Abstract
Studies in mouse and chick have shown that the 5′ HoxD genes play major roles in the development of the limbs and genitalia. In humans, mutations in HOXD13 cause the dominantly inherited limb malformation synpolydactyly (SPD). Haploinsufficiency for the 5′ HOXD genes has recently been proposed to underlie the monodactyly and penoscrotal hypoplasia in two children with chromosomal deletions encompassing the entire HOXD cluster. Similar deletions, however, have previously been associated with split–hand/foot malformation (SHFM), including monodactyly. Here we report a father and daughter with SPD who carry a 117-kb microdeletion at the 5′ end of the HOXD cluster. By sequencing directly across the deletion breakpoint, we show that this microdeletion removes only HOXD9–HOXD13 and EVX2. We also report a girl with bilateral split foot and a chromosomal deletion that includes the entire HOXD cluster and extends ∼5 Mb centromeric to it. Our findings indicate that haploinsufficiency for the 5′ HOXD genes causes not SHFM but SPD and point to the presence of a novel locus for SHFM in the interval between EVX2 and D2S294. They also suggest that there is a regulatory region, upstream of the HOXD cluster, that is responsible for activating the cluster as a whole.
The HOX genes encode a highly conserved family of transcription factors that are fundamentally important for morphogenesis in virtually all multicellular organisms (Krumlauf 1994). Humans, like most vertebrates, have 39 HOX genes, arranged in four clusters (HOXA, HOXB, HOXC, and HOXD). Expression analysis, targeted mutagenesis, and overexpression studies in mouse and chick have shown that the 5′ HoxD genes (Hoxd9–Hoxd13) are critical for limb and genital tract development (reviewed by Zákány and Duboule [1999]). A similar role for the 5′ HOXD genes in humans has been confirmed by the discovery that specific mutations in HOXD13 cause the dominantly inherited limb malformation synpolydactyly (SPD [MIM 186000]) (Akarsu et al. 1996; Muragaki et al. 1996; Goodman et al. 1997, 1998). This malformation is characterized by syndactyly between the third and fourth fingers and between the fourth and fifth toes, with variable digit duplication in the syndactylous web. Severely affected males may also have hypospadias (Goodman et al. 1997).
Del Campo et al. (1999) recently reported striking limb and genital abnormalities in two unrelated boys with chromosomal deletions at 2q24.1-q31 and 2q31.1-q32.2. Both boys had a single bone in the zeugopod and monodactyly in the autopod of all four limbs, as well as penoscrotal hypoplasia and multiple other anomalies. Since these deletions removed the entire HOXD cluster, the limb and genital abnormalities were proposed to be due, at least in part, to haploinsufficiency for the 5′ HOXD genes. Similar deletions involving band 2q31.1, however, have previously been associated with limb abnormalities ranging from a wide cleft between toes 1 and 2 and brachysyndactyly of toes 2–5 to split hand, split foot, and monodactyly (reviewed by Boles et al. [1995] and Maas et al. [2000]). As first suggested by Boles et al. (1995), these abnormalities probably represent milder and more-severe forms, respectively, of split–hand/foot malformation (SHFM [MIM 183600]), and the monodactyly described by Del Campo et al. (1999) would fit well into the severe end of this spectrum.
Here we report a microdeletion at the 5′ end of the HOXD cluster in a father and daughter with SPD. This deletion removes just HOXD9 to EVX2 and extends only 85 kb upstream of HOXD13, implying that the phenotype caused by haploinsufficiency for the 5′ HOXD genes is not SHFM but SPD. We also report a cytogenetically visible deletion at 2q31-q33, in a girl with bilateral split foot, which encompasses the entire HOXD cluster but extends only a short distance centromeric to it. These results, together with those of Boles et al. (1995), suggest that the 5-Mb interval centromeric to EVX2 harbors a novel locus for SHFM.
