Summary
Direct reprogramming of adult somatic cells into alternative cell types has been shown for several lineages. We previously showed that GATA4, MEF2C, and TBX5 (GMT) directly reprogrammed nonmyocyte mouse heart cells into induced cardiomyocyte-like cells (iCMs) in vitro and in vivo. However, GMT alone appears insufficient in human fibroblasts, at least in vitro. Here, we show that GMT plus ESRRG and MESP1 induced global cardiac gene-expression and phenotypic shifts in human fibroblasts derived from embryonic stem cells, fetal heart, and neonatal skin. Adding Myocardin and ZFPM2 enhanced reprogramming, including sarcomere formation, calcium transients, and action potentials, although the efficiency remained low. Human iCM reprogramming was epigenetically stable. Furthermore, we found that transforming growth factor β signaling was important for, and improved the efficiency of, human iCM reprogramming. These findings demonstrate that human fibroblasts can be directly reprogrammed toward the cardiac lineage, and lay the foundation for future refinements in vitro and in vivo.
Highlights
-
•
Human fibroblasts can be directly induced toward a CM-like state by defined factors
-
•
Reprogramming of fibroblasts toward a CM state is epigenetically stable
-
•
Human and mouse in vitro iCMs display a comparable gene-expression shift
-
•
TGF-β signaling is important for human iCM reprogramming
Introduction
Cellular reprogramming of fibroblasts into specific cell types without passing through a progenitor state offers an alternative approach for generating lineages of interest compared with directed differentiation of pluripotent stem cells. In contrast to the ability of a single transcription factor, MYOD, to transdifferentiate fibroblasts to skeletal muscle cells (Davis et al., 1987), conversion of fibroblasts to neuronal-, hepatocyte-, or cardiomyocyte (CM)-like cells has required a combinatorial delivery of multiple transcription factors or microRNAs (miRNAs) (Huang et al., 2011; Ieda et al., 2010; Vierbuchen et al., 2010; Yoo et al., 2011). This feature is similar to reprogramming of fibroblasts into induced pluripotent stem (iPS) cells, as is the efficiency of direct reprogramming to specific cell types.
We previously reported that three developmental cardiac transcription factors (GATA4, MEF2C, and TBX5 [GMT]) can directly reprogram cultured mouse cardiac and dermal fibroblasts into CM-like cells (Ieda et al., 2010). These induced CM-like cells (iCMs) had global gene-expression profiles that were more similar to CMs than to fibroblasts, and many features of CMs, with a small subset of more fully reprogrammed iCMs exhibiting contractile activity.
Recently, we (Qian et al., 2012) and others (Inagawa et al., 2012; Song et al., 2012) showed that direct injection of GMT-encoding retrovirus into the mouse heart reprogrammed endogenous nonmyocytes (largely activated fibroblasts) into functional CMs in vivo after coronary artery ligation. More than half of the iCMs were more fully reprogrammed, displaying synchronous contractions with endogenous CMs and other iCMs (Qian et al., 2012). GMT induction in vivo resulted in decreased scar size and improved cardiac function. Addition of HAND2 was reported to improve GMT reprogramming of mouse fibroblasts in vitro and in vivo (Song et al., 2012), and Myocardin with TBX5 and MEF2C, rather than GATA4, also reprogrammed cells in vitro (Protze et al., 2012). Similarly, a cocktail of muscle-specific miRNAs generated CM-like cells in mice (Jayawardena et al., 2012). Thus, several strategies might reprogram cardiac fibroblasts, which comprise 50% of cells in the adult heart (Ieda et al., 2009), into iCMs that establish a self-reinforcing molecular network, and the in vivo environment may provide cues and/or mechanical forces to promote reprogramming.
Here, we sought to identify factors that directly reprogram human fibroblasts toward the CM lineage in vitro, with the notion that the in vivo environment may ultimately permit further reprogramming. Although we found that GMT was insufficient in human cells, adding ESRRG and MESP1 to GMT reprogrammed human fibroblasts derived from embryonic stem cells (ESCs), fetal heart, or neonatal skin into cells with CM-like gene expression and sarcomeres, albeit at low frequency. Further addition of Myocardin and ZFPM2 (FOG2) resulted in iCMs with more fully developed sarcomeres, rhythmic calcium transients, and (in some) action potentials. Finally, we found that transforming growth factor β (TGF-β) signaling was important for, and further improved, the efficiency of human iCM reprogramming.
Results
Screening for Human Cardiac Reprogramming Factors
We investigated whether GMT could reprogram human dermal fibroblasts (HDFs) or human cardiac fibroblasts (HCFs), but failed to detect upregulation of the cardiac-specific sarcomeric genes cardiac myosin heavy chain (MHC) or cardiac troponin T (cTNT). As an assay for cardiac markers, we used transgenic H9 human ESC (hESC)-derived fibroblasts (H9Fs), with mCherry driven by the mouse αMHC promoter (Kita-Matsuo et al., 2009). This tool allowed fluorescence-activated cell sorting (FACS) to detect cells that activated cardiac gene expression, with a plan for validation in human primary fibroblasts (Figure S1A available online). As described previously (Kita-Matsuo et al., 2009), mCherry was expressed in beating H9-derived CMs (H9-CMs), but not in other cells, and >96% of purified mCherry+ cells expressed cTNT (Figure S1).
To avoid contamination of CMs or cardiac progenitors in H9Fs, embryoid bodies (EBs) differentiated over 42 days in vitro or 3-month-old teratomas in mice were immunostained with antibodies to human THY1, a surface marker of fibroblasts (Hudon-David et al., 2007), and sorted by FACS. This established approach yields human fibroblasts for reprogramming studies (Hockemeyer et al., 2008; Ramkisoensing et al., 2011). Almost all purified THY1+/mCherry− cells expressed two more fibroblast markers, prolyl-4-hydroxylase β and vimentin, and none expressed cTNT (Figures S1D and S1E). Although there is no completely fibroblast-specific marker, these extra markers suggested that the pool was mostly fibroblasts. Furthermore, the THY1+/mCherry− H9Fs did not contain any c-KIT+ stem/progenitor cells.
As in the primary fibroblasts, no mCherry+ or cTNT+ cells were detected by FACS when GMT was overexpressed in H9Fs, despite high levels of expression. The addition of epigenetic modifiers (e.g., the histone deacetylase inhibitor Trichostatin A and the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine), which was previously reported to improve iPS cell reprogramming (Huangfu et al., 2008), did not induce reprogramming.
To identify factors that along with GMT would induce cardiac reprogramming in human cells, we selected 13 transcription factors or coactivators and three growth factors that were enriched in H9-CMs (Figure S1I). We used human GMT as the baseline cocktail for human cardiac reprogramming and screened for additional human factors that activated cardiac gene expression.
THY1+/mCherry− H9Fs were transduced with a mixture of retroviruses expressing all 19 factors, or GFP alone, as previously described (Ieda et al., 2010). A small number of mCherry+ cells were observed, and their proportion increased by 10 days posttransduction (Figure 1A). αMHC-mCherry+ cells could be quantitatively detected by FACS after 2 weeks (Figure 1H).
Figure 1.
