Abstract
The neuronal ceroid lipofuscinoses (NCLs) are a group of autosomal recessive neurodegenerative diseases characterized by the accumulation of autofluorescent lipopigment in various tissues and by progressive cell death in the brain and retina. The gene for variant late-infantile NCL (vLINCL), CLN6, was previously mapped to chromosome 15q21-23 and is predicted to be orthologous to the genes underlying NCL in nclf mice and in South Hampshire and Merino sheep. The gene underlying this disease has been identified with six different mutations found in affected patients and with a 1-bp insertion in the orthologous Cln6 gene in the nclf mouse. CLN6 encodes a novel 311–amino acid protein with seven predicted transmembrane domains, is conserved across vertebrates and has no homologies with proteins of known function. One vLINCL mutation, affecting a conserved amino acid residue within the predicted third hydrophilic loop of the protein, has been identified, suggesting that this domain may play an important functional role.
The neuronal ceroid lipofuscinoses (NCLs) are a group of autosomal recessive neurodegenerative diseases characterized by the accumulation of autofluorescent lipopigment in various tissues (Santavuori 1988). Clinical features include visual failure, seizures, developmental regression, and progressive mental and motor deterioration. Naturally occurring NCLs are found in many animal species, as well as humans, in whom at least seven genetically distinct subtypes are known (Goebel et al. 1999). Six NCL genes have been identified to date; three encode lysosomal enzymes (PPT1 [Vesa et al. 1995], TPP1 [Sleat et al. 1997; Vines and Warburton 1999], and CTSD [Tyynelä et al. 2000]), and three encode predicted transmembrane proteins (CLN3 [International Batten Disease Consortiums 1995], CLN5 [Savukoski et al. 1998], and CLN8 [Ranta et al. 1999]). We mapped CLN6, the gene for a variant late-infantile NCL (vLINCL [MIM 601780]) to chromosome 15q21-23, by homozygosity mapping, in two consanguineous families (Sharp et al. 1997). Further studies in additional families narrowed the critical region to <1 cM (Sharp et al. 1999, 2001).
A BAC contig across the refined CLN6 critical region (Sharp et al. 2001; R. B. Wheeler and J. D. Sharp, unpublished data) was constructed using a combination of conventional physical-mapping methods and an in silico approach. BLAST (Altschul et al. 1990), NIX, and the Human Genome Browser were used to identify overlapping BACs. Then, these BACs were analyzed using the Tandem Repeat Finder program (Benson 1999) to identify new polymorphic markers across the contig. Two of these markers (55782C and 90734.1) further refined the critical region, to a distance that was encompassed by seven BACs between AC055782 and AC090734 (see supplementary information and the Human Genome Browser). Transcripts were identified within the critical region, by use of homology searches and gene-prediction programs (NIX and the Human Genome Browser). These included four known genes (DDXBP1, FEM1B, ITGA11, and CLIPINC) and three novel genes with EST and cDNA homologies (associated with Unigene clusters Hs.239812, Hs.148759, and Hs.43654). On the basis of functional information, none of the seven genes could be prioritized as a disease candidate. Therefore, a systematic mutation-screening approach was adopted. For each of the transcripts, we obtained full-length cDNA sequence, identified exon-intron boundaries, and performed expression analysis and mutation screening (R. B. Wheeler and J. D. Sharp, unpublished data).
The cDNA AK000568 (Genome Database), coding for the hypothetical protein FLJ20561, was associated with Unigene cluster Hs.43654. This cDNA was mapped back to genomic sequence by use of BLAST to identify exon-intron boundaries (table 1). Genomic primers were designed for all seven exons (see supplementary information), and sequence analysis of patient genomic DNA revealed six different mutations in 15 families affected with vLINCL (table 2; fig. 1 identifying this gene as CLN6. AK000568 has a predicted open reading frame of 936 nucleotides and comprises seven exons which span a genomic region of ∼22 kb. One nonsense mutation and three frameshift mutations are predicted to result in the production of a truncated protein. A missense mutation and a mutation that results in the deletion of a single amino acid were identified in exon 4.