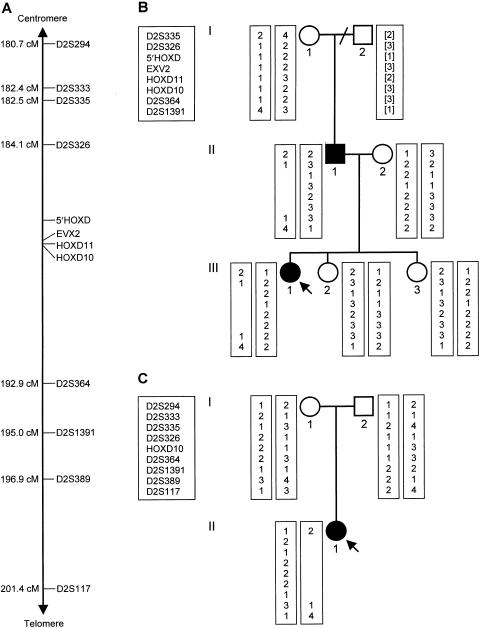
Family 1 (fig. 1B) was ascertained after referral, for genetic counseling, to the Institute of Human Genetics, University of Düsseldorf, Germany. The proband (III.1) was born with severe SPD in both hands, including complete cutaneous syndactyly between the third and fourth fingers, duplication of all three phalanges of the third fingers, and clinodactyly of the fifth fingers (fig. 2A). In the feet, she had broad halluces, bilateral partial cutaneous syndactyly between the second to fourth toes, hypoplastic second to fifth middle phalanges, short second to fifth metatarsals, and partial duplication of the bases of the second metatarsals in the first web spaces (fig. 2B). Her father (II.1) had milder hand involvement, with just bilateral partial cutaneous syndactyly between the third and fourth fingers. In the feet, however, he had abnormalities (fig. 2C) virtually identical to those of his daughter. Neither father nor daughter had any other medical problems, and a full skeletal survey revealed no additional malformations. The remaining family members were unaffected.
Figure 1.
Haplotype analysis in the two families. A, Relative positions of the 12 polymorphic markers used, with their approximate distances from the tip of the short arm of chromosome 2. D2S294, D2S333, D2S335, D2S326, D2S364, D2S1391, D2S389, and D2S117 are microsatellite repeats spanning a 20.7-cM interval in the vicinity of the HOXD cluster on chromosome 2q31 (Sheffield et al. 1995; Dib et al. 1996). HOXD10, originally reported as HOX4E (Rosen and Brown 1993), lies in the 3′ UTR of HOXD10. HOXD11 (a TG repeat in the intron of HOXD11), EVX2 (an AG repeat just telomeric to the 5′ UTR of EVX2), and 5′ HOXD (a CA repeat ∼60 kb centromeric to the HOXD cluster) are three new polymorphic markers identified in the present study (details available on request). B and C, Pedigree drawings of families 1 and 2, respectively (blackened symbols represent affected individuals; arrows indicate the probands), and results of haplotype analysis. The bracketed haplotype for individual I.2 from family 1, from whom DNA was not available, was inferred from the results for individuals I.1 and II.1.
Figure 2.
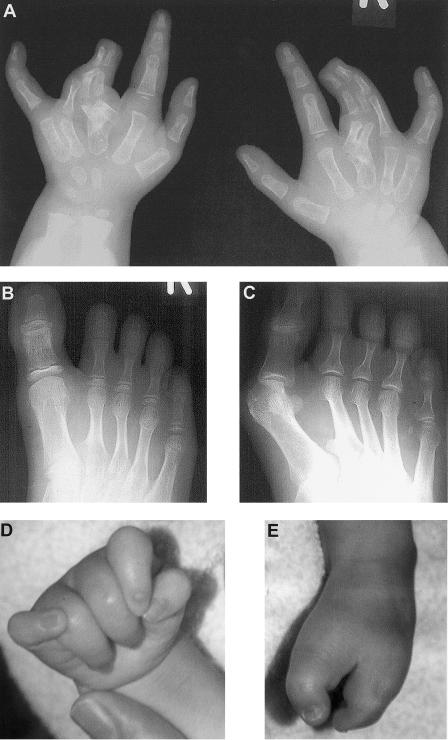
Limb abnormalities in the two families. A, Radiograph of both hands of individual III.1 from family 1 at age 2 years, showing complete cutaneous syndactyly between the third and fourth fingers, duplication of all three phalanges of the third fingers, broad third metacarpals, and hypoplastic fifth middle phalanges. B, Radiograph of right foot of individual III.1 from family 1 at age 11 years, showing broad hallux, partial cutaneous syndactyly between the second to fifth toes, fifth-toe brachydactyly, hypoplastic second to fifth middle phalanges, short second to fifth metatarsals, and partial duplication of the base of the second metatarsal in the first web space. C, Radiograph of right foot of individual II.1 from family 1 at age 35 years, showing broad hallux, partial cutaneous syndactyly between the second to fourth toes, fifth-toe brachydactyly, hypoplastic second to fifth middle phalanges, occasional symphalangism between the middle and distal phalanges, and partial duplication of the base of the second metatarsal in the first web space. D, Photograph of right hand of individual II.1 from family 2 at age 10 mo, showing clenched and overlapping fingers. E, Photograph of right foot of individual II.1 from family 2 at age 10 mo, showing severe “lobster-claw” deformity with absence of the middle three rays.