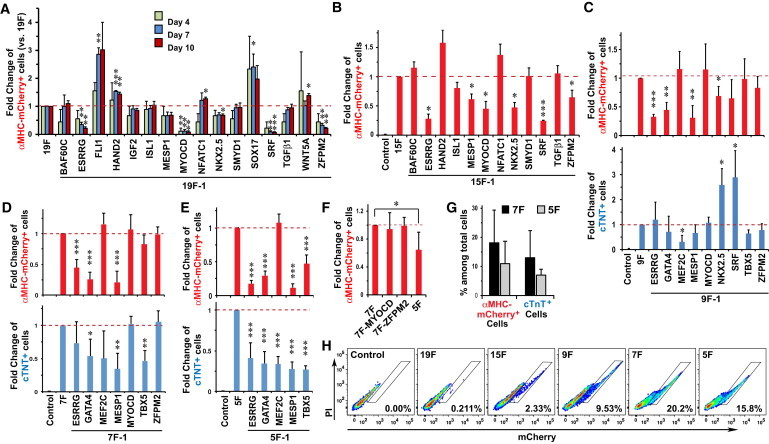
Identifying a Minimal Cocktail of Transcription Factors to Reprogram Human Fibroblasts toward CM-like Cells
(A) Screen results of αMHC-mCherry+ cell induction in H9Fs with 19 candidate reprogramming factors, and the effects of removing individual factors from the 19-factor (19F) pool on days 4, 7, and 10 after retroviral infection (n = 3).
(B) Effects of removing individual factors from the 15F pool on αMHC-mCherry+ cell induction, as assessed by FACS (n = 3).
(C–E) Effects of removing individual factors from the nine-factor (9F) (C, n = 4), 7F (D, n = 5), or 5F (E, n = 6) pool on αMHC-mCherry+ (top) and cTNT+ (bottom) cell induction by FACS.
(F) Removal of both MYOCD and ZFPM2, but not either one alone, from the 7F pool significantly decreased the induction of αMHC-mCherry+ cells by FACS (n = 6).
(G) Summary of results of αMHC-mCherry+ and cTNT+ induction 2 weeks after 5F (n = 11) or 7F (n = 13) retroviral infection.
(H) Representative FACS plots of αMHC-mCherry+ cells 2 weeks after infection with the indicated factors.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 compared with dashed lines.
Data represent the mean ± SD from independent experiments. See also Figures S1 and S2.
To identify dispensable factors, each factor was serially removed from the pool and the fold changes of αMHC-mCherry+ cells were recorded to normalize day-to-day variability. Removal of FLI1, SOX17, or WNT5A produced more mCherry+ cells (Figure 1A), suggesting that they inhibited cardiac reprogramming. Insulin growth factor 2 was also dispensable. Transduction with the remaining 15-factor (15F) pool resulted in a 10-fold increase in the percentage of mCherry+ cells to ∼2% (Figure 1H). Fourteen-factor pools lacking ESRRG, MESP1, MYOCD, NKX2.5, SRF, or ZFPM2 significantly decreased the yield of mCherry+ cells, so these genes were retained along with GMT. For the nine putative factors, we also examined expression of cTNT by FACS, because the expression of both genes is a more stringent criterion for cells that are shifting to a CM state. SRF and NKX2.5 were omitted because their absence increased cTNT+ cell numbers 3-fold. Removal of NKX2.5 resulted in a small but significant decrease in mCherry+ cells, but the dramatic increase in cTNT+ cells led us to exclude this factor for more double-positive cells.
Continued refinement showed that five factors (GATA4, MEF2C, TBX5, ESRRG, and MESP1) were sufficient to generate αMHC-mCherry+ and cTNT+ cells, and removing any one of these factors significantly decreased induction efficiency. MYOCD and ZFPM2 were dispensable in the seven-factor (7F) cocktail when removed individually, but the five-factor (5F) reprogramming efficiency was significantly lower than the 7F efficiency. The seven factors induced 18.1% ± 11.2% of total fibroblasts to activate the αMHC-mCherry reporter, and 59.0% ± 11.0% of these also expressed cTNT, resulting in 13.0% ± 9.3% of total fibroblasts becoming αMHC-mCherry+:cTNT+ 2 weeks after retroviral infection (Figure 1). Similar results were obtained with fibroblasts from 3-month-old hESC-derived teratomas.
Although no H9Fs expressed cardiac αMHC 2 weeks after GFP retroviral infection (Figure S2A), most mCherry+ cells induced by 5F or 7F reprogramming expressed cardiac αMHC protein by immunocytochemistry (Figure S2A). Four weeks after infection, more than half of the mCherry+ cells also expressed cTNT and sarcomeric α-actinin, and had begun to assemble sarcomeric structures. Well-organized sarcomeres were observed in 10-week reprogrammed αMHC-mCherry+ cells, similar to H9-CMs (Figure 2A). These reprogrammed αMHC-mCherry+ cells did not express calponin or smooth muscle actin, markers of smooth muscle cells (Figures S2B and S2C). Importantly, primary HDFs and HCFs that were infected with a lentivirus encoding αMHC-mCherry and 7F retrovirus could also be reprogrammed to express α-actinin and cTNT and assemble sarcomeres, although with lower frequency (1%–4%; Figures 2B, 2C, S3A, and S3B). The lower frequency may reflect the higher passage number of the primary fibroblasts utilized.
Figure 2.
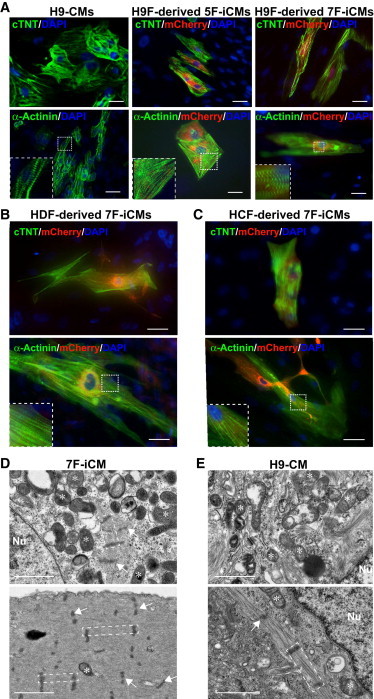
Human Fibroblasts Reprogrammed by Seven Factors Express Cardiac Genes and Form Sarcomere-like Structures
(A) Immunocytochemistry of αMHC-mCherry H9-CMs and H9Fs 10 weeks after retroviral infection with five or seven factors using mCherry, cTNT, or α-actinin antibodies, revealing partial sarcomeric organization in the cells indicated (highlighted in insets).
(B and C) Immunocytochemistry of HDFs and HCFs 6 weeks after 7F retroviral plus αMHC-mCherry lentiviral infection reveals sarcomeric gene expression in mCherry+ cells.
(D and E) Electron microscopic images of αMHC-mCherry+ 7F-iCMs (D, day 45 postinduction) reveal enriched mitochondria (star) and sarcomeres (dashed area) with Z lines (arrow) in 7F reprogrammed cells, which were similar to H9-CMs (E, day 22 postdifferentiation).
Scale bars, 20 μm (A–C) and 1 μm (D and E). Nu, nucleus. See also Figure S3.