Table 1.
Genomic Structure of CLN6, Showing Splice-Site Junctions
|
Splice Sitea |
||
| Exon | 3′ | 5′ |
| 1 | GCGCGCTCTCb | TGCAGGCCAGgtgggcgcgc |
| 2 | tccttcccagGCATGGCTCT | CATTGCCATGgtgagtgtga |
| 3 | tgctccgcagCTGGTATTCC | CTTGCTCAAGgtactgtccc |
| 4 | tgaaccccagCTCATCGAGC | GGAGACGCTGgtgaggccac |
| 5 | cctccctcagATCGACTCCT | ACTGCATGTGgtgagtgatg |
| 6 | cctgtcacagGTACATCCCC | TGTACTACTGgtgagtggac |
| 7 | gcctttccagGTACCTGGTC | ACCGAGAGCATGA |
Exon sequence is shown in uppercase; intron sequence is shown in lowercase.
The 3′ splice site for exon 1 is the start of cDNA sequence AK000568.
Table 2.
CLN6 Mutations in Patients with vLINCL[Note]
| Geographic Origin,Genotype, andNucleotide Change | No. ofFamilies | Mutation | Amino Acid Change/Predicted Consequence | Location(Exon) | Restriction-SiteChange |
| Greece: | |||||
| Homozygous: | |||||
| c.6delG | 1 | 1-bp deletion | Frameshift after E2 (extra 29 amino acids) | 1 | MnlI (loss) |
| Costa Rica: | |||||
| Heterozygousa | 1 | ||||
| Homozygous: | |||||
| c.214G→T | 6 | Nonsense | E72X | 3 | BfaI (gain) |
| c.368G→A | 1 | Missense | G123D | 4 | BanI (loss) |
| Pakistan: | |||||
| Heterozygousa,b | 1 | ||||
| Homozygous: | |||||
| c.316insCb | 2 | 1-bp insertion | Frameshift after P105 (extra 25 amino acids) | 4 | None |
| India: | |||||
| Homozygous: | |||||
| c.395_397delCT | 1 | 2-bp deletion | Frameshift after D131 (extra 17 amino acids) | 4 | HinfI (loss) |
| Portugal: | |||||
| Heterozygousa | 1 | ||||
| Homozygous: | |||||
| c.460_462delATC | 1 | 3-bp deletion | I154del | 4 | None |
Note.— For mutations in exons 1, 3, and 4, respectively, 26, 30, and 34 normal chromosomes were sequenced. Mutation nomenclature is as recommended by den Dunnen and Antonarakis (2001).
The second mutation has not been identified in these patients.
This mutation is identical to that in the nclf mouse.
Figure 1.

Electropherograms showing genomic nucleotide sequence across mutation sites in two patients and the nclf mouse. a, Homozygous 1-bp insertion in exon 4 in a Pakistani patient (c.316insC). b, Normal control sequence. c, A 3-bp deletion in exon 4 in a Portuguese patient (c.460_462delATC). d, Normal control sequence. e, A homozygous 1-bp insertion in exon 4 in the nclf mouse (c.301insC). f, Wild-type sequence. The nclf mouse DNA was obtained from The Jackson Laboratory (stock no. 002628). Mutations were detected by direct sequencing of genomic PCR products. PCR and sequencing primers are listed in the supplementary information. Each primer pair amplified the corresponding exon and the flanking splice sites. PCR products were sequenced using ABI dye terminator chemistry (Applied Biosystems) and were visualized on an ABI 373 sequencer (Applied Biosystems). Mutations were confirmed by sequencing in both directions.
Mouse ESTs homologous to CLN6 were identified by BLAST analysis, and a partial cDNA sequence was obtained which spans from exon 2 to the 3′ UTR. Additional 5′ sequence was identified from the mouse BLAT alignments in the Human Genome Browser, and the complete mouse cDNA was assembled. Five of the seven exons were present in trace sequencing archives, and genomic primers were designed for these (supplementary information for exon 4 primers). A corresponding mouse Unigene cluster was identified, Mm.30232, which maps to mouse chromosome 9 in the region of the nclf locus (Bronson et al. 1998). A 1-bp insertion (c.307insC, causing a frameshift after P102) was identified in exon 4 of Cln6 (fig. 1e) in the nclf mouse model (Bronson et al. 1998).