Family 2 (fig. 1C) was ascertained after referral, for genetic counseling, to the Wessex Clinical Genetics Service, Southampton, United Kingdom. The proband (II.1) was born with multiple abnormalities, including severe proportionate growth retardation, bilateral retinal colobomata, blepharophimosis, a right preauricular pit, and an anterior anus. In the hands, she had camptodactyly of all the fingers and clinodactyly of the fifth fingers (fig. 2D), whereas, in the feet, she had bilateral “lobster-claw” deformities, with absence of the middle three rays (fig. 2E). At age 3 mo, she developed seizures. She made almost no developmental progress and, sadly, died at age 2 years, after a chest infection. Her parents and older sister were not affected with congenital malformations.
Venous blood samples were obtained from individuals I.1, II.1, II.2, III.1, III.2, and III.3 in family 1 and from individuals I.1, I.2, and II.1 in family 2, with their or their parents’ consent and with the approval of the local ethical review board.
In the affected father and daughter from family 1, no mutations were identified in the entire coding region of HOXD13. At an A/G single-nucleotide polymorphism in exon 1 (Goodman et al. 1997), however, the father was found to carry an A residue, whereas the daughter was found to carry a G residue, suggesting that the other allele might be deleted. Although routine karyotype analysis was normal in both patients, haplotype analysis of the family (fig. 1B) demonstrated a submicroscopic interstitial deletion lying between D2S326 and D2S364 and removing at least EVX2 to HOXD10 together with >60 kb centromeric to EVX2. This deletion had arisen de novo on the father’s maternal chromosome 2q31 and had been transmitted to his affected daughter but not to his unaffected children.
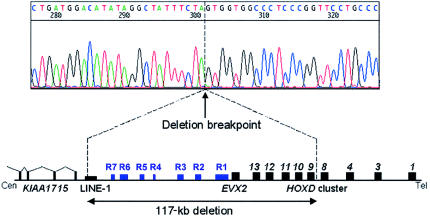
The approximate size of the deletion was confirmed in both patients by FISH studies with cosmids mapping to the region (Rossi et al. 1994; Simeone et al. 1994). Signal was obtained on only one chromosome 2 with cosmids P19 (containing EVX2) and G2 (containing EVX2 and HOXD10–HOXD13) but was obtained on both chromosomes 2 with cosmid D2 (containing DLX1 and DLX2), showing that the deletion encompasses EVX2 to HOXD10 but does not extend as far centromeric as DLX1 and DLX2. To define the telomeric end of the deletion, probes from the region HOXD8–HOXD10 were used for Southern blotting (details available on request). The junction fragments detected in individuals II.1 and III.1 indicated that the telomeric end lay in a 75-bp interval ∼650 bp downstream of HOXD. We therefore used inverse PCR to amplify a short genomic fragment containing the deletion breakpoint (details available on request), enabling us to sequence directly across the deletion (fig. 3). As predicted, the telomeric end of the deletion lies 658 bp downstream of the stop codon of HOXD9, between bases 108822 and 108823 of BAC RP11-387A (a fully sequenced 175-kb clone containing the entire HOXD cluster). The centromeric end proved harder to locate precisely. A BLASTN (NCBI BLAST Home Page) search of the Human Genome Project working draft sequence (International Human Genome Sequencing Consortium 2001) placed it within BAC RP11-514D19, a partially sequenced 208-kb clone that largely overlaps BAC RP11-387A1 but extends farther toward the centromere. A BLASTN search of the Celera human genome consensus sequence (Venter et al. 2001) placed it within an unfinished 342-kb scaffold segment (GA_x2HTBKMFJRS). To determine its exact position, we amplified and sequenced PCR products spanning the gaps in this scaffold segment (details available on request), allowing us to assemble ∼15 kb of continuous sequence (GenBank accession number AF415204) immediately upstream of the centromeric end of BAC RP11-387A1 (GenBank accession number AC009336). The centromeric breakpoint lies between bases 7044 and 7045 of this new sequence, 8,570 bp upstream of BAC RP11-387A1. The deletion in family 1 is thus 117,392 bp long.
Figure 3.
Precise extent of the deletion in family 1. The electropherogram shows the sequence across the deletion breakpoint. The telomeric end of the deletion is located 658 bp downstream of HOXD9. The centromeric end of the deletion is located within a LINE-1 element, ∼5 kb telomeric to the most 5′ known exon of KIAA1715. Inside the deleted segment, the seven highly conserved regions likely to represent regulatory elements are denoted by blue boxes (R1–R7).