By electron microscopy, enriched mitochondria and myofibrils were observed in 5F or 7F reprogrammed cells 6 weeks after retroviral infection. In 7F reprogrammed cells, myofibrils formed sarcomeres with clear Z lines, similar to ESC-CMs (Figures 2D, 2E, and S3C). Average sarcomere lengths were similar in reprogrammed cells (1.08 ± 0.32 μm) and ESC-derived CMs (1.21 ± 0.38 μm; Figure S3D). The range of myofibrillar organization was similar between human iCMs and ESC-CMs, both of which are incomplete and less organized than adult hCMs. Thus, cells with some CM features can be generated directly from human fibroblasts in vitro by seven defined transcription factors, and we will refer to these as human iCMs.
iCM Gene Expression Resembles that of Native CMs
αMHC-mCherry+ and cTNT+ cells gradually increased within the first 2 weeks, reaching the highest percentage at days 14–21 before declining (Figure S4A), likely because reprogrammed cells stopped proliferating, whereas nonreprogrammed cells continued to proliferate (Figure S4B). We studied the messenger RNA (mRNA) expression of pluripotency genes in H9Fs after 7F retroviral infection by qRT-PCR. Pluripotency genes were not significantly activated during reprogramming (Figure S4C). We sorted mCherry+ iCMs at 2, 3, and 4 weeks posttransduction with five or seven factors and compared the expression of cardiac-specific genes by quantitative PCR (qPCR). ACTC1, ACTN2, MYH6, MYL2, MYL7, TNNT2, NPPA, PLN, and RYR2 were significantly upregulated in 5F and 7F iCMs, but were not detected in H9Fs (Figure S5). Although the expression levels of many iCM genes were similar to those of H9-CMs, RYR2 and PLN levels were lower. In contrast, expression of fibroblast-enriched genes, including FN1, COL1A1, and COL5A2, was lower in reprogrammed mCherry+ cells, similar to H9-CMs (Figure S5B).
We compared the global gene expression patterns of 5F- or 7F-induced mCherry+/THY1− iCMs (4 and 12 weeks postinduction), H9-CMs, human fetal CMs (19 weeks old), and H9Fs by mRNA microarrays in triplicate. We found that 899 cardiac and 391 fibroblast-enriched genes were differentially expressed in H9Fs and human fetal CMs, with a false-discovery rate (FDR)-controlled p value < 0.01. When we compared these genes, we found that both 5F and 7F αMHC-mCherry+ cells shifted considerably from the H9F toward the H9-CM and human fetal CM pattern, with hundreds of genes coordinately altered (Figure 3A). We found that genes that are important for cardiac function were upregulated more highly in 7F than in 5F iCMs (Figure 3B), including CKM, MYH6, MYH7, TNNC1, and SLC8A1. A microarray assay also revealed the progressive repression of fibroblast genes between 4 and 12 weeks after induction of reprogramming factors (Figure 3A), which is consistent with the higher percentage of reprogrammed iCMs with well-organized sarcomeres 12 weeks after induction (Figures S5C and S5D).
Figure 3.
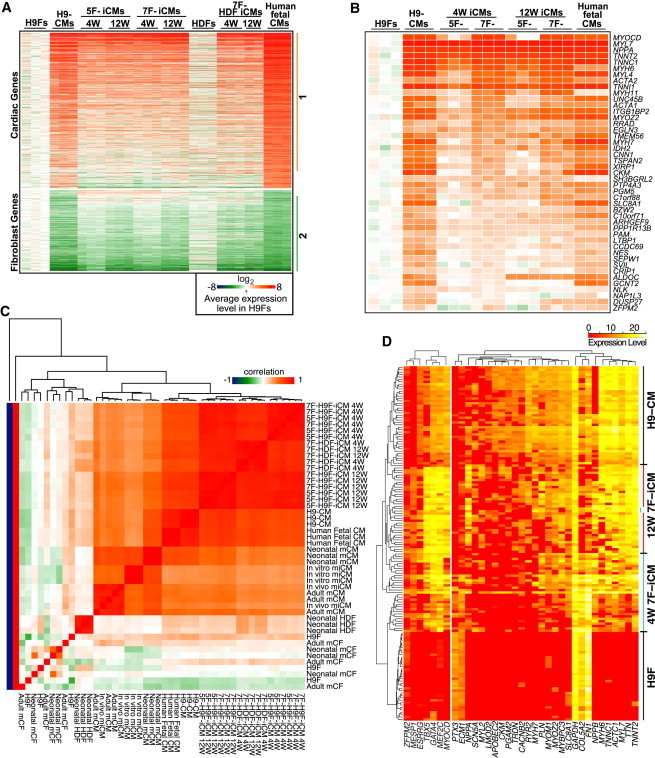
Human iCMs Are Transcriptionally Reprogrammed toward the CM State
(A) Heatmap image of microarray data illustrating gene expression profiles for the panel of genes that were differentially expressed between H9Fs and human fetal CMs evaluated in H9Fs, HDFs, human fetal CMs, H9-CMs, 5F- and 7F-iCMs, and HDF-derived iCMs (4 and 12 weeks postinduction); range of ±256-fold (log28) changes. The average level in three H9F samples was used as a baseline for comparison. Groups 1 and 2 include the genes that were upregulated or downregulated in CMs and iCMs compared with H9Fs.
(B) Heatmap of gene expression profiles for a panel of cardiac genes in group 1 of (A) that were more highly upregulated in 7F-iCMs compared with 5F-iCMs at 4 and 12 weeks after reprogramming.
(C) Correlation heatmap of orthologous gene expression for the panel of genes that were differentially expressed between fibroblasts and CMs evaluated in human iCMs reprogrammed by 5F or 7F and GMT-reprogrammed mouse (m) in vitro and in vivo iCMs. The correlation is indicated by color range; the scale along the values −1, 0, and 1 indicates perfect anticorrelation, no correlation, or perfect correlation, respectively.
(D) Heatmap of single-cell gene expression by qPCR using microfluidics (Fluidigm) in the cell types indicated. Each horizontal row represents an individual cell, and each vertical column represents the levels of a single gene. Gene-expression level is indicated by the color range.
See also Figures S4 and S5.
Primary fibroblasts (HDFs) were transduced with a lentivirus encoding αMHC-mCherry and the 7F retrovirus cocktail, so that the reprogrammed αMHC-mCherry+/THY1− HDF-iCMs could be purified for global gene expression assay at 4 and 12 weeks postinduction. Like the H9F-iCMs, the global gene-expression pattern in HDF-iCMs shifted toward the cardiac lineage; however, downregulation of fibroblast genes occurred even more rapidly than in H9F-iCMs (Figure 3A).
We next compared the degree of gene expression changes in human iCMs with published values for in vitro mouse iCM gene expression induced by GMT (Ieda et al., 2010). Analysis of orthologous gene expression indicated that, at the global gene-expression level, human fibroblasts were reprogrammed into CM-like cells by seven factors to a degree comparable to that observed for mouse iCMs reprogrammed by GMT in vitro (Figure 3C). Although we (Qian et al., 2012) and others (Song et al., 2012) reported that GMT can induce more complete reprogramming in vivo with resulting cardiac repair after injury, the global transcriptome of in vivo mouse iCMs has not been described. We therefore performed mRNA microarrays of isolated in vivo mouse iCMs and computationally analyzed the transcriptome changes as we did when we compared mouse and human in vitro iCMs. The isolated in vivo reprogrammed mouse iCMs and mouse adult CMs showed very similar global gene-expression patterns and thus were clustered into one group of cells (Figure 3C). This observation raises the possibility that the degree of reprogramming of human iCMs in vitro may result in more complete reprogramming in the in vivo environment.