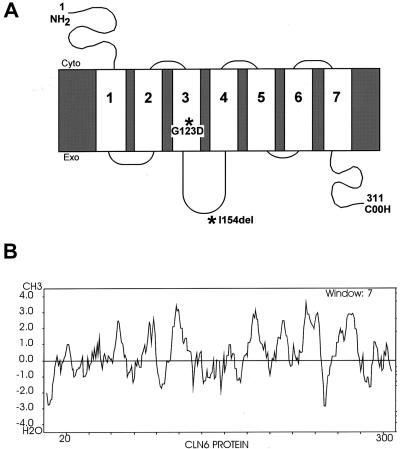
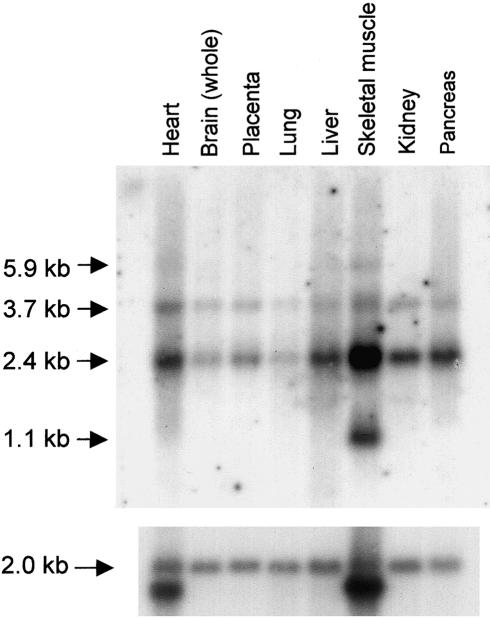
The CLN6 gene is predicted to encode a 311–amino acid protein with seven transmembrane domains (fig. 2) on the basis of TMHMM and TMAP transmembrane prediction programs and hydrophobicity profiles (Kyte and Doolittle 1982). Like other NCL membrane proteins (CLN3, CLN5, and CLN8), CLN6 has no homology with known proteins or functional domains, although the sequence is highly conserved across vertebrate species (data not shown). There is 90% identity between the human and mouse amino acid sequence (fig. 3). ESTs corresponding to human CLN6 originated from a wide variety of tissue libraries, and northern blot analysis confirmed ubiquitous expression of CLN6 (fig. 4). A transcript of 2.4 kb, consistent with the cDNA being nearly full length, was present in all tissues. In addition, a transcript of 3.7 kb was detected in all tissues, and transcripts of 1.1 kb and 5.9 kb were present in skeletal muscle.
Figure 2.
A, Schematic representation of CLN6 structure, showing the seven predicted transmembrane domains based on TMHMM and TMAP programs and hydrophobicity profiles. Single–amino-acid changes are indicated by asterisks (*). Exoplasmic and cytoplasmic surfaces are indicated. B, Kyte and Doolittle hydrophobicity plot of the predicted CLN6 protein. All prediction programs were run using the Biology Workbench.
Figure 3.
A comparison between human and mouse CLN6 amino acid sequences. Dark shading indicates identical amino acids, and gray shading indicates similar amino acids. Asterisks (*) denote the amino acid removed by the 3-bp deletion in a Portuguese patient (I154del) and the missense mutation in the Costa Rican patient (G123D). Human and mouse protein sequences were aligned using ClustalW (Thompson et al. 1997) and MacBoxshade 2.15E.
Figure 4.
Human multiple-tissue northern blot (Clontech) probed with a [32P]-labeled 808-bp fragment of CLN6 cDNA spanning from exon 3 to the 3′ UTR, according to the manufacturer's instructions. In all tissues, 2.4-kb and 3.7-kb transcripts were detected, with additional transcripts of 1.1 kb and 5.9 kb detected only in skeletal muscle. A β-actin probe was used as a control for lane loading.