Analysis of the deleted region by use of the program NIX (Intoduction to NIX Web site) revealed that the centromeric breakpoint occurs within a 1,165-bp LINE-1 fragment (L1MD2; fig. 3). Interstitial deletions occasionally can result from unequal homologous recombination between neighboring LINE-1 elements (Burwinkel and Kilimann 1998), but no such elements are present at the telomeric breakpoint. This is consistent with the very low density of interspersed repeats within the four HOX clusters (International Human Genome Sequencing Consortium 2001), almost certainly reflecting the need to avoid disruption of large-scale cis-acting regulatory elements. Several previously characterized chromosomal deletions and one translocation have LINE-1 sequence on one side and unrelated sequence on the other (Drechsler and Royer-Pokora 1996), as in the deletion reported here, and in all these cases the involvement of the LINE-1 element is probably incidental.
NIX analysis revealed, apart from HOXD9–HOXD13 and EVX2, three other potentially expressed sequences within the deleted area. Each matches only a single short cDNA clone (GenBank accession numbers AA620964, D61190, and AI214712), however, and therefore may not, in fact, be expressed. By contrast, sequences matching a 5.7-kb partial cDNA clone (GenBank accession number AB051502) were identified just beyond the deletion, ∼78 kb centromeric to EVX2. This cDNA, originally isolated from human brain (Nagase et al. 2000), matches >80 smaller cDNAs isolated from a wide variety of adult human tissues. The gene, KIAA1715, is transcribed in the opposite direction to the HOXD cluster and has at least 14 exons, of which the most 5′ so far identified lies 5.2 kb centromeric to the deletion (fig. 3). Analysis of its sequence by use of the program PIX (HGMP-RC PIX Web site) indicates that the encoded protein is probably membrane bound. Homologs are present in mouse, Drosophila, and Caenorhabditis elegans, and the mouse homolog lies upstream of the HoxD cluster, ∼77 kb centromeric to Evx2.
In mice, the area upstream of Evx2 is known to contain several regulatory elements for the HoxD cluster (van der Hoeven et al. 1996; Hérault et al. 1999; Kondo and Duboule 1999), although their sequence and exact position have not been determined. A BLAST comparison of the interval between EVX2 and the centromeric end of the deletion with the corresponding interval in mouse reveals a high level of sequence homology between a region 13.7–19.5 kb upstream of human HOXD13 (bases 57342–63193 of BAC RP11-387A1; region 1 [R1] in fig. 3) and a region 13.6–19.3 kb upstream of mouse Hoxd13 (bases 85879–91558 of BAC RP23-400H17). This region includes five blocks, each >500 bp long, that are >90% identical in human and mouse. A similar comparison with the corresponding region upstream of the horn shark HoxD cluster shows that each block also contains 80–540-bp stretches that are >85% identical in human and horn shark. Moreover, within the first two blocks there are three short stretches >90% identical to the corresponding region upstream of the human HOXA cluster. This strongly conserved region almost certainly represents an important HOX-cluster regulatory element.
Centromeric to R1, there are six shorter stretches of striking sequence homology between human and mouse (R2–R7, fig. 3). R2 (bases 48073–48774 of BAC RP11-387A1) contains conserved 226-bp and 208-bp blocks 28 kb upstream of HOXD13; R3 (bases 37536–38289) contains conserved 332-bp and 306-bp blocks 38 kb upstream of HOXD13; R4 (bases 17857–18094) contains a conserved 237-bp block 58 kb upstream of HOXD13; R5 (bases 11792–12135) contains a conserved 343-bp block 64 kb upstream of HOXD13; R6 (bases 7253–9632) contains conserved 358-bp, 270-bp, and 887-bp blocks 68 kb upstream of HOXD13; and R7 (bases 1998–2452) contains conserved 298-bp and 67-bp blocks 74 kb upstream of HOXD13. Some of these regions, which are all removed by the deletion in family 1, probably represent regulatory elements for the HOXD cluster, although the most centromeric of them could, alternatively, represent regulatory elements for KIAA1715. No conserved regions are identifiable, however, in the 5-kb interval between the centromeric breakpoint and the 5′ end of KIAA1715.