Given the heterogeneity of pooled iCMs, we sought to examine individual cells to correlate the expression of reprogramming factors with the degree of reprogramming. Using microfluidic technology, we isolated individual αMHC-mCherry+ iCMs at 4 or 12 weeks and assayed each reprogramming factor and a panel of cardiac- or fibroblast-enriched genes at the single-cell level. Although the degree to which gene expression shifted toward the CM lineage varied, most cells were more similar to H9-CMs (Figure 3D). Cardiac gene expression was higher after 12 weeks, and fibroblast genes were strikingly downregulated between 4 and 12 weeks. Importantly, virtually all of the αMHC-mCherry+ iCMs had high levels of GATA4, MEF2C, TBX5, and MYOCD, but many reprogrammed cells expressed no ESRRG or ZFPM2. MESP1 was expressed in most cells, but at lower levels. Thus, single-cell studies may reveal the essential factors for an individual cell to shift toward a CM state.
Human iCMs Are Epigenetically Reprogrammed to a CM-like State
To identify epigenetic modification patterns, we analyzed the DNA and histone methylation status of loci that reflect reprogramming from cardiac fibroblasts to iCMs. Bisulfite genomic sequencing in the promoter regions of MYH6, MYH7, and MYL7 in H9Fs, 2-week reprogrammed αMHC-mCherry+ iCMs, and αMHC-mCherry+ H9-CMs revealed that the three promoters were hypermethylated in H9Fs, as expected, but relatively demethylated in iCMs, similar to H9-CMs (Figure 4A). The 7F iCMs had slightly less methylation at the MYH7 and MYL7 promoters than the 5F iCMs.
Figure 4.
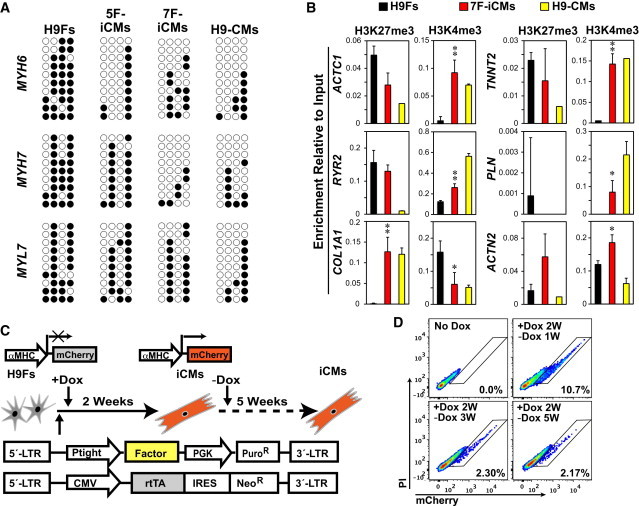
Human Fibroblasts Are Stably Reprogrammed into iCMs
(A) Bisulfite genomic sequencing for MYH6, MYH7, and MYL7 promoter methylation status in H9Fs, 5F- and 7F-iCMs (2 weeks postinfection), and H9-CMs. Open circles indicate unmethylated CpG dinucleotides; closed circles indicate methylated CpGs.
(B) The promoters of ACTC1, ACTN2, RYR2, TNNT2, PLN, and COL1A1 were analyzed by ChIP for trimethylation status of histone H3 of lysine 27 or 4 (H3K27me3 or H3K4me3) in H9Fs, 7F-iCMs, and H9-CMs. n = 3, ∗p < 0.05, ∗∗p < 0.01 versus H9Fs. Data represent the mean ± SD from independent experiments.
(C) Strategy to determine the temporal requirement of reprogramming factors for human cardiac reprogramming.
(D) FACS plots of αMHC-mCherry+ cells after retroviral infection without Dox (no Dox) or with 2-week Dox induction (+Dox 2W) followed by Dox withdrawal (−Dox) for 1, 3, and 5 weeks.
See also Figure S6.
Enrichment of trimethylated histone H3 of lysine 27 (H3K27me3) and lysine 4 (H3K4me3), which mark transcriptionally inactive or active chromatin, respectively, was analyzed in H9Fs, 2-week 7F αMHC-mCherry+ iCMs, and αMHC-mCherry+ H9-CMs. In αMHC-mCherry+ iCMs, H3K4me3 was significantly enriched at the promoters of all cardiac-specific genes tested, and depleted at the COL1A1 promoter, at which H3K27me3 increased to levels comparable to those observed in αMHC-mCherry+ H9-CMs (Figure 4B).
Using a doxycycline (Dox)-inducible retroviral system, αMHC-mCherry was induced in H9Fs as early as 3 days after infection with seven factors and Dox administration, whereas no mCherry+ cells were seen without Dox (Figures 4C, 4D, and S6). After 2 weeks, Dox was withdrawn. One week later, reprogrammed iCMs maintained αMHC-mCherry expression with an efficiency comparable to that observed for the noninducible system at 3 weeks (∼10%; Figure 4D), and they also continued to develop early sarcomeric structures (Figure S6C). The percentage of iCMs in culture decreased over the first several weeks as nonreprogrammed fibroblasts continued to divide, resulting in dilution of iCMs that exited the cell cycle. However, as fibroblasts started to senesce, the percentage of iCMs remained stable between 3 and 5 weeks after Dox withdrawal (Figure 4D). Expression of αMHC-mCherry and sarcomeric α-actinin were maintained in reprogrammed cells even 3 weeks after Dox withdrawal (Figure S6D), suggesting that these iCMs were stably reprogrammed, although this time point is too early for more mature sarcomeric structure.
Human iCMs Exhibit Some Cardiac Physiological Features
Electrical field stimulation triggered ∼20% of 4-week-induced mCherry+ iCMs to generate regular Ca2+ transients similar to those observed in H9-CMs (Figures 5A; Movie S1). Reprogrammed 7F-iCMs responded to caffeine with large Ca2+ transients similar to those observed in H9-CMs (Figure 5B). HDF-derived iCMs also exhibited calcium transients (Figure 5C; Movie S2). Ten weeks after induction, a resting membrane potential of −73.4 ± 8.4mV (n = 6) was detectable in αMHC-mCherry+ iCMs, similar to the hyperpolarized level of adult quiescent CMs (Wang et al., 1993). Action potentials were elicited in some iCMs (n = 3 out of >200 recorded cells), suggesting that this combination of factors can induce some cells to be more fully reprogrammed even in vitro, but most are partially reprogrammed (Figure 5D).
Figure 5.
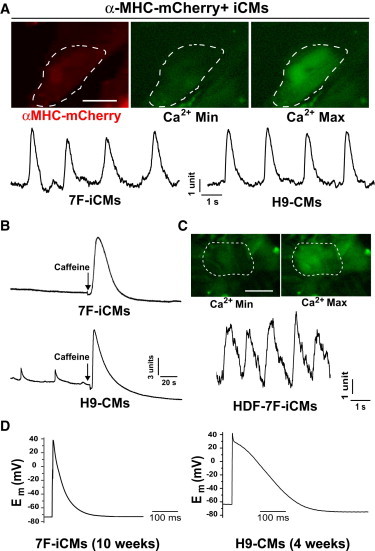
Human iCMs Exhibit Ca2+ Flux and Action Potentials
(A) Top: Electrical field stimulation induces Ca2+ transients, as shown by Ca2+ imaging (minimal (Min) and maximal (Max) levels), in αMHC-mCherry+ 7F-iCMs 4 weeks after retroviral infection. Bottom: A representative trace of Ca2+ transients recorded from reprogrammed αMHC-mCherry+ 7F-iCMs (left, n = 9) is similar to the trace recorded from 3-week-old H9-CMs (right).
