMed] [Google Scholar]
- Groop L, Forsblom C, Lehtovirta M, Tuomi T, Karanko S, Nissén M, Ehrnström B-O, Forsén B, Isomaa B, Snickars B, Taskinen M-R (1996) Metabolic consequences of a family history of NIDDM (The Botnia Study). Diabetes 45:1585–1593 [DOI] [PubMed] [Google Scholar]
- Groop L, Tuomi T (1997) Non-insulin dependent diabetes mellitus—a collision between thrifty genes and an affluent society. Ann Med 29:37–53 [DOI] [PubMed] [Google Scholar]
- Hanis CL, Boerwinkle E, Chakraborty R, Ellsworth DL, Concannon P, Stirling B, Morrison VA, et al (1996) A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet 13:161–166 [DOI] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC (1998) An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet 63:1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lindgren CM, Daly MJ, Kirby A, Schaffner SF, Burtt NP, Altshuler D, Parker A, Rioux JD, Platko J, Gaudet D, Hudson TJ, Groop LC, Lander ES (2001) Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am J Hum Genet 69:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, Kuroki H, Kasahara T, Iwamoto Y, Bell GI (1997) Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat Genet 17:384–385 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Imperatore G, Hanson RL, Pettitt DJ, Kobes S, Bennett PH, Knowler WC, Pima Diabetes Gene Group (1998) Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Diabetes 47:821–830 [DOI] [PubMed] [Google Scholar]
- Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, Martin L, Blangero J, Comuzzie AG (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA 97:14478–14483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos KL, Kardia SL, Ferrell RE, Turner ST, Boerwinkle E, Sing CF (2001) Genome-wide linkage analysis reveals evidence of multiple regions that influence variation in plasma lipid and apolipoprotein levels associated with risk of coronary heart disease. Arterioscler Thromb Vasc Biol 21:971–978 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Luo TH, Zhao Y, Li G, Yuan WT, Zhao JJ, Chen JL, Huang W, Luo M (2001) A genome-wide search for type II diabetes susceptibility genes in Chinese Hans. Diabetologia 44:501–506 [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, Kanninen T, Kirby A, Kruglyak L, Munnelly K, Parkkonen M, Reeve-Daly MP, Weaver A, Brettin T, Duyk G, Lander ES, Groop LC (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Cole SA, Hsueh WC, Comuzzie AG, Blangero J, MacCluer JW, Hixson JE (2000) Linkage of serum insulin concentrations to chromosome 3p in Mexican Americans. Diabetes 49:513–516 [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, ÓConnell P, Harrison CR, Manire M, Shipman P, Moyer MP, Stern MP, Frazier ML (1995) Evidence for linkage of postchallenge insulin levels with intestinal fatty acid-binding protein (FABP2) in Mexican-Americans. Diabetes 44:1046–1053 [DOI] [PubMed] [Google Scholar]
- Norman RA, Thompson DB, Foroud T, Garvey WT, Bennett PH, Bogardus C, Ravussin E, Pima Diabetes Gene Group (1997) Genomewide search for genes influencing percent body fat in Pima Indians: suggestive linkage at chromosome 11q21-q22. Am J Hum Genet 60:166–173 [PMC free article] [PubMed] [Google Scholar]
- Parker A, Meyer J, Lewitzky S, Rennich JS, Chan G, Thomas JD, Orho-Melander M, Lehtovirta M, Forsblom C, Hyrkko A, Carlsson M, Lindgren C, Groop LC (2001) A gene conferring susceptibility to type 2 diabetes in conjunction with obesity is located on chromosome 18p11. Diabetes 50:675–680 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Permutt MA, Wasson JC, Suarez BK, Lin J, Thomas J, Meyer J, Lewitzky S, Rennich JS, Parker A, DuPrat L, Maruti S, Chayen S, Glaser B (2001) A genome scan for type 2 diabetes susceptibility loci in a genetically isolated population. Diabetes 50:681–685 [DOI] [PubMed] [Google Scholar]
- Pratley RE, Thompson DB, Prochazka M, Baier L, Mott D, Ravussin E, Sakul H, Ehm MG, Burns DK, Foroud T, Garvey WT, Hanson RL, Knowler WC, Bennett PH, Bogardus C (1998) An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J Clin Invest 101:1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SS (1990) Mapping genes in diabetes: genetic epidemiological perspective. Diabetes 39:1315–1319 [DOI] [PubMed] [Google Scholar]
- Saad MF, Knowler WC, Pettitt DJ, Nelson RG, Mott DM, Bennett PH (1988) The natural history of impaired glucose tolerance in the Pima Indians. N Engl J Med 319:1500–1506 [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, et al (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed] [Google Scholar]
- Shaw JT, Lovelock PK, Kesting JB, Cardinal J, Duffy D, Wainwright B, Cameron DP (1998) Novel susceptibility gene for late-onset NIDDM is localized to human chromosome 12q. Diabetes 47:1793–1796 [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Ferrer J, Clarke WL, Habener JF (1997) Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet 17:138–139 [DOI] [PubMed] [Google Scholar]
- Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepretre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P (2000) Genomewide search for type 2 diabetes–susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2–diabetes locus on chromosome 1q21-q24. Am J Hum Genet 67:1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O'Rahilly S, et al (2001) A genomewide scan for loci predisposing to type 2 diabetes in a UK population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet 69:553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI (1996a) Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384:458–460 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, et al (1996b) Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384:455–458 [DOI] [PubMed] [Google Scholar]