In family 2, karyotype analysis of the proband and her parents revealed that she carried a de novo interstitial deletion removing chromosomal bands 2q31.3-2q33.3. Haplotype analysis (fig. 1C) showed that this deletion arose on her paternal chromosome 2. Its centromeric end lies between D2S294 and D2S333, whereas its telomeric end lies between D2S1391 and D2S389. The centromeric breakpoints in two similar interstitial deletions (at 2q31.1-q31.3 and 2q24.3-q31.3) also lie within the 1.8-cM interval between D2S294 and D2S335 (Slavotinek et al. 1999), and all three breakpoints may have occurred at the known fragile site FRA2G (Limongi et al. 2000). The deletion in the proband thus spans a genetic distance of 12.6–16.2 cM and removes DLX1 and DLX2, as well as the entire HOXD cluster and EVX2.
SPD has previously been shown to be caused by mutations in HOXD13 (Akarsu et al. 1996; Muragaki et al. 1996; Goodman et al. 1997, 1998). We have now demonstrated that it can also result from a microdeletion at the 5′ end of the HOXD cluster. This probably reflects the consequences of haploinsufficiency for one or more of the deleted genes, rather than disruption or perturbed regulation of genes flanking the deletion. On the centromeric side, the nearest gene, KIAA1715, may not be affected by the deletion, although a hitherto-unidentified 5′ exon or regulatory element may be removed. Haploinsufficiency for a widely expressed membrane-bound protein, however, is highly unlikely to produce a perfect phenocopy of SPD. On the telomeric side, the four remaining HOXD genes (HOXD1, HOXD3, HOXD4, and HOXD8) are not normally expressed in the developing limbs. Nevertheless, they could contribute to the SPD phenotype if the deletion placed them under the control of a regulatory element that could misdirect their expression to the autopod, such as the digit enhancer thought to regulate the 5′ Hoxd genes and Evx2 in mouse (van der Hoeven et al. 1996; Hérault et al. 1999; Kondo and Duboule 1999). As mentioned above, however, the microdeletion appears to remove all conserved regions potentially representing regulatory elements between EVX2 and the 5′ end of KIAA1715, and HOXD regulatory sequences are unlikely to be located more centromeric than this, since they would then lie within KIAA1715. The SPD caused by the microdeletion thus is most simply explained by haploinsufficiency for one or more of the deleted genes, five of which (HOXD10–HOXD13 and EVX2) are known to be important in autopod development and one of which (HOXD13) is specifically implicated in SPD.
Two types of HOXD13 mutation have previously been identified in SPD. Most cases are caused by expansions of an N-terminal polyalanine tract (Akarsu et al. 1996; Muragaki et al. 1996; Goodman et al. 1997). Genetic complementation experiments in the synpolydactyly homolog mouse (Bruneau et al. 2001) recently have confirmed that the resulting mutant protein exerts a “super” dominant negative effect, by interfering with the function of the remaining wild-type HOXD13 and other 5′ HOXD proteins. An atypical form of SPD, associated with a distinctive foot phenotype (broad halluces, hypoplastic middle phalanxes, and partial duplication of the bases of the second metatarsals), is caused by different frameshifting deletions in HOXD13 (Goodman et al. 1998; Calabrese et al. 2000), all of which are thought to inactivate the resulting protein. The patients with the microdeletion have a foot phenotype almost identical to that produced by these frameshift mutations, suggesting that the chief cause of their abnormalities is haploinsufficiency for HOXD13. Mice homozygous for a targeted deletion that removes Hoxd11–Hoxd13 have digital abnormalities closely resembling human SPD (Zákány and Duboule 1996), although heterozygotes have only mild shortening of the second and fifth digits. The more severe phenotype in the patients with the microdeletion may reflect the additional effects of haploinsufficiency for HOXD10 and EVX2 and/or a greater sensitivity in humans than in mice to reduction in HOXD11–HOXD13 gene dosage.
Although HOXD10–HOXD13 are expressed in the zeugopod and HOXD9 is expressed in the stylopod during development, the patients with the microdeletion have no limb abnormalities outside the autopod. Similarly, no zeugopod or stylopod malformations have been observed in mice heterozygous for targeted loss-of-function mutations in single or multiple 5′ Hoxd genes (reviewed by Rijli and Chambon [1997]). The deleted genes are also expressed in the developing vertebral column, gastrointestinal and urogenital tracts, and genital tubercle, yet neither the patients with the microdeletion nor the corresponding heterozygous knockout mice have any detectable malformations of these structures. Loss-of-function mutations in the HOXD9–HOXD12 and EVX2 genes thus are unlikely to cause significant malformations in humans. The same may be true of most HOXA genes, since a patient hemizygous for the entire HOXA cluster has a phenotype attributable to haploinsufficiency for just HOXA3 and HOXA13 (Devriendt et al. 1999). Gain-of-function mutations in the deleted genes, however, including mutations affecting their regulation, may well underlie rare human phenotypes (Goodman and Scambler 2001; Goodman et al. 2001).