(B) A representative trace of caffeine (20 mM)-induced Ca2+ transients recorded from reprogrammed 8-week αMHC-mCherry+ 7F-iCMs (top, n = 15) is similar to the trace recorded from 4-week-old H9-CMs (below).
(C) Electrical field stimulation induces Ca2+ transients, as shown by Ca2+ imaging (Min and Max level, top) and a representative trace (bottom), in HDF-derived iCMs 6 weeks after 7F retroviral infection. (D) Typical action potential measured in 7F-iCMs (n = 3) 10 weeks after induction in comparison with H9-CMs (right).
TGF-β Signaling Promotes Human Cardiac Reprogramming
To probe additional biological pathways that may regulate human cardiac reprogramming, we tested eight compounds that affect iPS cell reprogramming (Huangfu et al., 2008; Lin et al., 2009) for their capacity to influence 7F-mediated cardiac reprogramming. Although none increased the yield of αMHC-mCherry+ cells, SIS3 inhibited induction of αMHC-mCherry+ cells (Figure 6A). SIS3 specifically inhibits SMAD3 (Jinnin et al., 2006), a transcription factor that is activated downstream of TGF-β signaling, suggesting that activation of the TGF-β signaling pathway is important for 7F human cardiac reprogramming. Adding activin A or TGF-β1 did not enhance 7F reprogramming, but TGF-β1 more than doubled the number of αMHC-mCherry+ cells generated by 5F reprogramming, similar to the levels observed with 7F reprogramming (Figure 6B). SIS3 (10 μM) reversed the effect. We did not find any evidence that the increase in iCM numbers was the result of increased proliferation of iCMs in a dye-dilution assay (Figure 6C), indicating that TGF-β1 likely plays an early inductive role in the reprogramming process.
Figure 6.
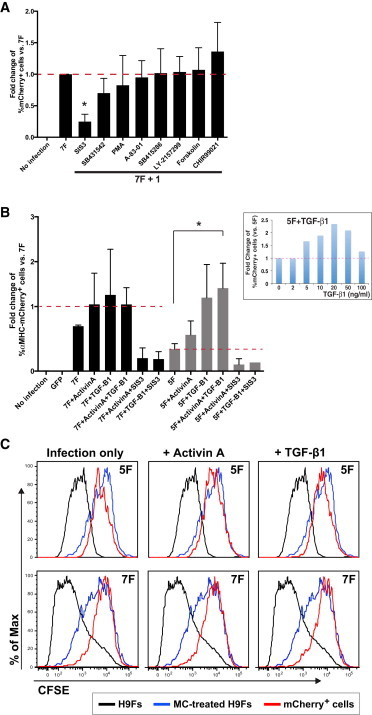
Human iCM Reprogramming Involves TGF-β1 Signaling
(A) Results from exposure to eight small molecules show the relative degree of αMHC-mCherry+ cell induction 2 weeks after 7F retroviral infection (n = 3). Cells were exposed to individual compounds from days 1–14 after 7F infection at the following doses: A-83-01 (0.5 μM), Chir99201 (3 μM), forskolin (5 μM), LY-2157299 (300 nM), PMA (100 nM), SIS3 (10 μM), SB415286 (10 μM), and SB431542 (10 μM). Data represent the mean ± SD from independent experiments.
(B) Effects of activin A (20 ng/ml) and TGF-β1 (20 ng/ml) on αMHC-mCherry+ cell induction 2 weeks after retroviral infection with five or seven factors (n = 3). The inset panel represents the effects of TGF-β1 at different doses. ∗p < 0.001. The SIS3 dose was 10 μM. Data represent the mean ± SD from independent experiments.
(C) A cell proliferation assay using CFSE revealed that activin A and TGF-β1 did not increase the proliferation of iCMs. Noninfected H9Fs (black line) were used as a positive control of cell division, as indicated by decreased fluorescence intensity. Mitomycin C (MC)-treated H9Fs (gray line) were used as negative controls for proliferation and demonstrated high fluorescence intensity. H9Fs were treated with activin A or TGF-β1 from days 1–14 after 5F or 7F infection. Images are representative of three independent experiments.
Discussion
Here, we have shown that GATA4, MEF2C, TBX5, ESRRG, MESP1, MYOCD, and ZFPM2 together reprogram human fibroblasts toward a CM-like state. Human iCMs upregulated hundreds of CM-enriched genes, downregulated hundreds of fibroblast-enriched transcripts, and assembled sarcomeres. Reprogrammed cells were epigenetically similar to CMs at the loci tested. Although most cells were partially reprogrammed, ∼20% generated functional Ca2+ transients, and some fired action potentials similar to those of human CMs. TGF-β signaling was important for, and improved the efficiency of, human iCM reprogramming.
In addition to GMT, which coactivates the expression of many cardiac genes (Garg et al., 2003; Ghosh et al., 2009; He et al., 2011), ESRRG and MESP1 also promoted human cardiac reprogramming. ESRRG (also known as nuclear receptor NR3B3) is a key regulator of mitochondrial biogenesis. Overexpressing ESRRG in rat neonatal cardiac myocytes activates transcripts encoding sarcoplasmic reticulum Ca2+-handling proteins and contractile protein isoforms that are specifically expressed in adult ventricles (Dufour et al., 2007). MESP1 acts as a key transcriptional regulator of cardiovascular specification during development and resides at the top of a gene network that determines cardiovascular cell fate (Bondue et al., 2008). Interestingly, MESP1 and ETS2 together reprogram HDFs into cardiac progenitor-like cells (Islas et al., 2012), highlighting the central role of MESP1 in cardiac cell fate.
Adding MYOCD and ZFPM2 to the 5F mix quantitatively and qualitatively improved human cardiac reprogramming. Myocardin, encoded by MYOCD, is a smooth- and cardiac-muscle-enriched transcriptional coactivator of serum response factor (Wang et al., 2001). It maintains CM structure and sarcomeric organization, and its cell-autonomous loss in CMs triggers programmed cell death (Huang et al., 2009). Although Myocardin promotes the smooth muscle fate in mouse fibroblasts (Chen et al., 2002), in the context of other cardiac transcription factors shown here, it appears to activate cardiac rather than smooth muscle genes. The zinc-finger protein ZFPM2 (also known as friend of Gata-2 [FOG2]) is a cofactor of GATA transcription factors and is essential in heart morphogenesis (Tevosian et al., 2000; Svensson et al., 2000). ZFPM2 activates and represses the expression of GATA target genes (Lu et al., 1999). Single-cell analyses suggest that MYOCD and MESP1, with GMT, were most consistently activated in reprogrammed cells, and highlight the value of interrogating individual cells among heterogeneous reprogrammed cells. These transcription factors likely function in interrelated reinforcing networks to regulate gene expression. Addition of TGF-β1 may substitute for MYOCD and ZFPM2, and future studies will explore this key signaling cascade in cardiac reprogramming.