The majority of mutations result in a frameshift or nonsense change, with the introduction of a premature stop codon (table 2). However, one Portuguese patient was homozygous for a 3-bp deletion, in exon 4 (c.460_462delATC), that is predicted to remove a single amino acid (I154del) within the predicted third hydrophilic loop of the protein (fig. 2a). This residue is within a region of the protein that is highly conserved across at least five species (human, mouse, cow, pig, and chicken), suggesting that it is likely to have an important role in the function of the protein. In addition, a Costa Rican patient was homozygous for an exon 4 missense mutation (c.368G→A) that changes glycine to aspartic acid (G123D) within the predicted third transmembrane domain. The introduction of a charged amino acid is predicted to disrupt this domain.
A homologous region of exon 4 is mutated in the nclf mouse and in three families of Pakistani origin, creating an identical gene defect (see table 2). The genomic mutation creates an insertion of a single cytosine within a run of six cytosine residues, suggesting that it arises from a common mutation mechanism: slipped mispairing during DNA replication.
Unlike other forms of NCL, there is no evidence to date for a common CLN6 mutation. We previously speculated on a common ancestry between Romany Gypsies and natives of India and Portugal (Sharp et al. 1999). However, the current data do not support this hypothesis, since no exon 4 mutations have been detected in a subset of Czech Romany Gypsy patients. A local founder effect is present in Costa Rican subjects, with six of eight patients homozygous for the same mutation in exon 3 (c.214G→T, E72X). However, one family was heterozygous for this mutation, and a second family was homozygous for a different mutation in exon 4 (c.368G→A, G123D). The higher-than-expected number of mutations within this small inbred population provides evidence that CLN6 may be a highly mutable gene. Two distinct mutations have also been identified in three Pakistani families, providing further evidence to support this observation (table 2).
CLN6 is the second gene to be identified with a corresponding mutation in the murine ortholog (Ranta et al. 1999). In this case, it is also predicted to be orthologous to NCL-causing genes in two large animal models, the South Hampshire sheep (Broom and Zhou 2001) and the Merino sheep (Tammen et al. 2001), which will be important in understanding the etiology of the disease. The availability of these animal models should allow exploration of the reasons for phenotypic differences between mammalian species carrying mutations at one particular locus.
With the identification of the CLN6 gene, NCL-associated mutations are now defined in four novel membrane proteins (CLN3, CLN5, CLN6, and CLN8) that lead to a common cellular phenotype. These proteins may all participate in an important basic biological pathway that remains to be elucidated.
Acknowledgments
This work would not have been possible without the help and cooperation of the patients and their parents and physicians. We especially thank Mrs. Paula Mendes, for providing DNA and clinical information on a subset of Portuguese families. We thank Professor Brian Lake and Professor Milan Elleder, for ultrastructural analysis on patients. This work was funded by The Wellcome Trust (grant 054606/Z/98/Z/JRS/JP/JAT), The Medical Research Council (grant G9706148), The Children’s Brain Diseases Foundation, and the Batten Disease Support and Research Association. We also thank Mr. Keith Parker and Dr. Alicia White, for expert technical assistance, and Dr. Hannah Mitchison, for critical reading of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Biology Workbench, http://workbench.sdsc.edu/ (for TMHMM and TMAP transmembrane prediction programs)
- Genome Database, http://gdbwww.gdb.org/ (for cDNA coding for the hypothetical protein FLJ20561; accession number AK000568)
