Abstract
We have analyzed data from 573 pedigrees from the United Kingdom for evidence for linkage to loci influencing adult stature. Our data set comprised 1,214 diabetic and 163 nondiabetic siblings for whom height data were available. We used variance-components analysis implemented in GENEHUNTER 2 and a modification of the Haseman-Elston regression method, HE-COM. We found evidence for a locus on 3p26 (LOD score 3.17) influencing height in this adult sample, with less-significant evidence for loci on chromosomes 7, 10, 15, 17, 19, and 20. Our findings extend similar recent studies in Scandinavian and Quebecois populations, adding further evidence that height is indeed under the control of multiple genes.
Stature—adult height—has been the focus of recent attention as a paradigm of complex traits capable of genetic dissection—highly heritable (⩾80%), easily and accurately measured, and readily available—and recent analyses in Finnish, Quebecois, and Swedish populations have confirmed it to have a significant genetic component (Hirschhorn et al. 2001; Perola et al. 2001). To contribute further to these studies, we have undertaken an analysis of height data in a set of 573 pedigrees previously analyzed for type 2 diabetes as part of the Diabetes U.K. Warren 2 Consortium (Wiltshire et al. 2001). These pedigrees are of European descent and exclusively British/Irish origin for three generations. They were genotyped for 418 autosomal microsatellite markers (mean spacing 9.26 cM [Haldane {H}]), taken predominantly from the ABI Prism MD-10 set, as described elsewhere (Wiltshire et al. 2001). Full details of these markers and the genotyping procedures can be found elsewhere (Wiltshire et al. 2001; Warren 2 Project Information Web site).
Of a total of 1,377 siblings (665 women and 712 men) for whom height data were available, 1,214 had diabetes. The mean height for women was 1.597 m (diabetic women 1.595 m, nondiabetic women 1.607 m; P=.07): mean height for men was 1.739 m (diabetic men 1.740 m, nondiabetic men 1.734 m; P=.60). These compare closely with contemporary U.K. population means of 1.599 m for women and 1.733 m for men for the same age range (Erens and Primatesta 2001).
A square-root transformation was applied to the height data, followed by adjustment for the effects of age, sex, and diabetic status by linear regression and standardization of residuals. Transformed and adjusted data were normally distributed, according to Shapiro-Wilks and Kolmogorov-Smirnov statistics.
Data were analyzed with the variance-components method implemented in GENEHUNTER 2 (Pratt et al. 2000), assuming an additive model and applying the all-possible-sib-pairs (weighted) analysis option. Pointwise and genomewide empirical significance levels for all variance-components LOD scores ⩾1.18 (nominal P=.01) were determined from 10,000 and 1,000 replicates of the data, respectively. For these calculations, marker genotypes were simulated (using SIMULATE; see the Laboratory of Statistical Genetics Web site) to match the marker characteristics seen in the original analyses.
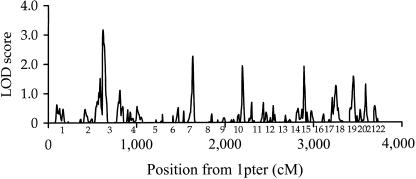
The heritability of height in our data set was 89%. The results from the genomewide scan are displayed in figure 1. All regions generating LOD scores ⩾1.18 (and associated empirical pointwise significance levels) are listed in table 1.
Figure 1 .
Multipoint genomewide variance-components linkage analysis of adult stature, with 418 autosomal microsatellite markers. The analysis was performed on a sample of 573 nuclear pedigrees containing 1,136 phenotyped sib pairs, with GENEHUNTER 2. Chromosome numbers on the X-axis are placed at the midpoint of the respective chromosomes.
Table 1.
Results of Variance-Components Linkage Analysis
| Chromosome | Marker or Interval | Position ofMaximumLOD Scorea | LODScore | NominalPointwiseP Value | EmpiricalPointwiseP Valueb |
| 2q37 | D2S206 | 271.0 | 1.51 | .0042 | .0082 |
| 3p26 | D3S1297–D3S1304 | 8.9 | 3.17 | .000067 | <.0001 |
| 7q11-21 | D7S669–D7S630 | 103.1 | 2.26 | .00062 | .0023 |
| 10q23 | D10S1686 | 119.9 | 1.93 | .0014 | .0033 |
| 15q12 | D15S1002 | 15.6 | 1.90 | .0016 | .0036 |
| 17qter | D17S785–D17S784 | 122.3 | 1.24 | .0083 | .016 |
| 19p13 | D19S221–D19S226 | 44.5 | 1.56 | .0036 | .0096 |
| 20q12-13 | D20S119–D20S178 | 65.8 | 1.29 | .0074 | .013 |
Distance in centimorgans (H) from the most p-terminal marker in the CEPH/Généthon map (Dib et al. 1996).
Estimated from 10,000 replicates.