Although the conversion of human fibroblasts to cells with broad gene expression shifts and development of sarcomeres is encouraging, it takes longer to reprogram human cells than mouse cells, and the process remains inefficient. Very recently, it was reported that human fibroblasts could be reprogrammed toward a cardiac cell fate by four cardiac transcription factors and two miRNAs, but only rare beating cells were observed after a long period in culture (Nam et al., 2013). Consistent with this, most cells in our study were also only partially reprogrammed, with no visible contractile activity after a long time in culture. We found that the resting membrane potential in human iCMs was stably hyperpolarized, similar to adult quiescent CMs but in contrast to the less mature ESC-derived fetal CMs (Fu et al., 2011). Since some cells could elicit good cardiac action potentials following electrical stimulation, it will be important to identify the epigenetic barriers that prevent more complete reprogramming. Efforts to develop functional screens with small molecules and secreted proteins that may improve reprogramming in vitro are under way. Nevertheless, our identification of a cocktail of factors that globally shift fibroblasts toward a CM-like state is an important step. Based on the comparable degree of reprogramming we observed between human and mouse in vitro iCMs, we speculate that the human factors reported here may function better in vivo, as is the case for GMT-induced reprogramming in mice (Qian et al., 2012; Song et al., 2012). It will be important to determine whether the five or seven factors described here more fully reprogram iCMs in vivo, perhaps in a nonhuman primate model.
Experimental Procedures
hESC Culture and Differentiation
hESCs and discarded and deidentified human tissues were used in accordance with the policies of the University of California, San Francisco, Institutional Review Board. Stable αMHC-mCherry-expressing H9 hESCs (Kita-Matsuo et al., 2009) were grown on Matrigel (BD) in mTeSR1 media (StemCell Technologies). CMs were differentiated using the matrix sandwich method (Zhang et al., 2012) and harvested at 3 weeks postdifferentiation by FACS Aria II (BD Biosciences).
For fibroblast differentiation from αMHC-mCherry H9 hESCs, EBs were formed from enzymatically dispersed hESCs suspended in low-attachment plates for 7 days, and then plated on gelatin-coated dishes and cultured in medium (Dulbecco’s modified Eagle’s medium [DMEM]/20% fetal bovine serum [FBS]) for another 14 days. Alternatively, αMHC-mCherry H9 cells were subcutaneously transplanted into NOD-SCID mice to form teratomas as previously described (Thomson et al., 1998). Teratomas were removed 3 months after cell injection, minced, and cultured in medium (DMEM/20% FBS). The migrated cells were harvested and filtered with 40 μm cell strainers (BD). The screening experiments to identify the minimal combination of reprogramming factors for human cardiac reprogramming were done only in EB-differentiated fibroblasts, and other results of characterization were collected from both types of H9Fs, which showed no significant difference.
For αMHC-mCherry−/THY1+ human-cell sorting, cells were incubated with allophycocyanin (APC)-conjugated anti-human THY1 antibody (eBioscience) and sorted by FACS Aria II (BD Biosciences). The purified fibroblasts were cultured and expanded in M106 with a 1:3 split ratio, and then frozen for reprogramming experiments. A total of 33 batches of fibroblasts were differentiated from EBs or teratomas. Eight of these batches did not proliferate well after purification by FACS with THY1 staining and were not used in our studies.
There was some variance of reprogramming efficiency among different batches of fibroblasts; therefore, the data for each experiment were collected from more than three different batches of fibroblasts. We observed that the reprogramming efficiency was dramatically decreased in fibroblasts with a high passage number (n > 13); therefore, fibroblasts with a low passage number (n = 6–12) were used in the current study. Fibroblasts were reseeded on gelatin-coated plates at a density of 104 cells/cm2 1 day before retroviral infection.
HDFs were purchased from PromoCell. HCFs were isolated from discarded and deidentified human heart tissue, and THY1+ HCFs were purified by FACS and expanded as described above.
Molecular Cloning and Retroviral Infection
To construct pMXs retroviral vectors, we amplified the coding regions of candidate genes by PCR and subcloned them into pMXs vector. The pMXs retroviral vectors and retroviral packaging vectors (Alstem) were transfected into HEK293 FT cells using FuGENE 6 (Promega) to generate viruses with ∼3 × 106 IFU/ml titer. The viruses yielded a transduction efficiency of 80%–90% in human fibroblasts, as indicated by GFP retroviral infection. Virus-containing supernatants were pooled and used for transduction of human fibroblasts. The virus-containing medium was replaced with 10% FBS of DMEM/M199 (4:1) medium 24 hr after retroviral infection and changed every 3 days. RPMI1640 with B27 was used 2 weeks after retroviral infection to curb proliferative cell overgrowth in some long-time-cultured iCMs.
For the Dox-inducible system, we used the Retro-X-Tet-ON Advanced Systems (Clontech) according to the manufacturer’s recommendations. To construct pRetro-X-Tight-cDNA vectors, we amplified the coding regions of EGFP, ESRRG, GATA4, MEF2C, MESP1, TBX5, MYOCD, and ZFPM2 by PCR and subcloned them into pRetro-X-Tight-Puro vector. The pRetro-X-Tet-On Advanced-Neo or pRetro-X-tight-cDNA retroviral vectors and packaging constructs (Alstem) were transfected into HEK293 FT cells with FuGene 6 (Promega) to generate viruses, and virus-containing supernatants were collected after 48 hr. Cells were transduced with Retro-X-Tet ON and Retro-X-Tight-cDNA overnight supplemented with 5 μg/ml polybrene, and purified by positive selection with puromycin and neomycin. Dox (100 ng/ml) was used for gene induction.
FACS Analyses and Sorting
For mCherry expression analyses, cultured cells were dissociated and analyzed on the LSR-II (BD) with FlowJo software. αMHC-mCherry−/THY1+ H9Fs were purified with APC-conjugated anti-human THY1 antibody (eBioscience) (Ieda et al., 2009). For αMHC-mCherry+/cTNT+ assay, cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.5% w/v saponin, and stained with anti-dsRed (AbCam) and anti-cTNT (Thermo Scientific) antibodies, followed by secondary antibodies conjugated to Alexa-594 and Alexa-647 (Invitrogen). For c-KIT analysis, cells were incubated with PECy5.5-conjugated anti-c-KIT antibody. We used undifferentiated H9 ESCs as a positive control for c-KIT staining.
Immunocytochemistry and Histology
Cells were fixed in 4% PFA for 15 min at room temperature, permeabilized with 0.5% w/v saponin, blocked in 5% normal goat serum, and incubated with primary antibodies against red fluorescent protein (AbCam), cardiac αMHC (AbCam), cTNT (Thermo Scientific), sarcomeric α-actinin (Sigma Aldrich), calponin (Sigma Aldrich), smooth muscle actin (Sigma Aldrich), and vimentin (Progen). We used secondary antibodies conjugated to Alexa-488 or -594 (Molecular Probes) and costained cells with DAPI (Invitrogen) during the secondary incubation step.
Transmission Electron Microscopy
For transmission electron microscopy, reprogrammed αMHC-mCherry+ H9Fs (D45 after 7F induction) and H9-CMs (D22 postdifferentiation) were fixed in 1% formaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4, postfixed in 2% osmium tetroxide in the same buffer, block stained with 2% aqueous uranyl acetate, dehydrated in acetone, infiltrated, and embedded in LX-112 resin (Ladd Research Industries). Samples were ultrathin sectioned on a Reichert Ultracut S ultramicrotome and counterstained with 0.8% lead citrate. Grids were examined on a JEOL JEM-1230 transmission electron microscope (JEOL USA) and photographed with a Gatan Ultrascan 1000 digital camera (Gatan). The sarcomere structure was identified between two Z bands with continued myofibril connection, and the sarcomere length was quantified by Axiovision (Zeiss).
qRT-PCR
Total RNA was isolated from cells and qRT-PCR was performed on an ABI 7900HT (Applied Biosystems) with the TaqMan probes (Applied Biosystems) listed below. The mRNA levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA.