- Human Genome Browser, http://genome.ucsc.edu/goldenPath/hgTracks.html (for BAC sequences AC055782 to AC090734, homologies, and BLAT)
- MacBoxshade 2.15E, http://www.isrec.isb-sib.ch/ftp-server/boxshade/MacBoxshade/ (for human and mouse protein sequence alignment)
- NIX, http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/ (for homologies and gene predictions)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CLN6 [MIM 601780])
- Unigene, http://www.ncbi.nlm.nih.gov/UniGene/ (for human and mouse Unigene clusters)
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson RT, Donahue LR, Johnson KR, Tanner A, Lane PW, Faust JR (1998) Neuronal ceroid lipofuscinosis (nclf), a new disorder of the mouse linked to chromosome 9. Am J Med Genet 77:289–297 [DOI] [PubMed] [Google Scholar]
- Broom MF, Zhou C (2001) Fine mapping of ovine ceroid lipofuscinosis confirms orthology with CLN6. Europ J Paediatr Neurol Suppl A 5:33–36 [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE (2001) Nomenclature for the description of human sequence variations. Hum Genet 109:121–124 [DOI] [PubMed] [Google Scholar]
- Goebel HH, Mole SE, Lake BD (eds) (1999) The neuronal ceroid lipofuscinoses (Batten disease). IOS Press, Amsterdam [Google Scholar]
- International Batten Disease Consortium, The (1995) Isolation of a novel gene underlying Batten disease, CLN3. Cell 82:949–957 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathy character of a protein. J Mol Biol 157:105–132 [DOI] [PubMed] [Google Scholar]
- Ranta S, Zhang Y, Ross B, Lonka L, Takkunen E, Messer A, Sharp J, Wheeler R, Kusumi K, Mole S, Liu W, Soares MB, de Fatima Bonaldo M, Hirvasniemi A, Chapelle ADL, Gilliam TC, Lehesjoki AE (1999) The neuronal ceroid lipofuscinoses in human EPMR and mnd mutant mice are associated with mutations in CLN8. Nat Genet 23:233–236 [DOI] [PubMed] [Google Scholar]
- Santavuori P (1988) Neuronal ceroid lipofuscinosis in childhood. Brain Dev 10:80–83 [DOI] [PubMed] [Google Scholar]
- Savukoski M, Klockars T, Holmberg V, Santavuori P, Lander ES, Peltonen L (1998) CLN5, a novel gene encoding a putative transmembrane protein mutated in Finnish variant late infantile neuronal ceroid lipofuscinosis. Nat Genet 19:286–288 [DOI] [PubMed] [Google Scholar]
- Sharp JD, Wheeler RB, Lake BD, Fox M, Gardiner RM, Williams RE (1999) Genetic and physical mapping of the CLN6 gene on chromosome 15q21-23. Mol Genet Metab 66:329–331 [DOI] [PubMed] [Google Scholar]
- Sharp JD, Wheeler RB, Lake BD, Savukowski M, Järvelä IE, Peltonen L, Gardiner RM, Williams RE (1997) Loci for classical and a variant late infantile neuronal ceroid lipfuscinoses map to chromosomes 11p15 and 15q21-23. Hum Mol Genet 6:591–596 [DOI] [PubMed] [Google Scholar]
- Sharp JD, Wheeler RB, Schultz RA, Joslin JM, Mole SE, Williams RE, Gardiner RM (2001) Analysis of candidate genes in the CLN6 critical region using in silico cloning. Europ J Paediatr Neurol Suppl A 5:29–31 [DOI] [PubMed] [Google Scholar]
- Sleat DE, Donnelly RJ, Lackland H, Liu C-G, Sohar I, Pullarkat RK, Lobel P (1997) Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science 277:1802–1805 [DOI] [PubMed] [Google Scholar]
- Tammen I, Cook RW, Nicholas FW, Raadsma HW (2001) Neuronal ceroid lipofuscinosis in Australian Merino sheep: A new animal model. Europ J Paediatr Neurol Suppl A 5:37–41 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynelä J, Sohar I, Sleat DE, Gin RM, Donnelly RJ, Baumann M, Haltia M, Lobel P (2000) A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J 19:2786–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesa J, Hellsten E, Verkruyse LA, Camp LA, Rapola J, Santavuori P, Hofmann SL, Peltonen L (1995) Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376:584–587 [DOI] [PubMed] [Google Scholar]
- Vines DJ, Warburton MJ (1999) Classical late infantile neuronal ceroid lipofuscinosis fibroblasts are deficient in lysosomal tripeptidyl peptidase I. FEBS Lett 443:131–135 [DOI] [PubMed] [Google Scholar]