Evidence for a locus on 3p26 influencing height was observed (LOD score 3.17, D3S1297–D3S1304, empirical pointwise P<.0001, empirical genomewide P=.11). Removing outlying phenotypic values (>3 SD from the mean age-, sex-, and diabetic-status–adjusted height, although none was >3.8 SD from the mean) reduced the evidence for linkage slightly (LOD score 3.04). Evidence for linkage (LOD score ⩾1.18) was also detected on chromosomes 2q37, 7q11-21, 10q23, 15q12, 17qter, 19p13, and 20q12-13 (table 1). It is clear that estimates of locus-specific heritability, when derived from the same data as those used to detect evidence for linkage, are grossly inflated and largely meaningless (Göring et al. 2001); therefore, estimates are not presented. Power calculations (1,000 replicates of a data set with the same pedigree structure as our data and an average Warren 2 chromosome of 20 markers of 78% heterozygosity and 9.26 cM [H] spacing) demonstrate that, for a locus-specific additive heritability of 0.20 (i.e., a locus accounting for 20% of total trait variance), our data set has 92% power to detect evidence for linkage with LOD score ⩾0.59, 76% power with LOD score ⩾1.18, and 15% power with LOD score ⩾3.3.
Our empirical significance calculations suggest that the empirical pointwise P values associated with LOD scores observed with variance-components analysis in this study are slightly larger than the asymptotic pointwise P values, indicating possible deviation from the implicit assumptions of multivariate normality, due, for instance, to either the presence of a major gene or gene-environment interactions (Allison et al. 1999). Variance-components analysis is also known to be statistically liberal when applied to nonrandomly ascertained—that is, selected—data sets (Allison et al. 1999; Dolan et al. 1999; Sham and Purcell 2001). The inflation of type I error in selected samples may be addressed within the formal variance-components framework by conditioning the likelihood function on trait values (Sham et al. 2000). An alternative, a regression-based approach, is the recent modification of the original Haseman-Elston method, HE-COM (Sham and Purcell 2001), which regresses a weighted sum of the squared differences in sibling trait values (Haseman and Elston 1972) and the squared sums of sibling trait values (Drigalenko 1998) on the proportion of allele sharing identical by descent (IBD) at the locus. The weights applied are inversely proportional to the variances (as functions of the population sibling correlation coefficients) of the squared differences and squared sums. When sibling trait values are standardized against the population mean and variance, the method becomes a valid and powerful test for linkage in selected samples (Sham and Purcell 2001).
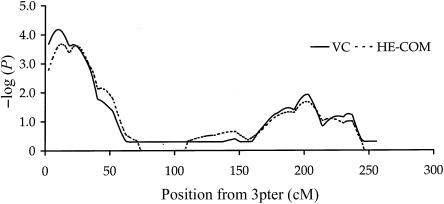
Therefore, to examine further this possibility that the selection of the Warren 2 sib pair repository according to diabetic status may have resulted in inflation of the evidence for linkage from our variance-components analyses, those chromosomes giving significant results were reexamined with HE-COM, implemented in the SAS macro QMS2 (Lessem and Cherny 2001). We adjusted and standardized our data against age- and sex-adjusted height data from the U.K. population, collected as part of the Health Survey for England 1998 (Erens and Primatesta 2001), and used a population sibling correlation estimate of 0.5. Multipoint allele-sharing IBD coefficients were generated with GENEHUNTER 2. The results of the HE-COM analysis are shown in table 2. With HE-COM, we observed evidence for linkage on every chromosome indicated by variance-components analysis, with largely comparable levels of significance. Specifically, we found evidence for linkage on chromosome 3p26, with nominal pointwise significance of P=.00013. Figure 2 shows both variance-components and HE-COM analysis results for chromosome 3p26. This linkage result was robust to the removal of extreme values of the dependent variable in the HE-COM regression (i.e., the weighted sum of squared sibling trait differences and squared sibling trait sums) for each value (0, 1, and 2) of the proportion of allele sharing (nominal pointwise significance of P=.00013). Our results suggest that selection of our data set on the basis of type 2 diabetes does not have a large effect on the underlying multivariate distribution of our data and that the variance-components analyses are valid.
Table 2.
Results of HE-COM Linkage Analysis
| Chromosome | Marker or Interval | Position of Maximum LOD Scorea | NominalPointwiseP Value |
| 2q37 | D2S206 | 271.0 | .033 |
| 3p26 | D3S1297–D3S1304 | 12.1 | .00013 |
| 7q11-21 | D7S669–D7S630 | 101.4 | .025 |
| 10q23 | D10S1686 | 119.9 | .031 |
| 15q12 | D15S1002 | 15.6 | .016 |
| 17qter | D17S785–D17S784 | 116.5 | .0068 |
| 19p13 | D19S221–D19S226 | 44.6 | .012 |
| 20q12-13 | D20S119–D20S178 | 66.8 | .038 |
Distance in centimorgans (H) from the most p-terminal marker in the CEPH/Généthon map (Dib et al. 1996).
Figure 2 .
Multipoint variance-components and HE-COM linkage analyses of adult stature, with 23 microsatellite markers on chromosome 3. For comparison between the two analyses, variance-components LOD scores are presented as −log10(nominal P value). QMS2 assigns P=1 for zero or negative values of the test statistic.