Cytogenetically visible deletions of the long arm of chromosome 2 have previously been described in >30 patients with multiple malformations, including abnormalities of the limbs (Boles et al. 1995; Maas et al. 2000). Deletions removing 2q24.3 are generally associated with flexion deformity of the fingers, as in individual II.1 from family 2, whereas deletions removing 2q31.1 are associated with a broad cleft between toes 1 and 2 and with brachysyndactyly of toes 2–5, suggesting that 2q24.3 and 2q31.1 harbor genes with different roles in limb development (Maas et al. 2000). Patients with deletions of 2q31.1 may also have more-severe limb defects, including bilateral split hand (Ramer et al. 1990), bilateral split foot (Benson et al. 1986; Boles et al. 1995), and monodactyly of the upper limbs with only a single forearm bone (Ramer et al. 1990), similar to that in the two boys described by Del Campo et al. (1999). Such monodactyly is a well-recognized manifestation of severe SHFM. These observations led Boles et al. (1995) to propose that SHFM “may represent the extreme end of the spectrum of digital anomalies associated with deletions in this region” (p. 158). Our findings in individual II.1 from family 2, who had a 2q31-q33 deletion and bilateral split foot, support this hypothesis.
The underlying gene is unlikely to be one or more of the 5′ HOXD genes, for the reasons given above. Moreover, the deletion in the patient with bilateral split foot reported by Boles et al. (1995) lies entirely centromeric to EVX2. Together with our haplotype analysis in individual II.1 from family 2, this suggests that an SHFM locus may lie between EVX2 and D2S294, an interval of only ∼5 Mb. Candidates genes in this interval include DLX1 and DLX2, homeobox genes whose mouse homologs are expressed in the apical ectodermal ridge and progress zone of developing limbs (Qiu et al. 1997). Two closely related genes, DLX5 and DLX6, are implicated in SHFM, since they lie in the 1.5-Mb critical interval in which to which SHFM1 (MIM 183600) has been mapped (Crackower et al. 1996), but limb abnormalities are not observed in heterozygous or homozygous mice carrying targeted deletions in Dlx1, Dlx2, or both Dlx1 and Dlx2 (Qiu et al. 1997).
If haploinsufficiency for HOXD9–HOXD13 and EVX2 causes SPD, as our findings in family 1 indicate, large deletions encompassing the HOXD cluster should produce similar abnormalities. Severe SPD need not always occur, however, since individual II.1 from family 1 has milder hand abnormalities than does individual III.1, and the phenotype produced by specific HOXD13 mutations is very variable (Goodman et al. 1997, 1998). Almost all of the large deletions reported elsewhere have been defined by cytogenetic rather than molecular analysis, and the HOXD cluster has been proven to be deleted in only six cases (Nixon et al. 1997; Del Campo et al. 1999; Slavotinek et al. 1999; Prieur et al. 2000). One individual has a limb phenotype very similar to that of individual II.1 from family 1 (Prieur et al. 2000), whereas three have minor limb abnormalities also found in mild SPD (Nixon et al. 1997; Slavotinek et al. 1999). The monodactyly in the boys described by Del Campo et al. (1999) and the bilateral split foot in individual II.1 from family 2 would mask any accompanying SPD.
The phenotype resulting from the microdeletion in family 1 also has interesting implications for regulation of the entire HOXD cluster. Kondo and Duboule (1999) described a regulatory element responsible for early repression of the mouse HoxD cluster, probably lying <28 kb upstream of Hoxd13, although they did not identify the exact sequences involved. Surviving mice heterozygous for a targeted deletion of this region together with Hoxd11–Hoxd13 and Evx2 displayed posterior homeotic transformations of the upper axial skeleton, including abnormalities of the cervical and thoracic vertebrae and sternum, due to premature and ectopic activation of the remaining 3′ Hoxd genes. If (as seems likely) a homologous regulatory element exists upstream of the human HOXD cluster, it would be removed by the microdeletion reported here, and individuals II.1 and III.1 from family 1 should have vertebral and sternal malformations due to premature and ectopic activation of HOXD1–HOXD8. Despite thorough clinical and radiological examination, however, we detected no such abnormalities, suggesting that the nondeleted 3′ HOXD genes are not expressed at all. This could happen if the deleted sequences included a major locus control region (LCR) necessary to initiate activation of the cluster as a whole. The six short conserved regions that we have identified 28–74 kb upstream of HOXD13 (R2–R7) represent good candidates for such an LCR, whereas the large conserved region 13.7–19.5 kb upstream of HOXD13 (R1) could well constitute the repressor element postulated by Kondo and Duboule (1999).