The following TaqMan probes (Applied Biosystems) were used: ACTC1 (Hs01109515_m1), ACTN2 (Hs00153809_m1), NPPA (Hs00383230_g1), MYH6 (Hs00411908_m1), MYL2 (Hs00166405_m1), MYL7 (Hs00221909_m1), TNNT2 (Hs00165960_m1), ATP2A2 (Hs01566028_g1), PLN (Hs00160179_m1), RYR2 (Hs00892842_m1), COL1A1 (Hs00164004_m1), COL5A2 (Hs00893878_m1), FN1 (Hs00365052_m1), and GAPDH (Hs02758991_g1), CNN1 (Hs00154543_m1), ACTA2 (Hs00426835_g1), OCT4 (Hs00742896_s1), NANOG (Hs02387400_g1), SOX2 (Hs01053049_s1), and TDGF1 (Hs02339496_g1).
For single-cell qPCR assay, purified H9Fs, H9-CMs, and reprogrammed αMHC-mCherry iCMs were harvested by the C1 Single-Cell Auto Prep System (Fluidigm) followed by reverse transcription and preamplification. qPCR was performed with the use of the BioMark HD System (Fluidigm) to profile gene expression in single cells, and analyzed with the SINGuLAR Analysis Toolset (Fluidigm).
Whole-Transcriptome Assay
After 4 or 12 weeks in culture, uninfected H9Fs and 5F and 7F αMHC-mCherry+:THY1− iCMs were collected by FACS. RNA was extracted using the PicoPure RNA Isolation Kit (Arcturus). RNA was hybridized to the Affymetrix Human Gene 1.0 ST Array. Microarrays were performed in triplicate from independent biological samples according to the standard Affymetrix GeneChip protocol. Data were analyzed using the Affymetrix Power Tool (APT, version 1.8.5). For data analysis, linear models were fitted for each gene on the sample group to derive estimated group effects and their associated significance with the limma package (Smyth, 2004) in R/Bioconductor. Moderated t statistics and the associated p values were calculated. We adjusted the p values for multiple testing by controlling for FDR by the Benjamini-Hochberg method. Gene annotations were retrieved from Affymetrix (version Nov. 12, 2007). Differential gene expression was defined using the statistics/threshold combination. Genes that were differentially expressed in at least one comparison (FDR-adjusted p < 0.01) are shown in Figure 4A.
For an orthologous comparison with gene expression from mouse in vivo iCMs, mouse adult CMs were isolated from three reprogrammed (GMT-injected) and three control (dsRed-injected) Periostin-Cre:Rosa-Tomato mice 4 weeks after coronary ligation and intramyocardial viral delivery, as previously described (Qian et al., 2012). Fifty reprogrammed iCMs (based on the presence of red fluorescence) or endogenous CMs from each GMT-injected or dsRed-injected heart were collected using a micropipette. The cells were processed with RNA isolation and purification using the PicoPure RNA Isolation Kit (Arcturus). The microarray assay was performed and data were analyzed as described above. For comparison of orthologous gene expression, genes with 1:1 orthologs between human and mouse that were differentially expressed in human fibroblasts and CMs were identified at an FDR of 0.05. The Pearson rho correlation was then calculated for expression of these genes across all data sets.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed on H9Fs, 7F-iCMs, and H9-CMs with the use of the Imprint Chromatin Immunoprecipitation Kit (Sigma). Antibodies against H3K27me3 and H3K4me3 were obtained from Active Motif, and normal rabbit immunoglobulin G was obtained from Cell Signaling Technology. Primers for qPCR custom TaqMan gene-expression assays (Applied Biosystems) were designed online to target the core promoters of cardiac genes. The primers for qPCR custom TaqMan gene-expression assays (Applied Biosystems) included ACTC1_CP (AIT95RY), ACTN2_CP (AIS07LQ), RYR2_CP (AIRR9FI), PLN_CP (AJ6RM9Q), TNNT2_CP (AIQJA9A), and COL1A1_CP (AJAAYRZ).
Bisulfite Genomic Sequencing
Bisulfite treatment was performed using the Epitect Bisulfite Kit (QIAGEN). Amplified products were cloned into pCR2.1-TOPO (Invitrogen). Nine to ten randomly selected clones were sequenced with the M13 forward and M13 reverse primers for each gene.
The following PCR primer sequences were used for bisulfite sequencing: MYH6-Sense: GGGAGGAATGTGTTTAAGGATTAAAAA; MYH6-Antisense: TACAAAACATCCCACCCCAAACCTC; MYH7-Sense: ATTGGTTGTTTGTGGTTTTGGTGGT; MYH7-Antisense: TAAAAACATTTCCCCCAAACTCCCCC; MYL7-Sense: TTGTTAGGATGGATAGATAGTAGGATGTTATA; and MYL7-Antisense: CCCAAAAACCAACACCTAAAAAACC.
Ca2+ Imaging and Electrophysiology
Cells were labeled with Fluo-4-AM (Invitrogen) for 30 min at 37°C, washed, and incubated for an additional 30 min at room temperature to allow de-esterification of the dye. Fluo-4-labeled cells were analyzed by Axio Observer (Zeiss) with MiCAM02 (SciMedia) at 37°C. Ca2+ transients were elicited in reprogrammed αMHC-mCherry+ cells by electrical field stimulation (20 V, 5 ms at 1 Hz or 0.5 Hz).
Electrophysiological analyses were performed on iCMs 4 or 10 weeks after 7F transduction, by whole-cell recording with an Axopatch 200B amplifier and pClamp10 software (Axon Instruments). Glass patch pipettes, with typical resistances of 2–4 MΩ, were directly attached onto single iCMs (identified by mCherry expression) for whole-cell recording in Tyrode’s bath solution. Using the current-clamp model, action potentials were elicited in single mCherry+ cells with stimulations of 2 nA for 2 ms.
Cell Proliferation Assay
Cell proliferation was investigated with the use of carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen). The day after retroviral infection, H9Fs were incubated with 5 μM CFSE, which passively diffuses into cells and yields highly fluorescent CFSE when the acetate groups are removed by cytoplasmic esterases. The CFSE is divided equally among daughter cells following division (Lyons et al., 2001). Cumulative cell proliferation from day 1 to day 14 can be evaluated by analyzing the fluorescence intensity 2 weeks after retroviral induction. H9Fs treated with mitomycin C (10 μg/ml) just before CFSE treatment were used as a negative control (higher fluorescence) and H9Fs were used as a positive control (lower fluorescence) for cell division.
Small-Molecule Compound Testing
H9Fs were treated with individual small-molecule compounds, activin A, or TGF-β1 the day after 5F or 7F retroviral infection, and media with fresh compounds were changed every 3 days for 2 weeks, at which time the reprogramming efficiency was evaluated by FACS. The following doses were used in the current study: A-83-01 (0.5 μM), Chir99201 (3 μM), forskolin (5 μM), LY-2157299 (300 nM), phorbol-12-myristate-13-acetate (PMA, 100 nM), SIS3 (10 μM), SB415286 (10 μM) SB431542 (10 μM), activin A (20 ng/ml), and TGF-β1 (20 ng/ml).