In summary, our study suggests that a novel locus on chromosome 3p26 influences adult height. Examination of the human genome sequence (International Human Genome Sequencing Consortium 2001) suggested several suitable candidate genes, including DEC1 (MIM 604256) on 3p26, a transcription factor involved in the regulation of chondrocyte differentiation. We have observed further, albeit weaker, evidence for linkage to loci on several other chromosomes, including 7q11-21. This region on chromosome 7q lies 60 cM centromeric to that detected in Scandinavian subjects (Hirschhorn et al. 2001; Perola et al. 2001) and therefore is unlikely to represent the same locus, despite the wide variation possible in location estimates (Hauser and Boehnke 1997; Roberts et al. 1999). Indeed, we observed no evidence for linkage (i.e., LOD score 0) at the region on chromosome 7q31-36 showing evidence for linkage in the Scandinavian studies. The region showing evidence for linkage on chromosome 20q12-13 lies 3.4 cM telomeric to that observed in Finnish subjects (Hirschhorn et al. 2001); we find no other instances of concordance between our study and those previously published. Possible ethnic differences between Finnish, Swedish, and Quebecois populations, in which the first genomewide scans have been conducted, and the U.K. population may account, in part, for differences between our study and those by Hirschhorn et al. (2001) and Perola et al. (2001). The effects of data sampling on the linkage statistic, as demonstrated by Hirschhorn et al. (2001) in their Swedish data set and by our own power calculations, may be an additional cause for the discrepant findings. Nevertheless, our power calculations with a locus of moderate size (20% heritability) demonstrate excellent power to detect evidence for linkage with a LOD score of 0.59, and we might, therefore, have expected to see some signal where substantial susceptibility loci were replicated between the different populations. However, any precise analysis of the power of our study to detect the chromosome 7q locus, for example, is thwarted by uncertainties over the true locus-effect sizes estimated from previous studies (Göring et al. 2001). Further studies in additional populations will provide a better understanding of the genetics of stature.
Acknowledgments
We thank Prof. Paola Primatesta, Department of Epidemiology & Public Health, University College London, for the data from the Health Survey for England 1998, and Dr. Stacey Cherny, Wellcome Trust Centre for Human Genetics, Oxford, for useful discussion on HE-COM. This work was supported by Diabetes U.K.
Electronic Database Information
- Laboratory of Statistical Genetics (Rockefeller University) Web site, ftp://linkage.rockefeller.edu/software/simulate/ (for SIMULATE)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DEC1 [MIM 604256])
- Warren 2 Project Information (Wellcome Trust Centre for Human Genetics, Oxford), http://www.well.ox.ac.uk/warren2/ (for Warren 2 scan marker information)
References
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci–mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Fauré S, Fizames C, Samson D, Druout N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dolan VC, Boomsma DI, Neale MC (1999) A simulation study of the effects of assignment of prior identity-by-descent probabilities to unselected sib pairs, in covariance-structure mapping of a quantitative-trait locus. Am J Hum Genet 64:268–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigalenko E (1998) How sib pairs reveal linkage. Am J Hum Genet 63:1242–1245 [PMC free article] [PubMed] [Google Scholar]
- Erens B, Primatesta P (2001) Health Survey for England 1998, The Stationary Office, London [Google Scholar]
- Göring HHH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M (1997) Confirmation of linkage results in affected-sib-pair linkage analysis for complex genetic traits. Am J Hum Genet Suppl 61:A278 [Google Scholar]
- Hirschhorn JN, Lindgren CM, Daly MJ, Kirby A, Schaffner SF, Burtt NP, Altshuler D, Parker A, Rioux JD, Platko J, Gaudet D, Hudson TJ, Groop LC, Lander ES (2001) Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am J Hum Genet 69:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Lessem JM, Cherny SS (2001) DeFries-Fulker multiple regression of sibship QTL data: a SAS macro. Bioinformatics 17:371–372 [DOI] [PubMed] [Google Scholar]
- Perola M, Öhman M, Hiekkalinna T, Leppävuori J, Pajukanta P, Wessman M, Koskenvuo M, Palotie A, Lange K, Kaprio J, Peltonen L (2001) Quantitative-trait-locus analysis of body-mass index and of stature, by combined analysis of genome scans of five Finnish study groups. Am J Hum Genet 69:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell S (2001) Equivalence between Haseman-Elston and variance-components linkage analyses for sib pairs. Am J Hum Genet 68:1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Zhao JH, Cherny SS, Hewitt JK (2000) Variance-components QTL linkage analysis of selected and non-normal samples: conditioning on trait values. Genet Epidemiol 10 Suppl 1:S22–S28 [DOI] [PubMed] [Google Scholar]
- Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O'Rahilly S, Frayling TM, Bell JI, Lathrop GM, Bennett A, Dhillon R, Fletcher G, Groves CJ, Jones E, Prestwich P, Simecek N, Subba Rao PV, Wishart M, Foxon R, Howell S, Smedley S, Cardon LR, Menzel S, McCarthy MI (2001) A genomewide scan for loci predisposing to type 2 diabetes in a U.K. population (The Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet 69:553–569 (erratum: 69:922) [DOI] [PMC free article] [PubMed] [Google Scholar]