Acknowledgments
We are most grateful to the families for their participation in our study and to Dr. Christine Hall for her help in assessing the radiographs. This work was supported by a Clinician Scientist Fellowship from the Medical Research Council, United Kingdom (to F.R.G.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the sequence of HOXD13 [accession numbers AF005219 and AF005220)]; BAC RP11-387A1, containing the human HOXD cluster [accession number AC009336]; the ∼15-kb region centromeric to BAC RP11-387A1 [accession number AF415204]; BAC RP11-514D19 [accession number AC016915]; PAC RP1-170O19, containing the human HOXA cluster [accession number AC004080]; BAC RP23-400H17, containing the mouse HoxD cluster [accession number AC015584]; the horn shark HoxD cluster [accession number AF224263]; and cDNA clones [accession numbers AA620964, D61190, AI214712, and AB051502])
- HGMP-RC PIX, http://www.hgmp.mrc.ac.uk/Registered/Webapp/pix/ (for analysis of peptide sequences)
- Introduction to NIX, http://www.hgmp.mrc.ac.uk/NIX/ (for analysis of nucleic acid sequences)
- NCBI BLAST Home Page, http://www.ncbi.nlm.nih.gov/BLAST/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SPD [MIM 186000] and SHFM1 [MIM 183600])
References
- Akarsu AN, Stoilov I, Yilmaz E, Sayli BS, Sarfarazi M (1996) Genomic structure of HOXD13 gene: a nine polyalanine duplication causes synpolydactyly in two unrelated families. Hum Mol Genet 5:945–952 [DOI] [PubMed] [Google Scholar]
- Benson K, Gordon M, Wassman ER, Tsi C (1986) Interstitial deletion of the long arm of chromosome 2 in a malformed infant with karyotype 46,XX,del(2)(q31q33). Am J Med Genet 25:405–411 [DOI] [PubMed] [Google Scholar]
- Boles RG, Pober BR, Gibson LH, Willis CR, McGrath J, Roberts DJ, Yang-Feng TL (1995) Deletion of chromosome 2q24-q31 causes characteristic digital anomalies: case report and review. Am J Med Genet 55:155–160 [DOI] [PubMed] [Google Scholar]
- Bruneau S, Johnson KR, Yamamoto M, Kuroiwa A, Duboule D (2001) The mouse Hoxd13spdh mutation, a polyalanine expansion similar to human type II synpolydactyly (SPD), disrupts the function but not the expression of other Hoxd genes. Dev Biol 237:345–353 [DOI] [PubMed] [Google Scholar]
- Burwinkel B, Kilimann MW (1998) Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol 277:513–517 [DOI] [PubMed] [Google Scholar]
- Calabrese O, Bigoni S, Gualandi F, Trabanelli C, Camera G, Calzolari E (2000) A new mutation in HOXD13 associated with foot pre-postaxial polydactyly. Eur J Hum Genet 8 Suppl 1:140 [Google Scholar]
- Crackower MA, Scherer SW, Rommens JM, Hui CC, Poorkaj P, Soder S, Cobben JM, Hudgins L, Evans JP, Tsui L-C (1996) Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Genet 5:571–579 [DOI] [PubMed] [Google Scholar]
- Del Campo M, Jones MC, Veraksa AN, Curry CJ, Jones KL, Mascarello JT, Ali-Kahn-Catts Z, Drumheller T, McGinnis W (1999) Monodactylous limbs and abnormal genitalia are associated with hemizygosity for the human 2q31 region that includes the HOXD cluster. Am J Hum Genet 65:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Jaeken J, Matthijs G, Van Esch H, Debeer P, Gewillig M, Fryns J-P (1999) Haploinsufficiency of the HOXA gene cluster, in a patient with hand-foot-genital syndrome, velopharyngeal insufficiency and persistent patent ductus Botalli. Am J Hum Genet 65:249–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morisette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Drechsler M, Royer-Pokora B (1996) A LINE element is present at the site of a 300-kb deletion starting in intron 10 of the PAX6 gene in a case of familial aniridia. Hum Genet 98:297–303 [DOI] [PubMed] [Google Scholar]
- Goodman FR, Bacchelli C, McKeown CME, Scambler PJ (2001) An amino acid substitution in the HOXD13 homeodomain causes a novel brachydactyly-polydactyly syndrome. Eur J Hum Genet 9 Suppl 1:179 [Google Scholar]