Statistical Analyses
Data were collected from independent biological replicates with the indicated n values and are expressed as mean ± SD. Differences between groups were examined for statistical significance using Student’s t test, ANOVA, or Kruskal-Wallis followed by Tukey’s test for multiple comparisons; p values < 0.05 were regarded as significant.
Acknowledgments
We thank members of the Srivastava lab for critical discussions and comments on the manuscript, A. Lucido and G. Howard for editorial assistance, B. Taylor for manuscript preparation, and R. Chadwick and Y. Hao (Gladstone Genomics Core), J. Wong (Gladstone Electron Microscopy Core), and S. Thomas and A. Williams (Gladstone Bioinformatics Core) for their assistance. We also thank M. Mercola for providing the αMHC-mCherry H9 ESCs. This work was supported by grants from NHLBI/NIH (U01HL100406 and P01HL089707, to D.S. and B.G.B.), the California Institute of Regenerative Medicine (RB3-05174 to D.S. and RN2-00903-1 to B.G.B.), the Younger Family Foundation, and the Eugene Roddenberry Foundation. D.S. is also supported by the Whittier Foundation. B.G.B. holds the Lawrence J. and Florence A. DeGeorge Charitable Trust/American Heart Association (AHA) Established Investigator Award. J.D.F. is supported by a Scientist Development Grant from the AHA. N.R.S. is supported by a graduate research fellowship from the NSF. P.D.O was a Scholar of the California Institute for Regenerative Medicine. The J. David Gladstone Institutes received support from the National Center for Research Resources (grant RR18928-01).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
All microarray gene expression data reported in this study are available from the NCBI Gene Expression Omnibus (GEO), accession number GSE49192.
Supplemental Information
Electrical field stimulation induced Ca2+ transients in αMHC-mCherry+ 7F-iCMs 4 weeks after retroviral infection.
Electrical field stimulation induced Ca2+ transients in HDF-derived iCMs 6 weeks after 7F retroviral infection.
Note Added in Proof
During the review of this paper, Wada et al. reported that a combination of GMT plus Myocardin and MESP1 could also reprogram human fibroblasts toward the cardiomyocyte phenotype.
Wada, R., Muraoka, N., Yamakawa, H., Miyamoto, K., Sadahiro, T., Umei, T. Kaneda, R., Suzuki, T., Kamiya, K., Tohyama, S., Yuasa, S. Kokaji, K., Aeba, R., Yozu, R., Yamagishi, H., Kitamura, T., Fukuda, K., Ieda, M. (2013) Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA. 110, 12667–12672.
References
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M., Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Chen J., Kitchen C.M., Streb J.W., Miano J.M. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dufour C.R., Wilson B.J., Huss J.M., Kelly D.P., Alaynick W.A., Downes M., Evans R.M., Blanchette M., Giguère V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Fu J.D., Rushing S.N., Lieu D.K., Chan C.W., Kong C.W., Geng L., Wilson K.D., Chiamvimonvat N., Boheler K.R., Wu J.C. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2011;6:e27417. doi: 10.1371/journal.pone.0027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V., Kathiriya I.S., Barnes R., Schluterman M.K., King I.N., Butler C.A., Rothrock C.R., Eapen R.S., Hirayama-Yamada K., Joo K. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Ghosh T.K., Song F.F., Packham E.A., Buxton S., Robinson T.E., Ronksley J., Self T., Bonser A.J., Brook J.D. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol. Cell. Biol. 2009;29:2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A., Kong S.W., Ma Q., Pu W.T. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Cook E.G., Gao Q., Mitalipova M., Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Min Lu M., Cheng L., Yuan L.J., Zhu X., Stout A.L., Chen M., Li J., Parmacek M.S. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc. Natl. Acad. Sci. USA. 2009;106:18734–18739. doi: 10.1073/pnas.0910749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A.E., Melton D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudon-David F., Bouzeghrane F., Couture P., Thibault G. Thy-1 expression by cardiac fibroblasts: lack of association with myofibroblast contractile markers. J. Mol. Cell. Cardiol. 2007;42:991–1000. doi: 10.1016/j.yjmcc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Ieda M., Tsuchihashi T., Ivey K.N., Ross R.S., Hong T.T., Shaw R.M., Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagawa K., Miyamoto K., Yamakawa H., Muraoka N., Sadahiro T., Umei T., Wada R., Katsumata Y., Kaneda R., Nakade K. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 2012;111:1147–1156. doi: 10.1161/CIRCRESAHA.112.271148. [DOI] [PubMed] [Google Scholar]
- Islas J.F., Liu Y., Weng K.C., Robertson M.J., Zhang S., Prejusa A., Harger J., Tikhomirova D., Chopra M., Iyer D. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc. Natl. Acad. Sci. USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnin M., Ihn H., Tamaki K. Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Molecular pharmacology. 2006;69:597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- Kita-Matsuo H., Barcova M., Prigozhina N., Salomonis N., Wei K., Jacot J.G., Nelson B., Spiering S., Haverslag R., Kim C. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS ONE. 2009;4:e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H.S., Hao E., Hayek A., Ding S. A chemical platform for improved induction of human iPSCs. Nat. Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.R., McKinsey T.A., Xu H., Wang D.Z., Richardson J.A., Olson E.N. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons A.B., Hasbold J., Hodgkin P.D. Flow cytometric analysis of cell division history using dilution of carboxyfluorescein diacetate succinimidyl ester, a stably integrated fluorescent probe. Methods Cell Biol. 2001;63:375–398. doi: 10.1016/s0091-679x(01)63021-8. [DOI] [PubMed] [Google Scholar]
- Nam Y.J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J.A., DiMaio J.M., Baker L.A., Bassel-Duby R., Olson E.N. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S., Khattak S., Poulet C., Lindemann D., Tanaka E.M., Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell. Cardiol. 2012;53:323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkisoensing A.A., Pijnappels D.A., Askar S.F., Passier R., Swildens J., Goumans M.J., Schutte C.I., de Vries A.A., Scherjon S., Mummery C.L. Human embryonic and fetal mesenchymal stem cells differentiate toward three different cardiac lineages in contrast to their adult counterparts. PLoS ONE. 2011;6:e24164. doi: 10.1371/journal.pone.0024164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson E.C., Huggins G.S., Lin H., Clendenin C., Jiang F., Tufts R., Dardik F.B., Leiden J.M. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat. Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- Tevosian S.G., Deconinck A.E., Tanaka M., Schinke M., Litovsky S.H., Izumo S., Fujiwara Y., Orkin S.H. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Chang P.S., Wang Z., Sutherland L., Richardson J.A., Small E., Krieg P.A., Olson E.N. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang Z., Fermini B., Nattel S. Delayed rectifier outward current and repolarization in human atrial myocytes. Circ. Res. 1993;73:276–285. doi: 10.1161/01.res.73.2.276. [DOI] [PubMed] [Google Scholar]
- Yoo A.S., Sun A.X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R.E., Tsien R.W., Crabtree G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Klos M., Wilson G.F., Herman A.M., Lian X., Raval K.K., Barron M.R., Hou L., Soerens A.G., Yu J. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electrical field stimulation induced Ca2+ transients in αMHC-mCherry+ 7F-iCMs 4 weeks after retroviral infection.
Electrical field stimulation induced Ca2+ transients in HDF-derived iCMs 6 weeks after 7F retroviral infection.