- Goodman FR, Giovannucci-Uzielli M-L, Hall C, Reardon W, Winter R, Scambler P (1998) Deletions in HOXD13 segregate with an identical, novel foot malformation in two unrelated families. Am J Hum Genet 63:992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli ML, Lapi E, Majewski F, McGaughran J, McKeown C, Reardon W, Upton J, Winter RM, Olsen BR, Scambler PJ (1997) Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci USA 94:7458–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR, Scambler PJ (2001) Human HOX gene mutations. Clin Genet 59:1–11 [DOI] [PubMed] [Google Scholar]
- Hérault Y, Beckers J, Gerard M, Duboule D (1999) Hox gene expression in limbs: colinearity by opposite regulatory controls. Dev Biol 208:157–165 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Kondo T, Duboule D (1999) Breaking colinearity in the mouse HoxD complex. Cell 97:407–417 [DOI] [PubMed] [Google Scholar]
- Krumlauf R (1994) Hox genes in vertebrate development. Cell 78:191–201 [DOI] [PubMed] [Google Scholar]
- Limongi MZ, Pelliccia F, Gaddini L, Rocchi A (2000) Clustering of two fragile sites and seven homeobox genes in human chromosome region 2q31-q32.1. Cytogenet Cell Genet 90:151–153 [DOI] [PubMed] [Google Scholar]
- Maas SM, Hoovers JM, van Seggelen ME, Menzel DM, Hennekam RC (2000) Interstitial deletion of the long arm of chromosome 2: a clinically recognisable microdeletion syndrome? Clin Dysmorphol 9:47–53 [DOI] [PubMed] [Google Scholar]
- Muragaki Y, Mundlos S, Upton J, Olsen BR (1996) Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 272:548–551 [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O (2000) Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 7:347–355 [DOI] [PubMed] [Google Scholar]
- Nixon J, Oldridge M, Wilkie AO, Smith K (1997) Interstitial deletion of 2q associated with craniosynostosis, ocular coloboma, and limb abnormalities: cytogenetic and molecular investigation. Am J Med Genet 70:324–327 [PubMed] [Google Scholar]
- Prieur M, Lapierre J, Le Lorch M, Ozilou C, Amiel J, Sanlaville D, Vekemans M, Turleau C, Romana SP (2000) HOXD gene cluster haploinsufficiency does not generate gross limb abnormalities. Eur J Hum Genet 8 Suppl 1:74 [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JLR (1997) Role of the Dlx homeobox genes in proximo-distal patterning of the branchial arches: mutations of Dlx1, Dlx2, and Dlx1 and 2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol 185:165–184 [DOI] [PubMed] [Google Scholar]
- Ramer JC, Mowrey PN, Robins DB, Ligato S, Towfighi J, Ladda RL (1990) Five children with del (2)(q31q33) and one individual with dup (2)(q31q33) from a single family: review of brain, cardiac, and limb malformations. Am J Med Genet 37:392–400 [DOI] [PubMed] [Google Scholar]
- Rijli FM, Chambon P (1997) Genetic interactions of Hox genes in limb development: learning from compound mutants. Curr Op Genet Devel 7:481–487 [DOI] [PubMed] [Google Scholar]
- Rosen DR, Brown RH (1993) Dinucleotide repeat polymorphism in the HOX4E locus. Hum Mol Genet 2:617 [DOI] [PubMed] [Google Scholar]
- Rossi E, Faiella A, Zeviani M, Labeit S, Floridia G, Brunelli S, Cammarata M, Boncinelli E, Zuffardi O (1994) Order of six loci at 2q24-q31 and orientation of the HOXD locus. Genomics 24:34–40 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, Pulido JC, Yandava C, Sunden SL, Mattes G, Businga T, McClain A, Beck J, Scherpier T, Gilliam J, Zhong J, Duyk GM (1995) A collection of tri- and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet 4:1837–1844 [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Pannese M, D'Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K, Boncinelli E (1994) Cloning and characterization of two members of the vertebrate Dlx gene family. Proc Natl Acad Sci USA 91:2250–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek A, Schwarz C, Getty JF, Stecko O, Goodman F, Kingston H (1999) Two cases with interstitial deletions of chromosome 2 and sex reversal in one. Am J Med Genet 86:75–81 [DOI] [PubMed] [Google Scholar]
- van der Hoeven F, Zákány J, Duboule D (1996) Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell 85:1025–1035 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–130511181995 [Google Scholar]
- Zákány J, Duboule D (1996) Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 384:69–71 [DOI] [PubMed] [Google Scholar]
- ——— (1999) Hox genes in digit development and evolution. Cell Tissue Res 296:19–25 [DOI] [PubMed] [Google Scholar]