Abstract
Background
Yellow mustard (Sinapis alba L.) is an important condiment crop for the spice trade in the world. It has lagged behind oilseed Brassica species in molecular marker development and application. Intron length polymorphism (ILP) markers are highly polymorphic, co-dominant and cost-effective. The cross-species applicability of ILP markers from Brassica species and Arabidopsis makes them possible to be used for genetic linkage mapping and further QTL analysis of agronomic traits in yellow mustard.
Results
A total of 250 ILP and 14 SSR markers were mapped on 12 linkage groups and designated as Sal01-12 in yellow mustard. The constructed map covered a total genetic length of 890.4 cM with an average marker interval of 3.3 cM. The QTL for erucic content co-localized with the fatty acid elongase 1 (FAE1) gene on Sal03. The self-(in)compatibility gene was assigned to Sal08. The 4-hydroxybenzyl, 3-indolylmethyl and 4-hydroxy-3-indolylmethyl glucosinolate contents were each controlled by one major QTL, all of which were located on Sal02. Two QTLs, accounting for the respective 20.4% and 19.2% of the total variation of 2-hydroxy-3-butenyl glucosinolate content, were identified and mapped to Sal02 and Sal11. Comparative synteny analysis revealed that yellow mustard was phylogenetically related to Arabidopsis thaliana and had undergone extensive chromosomal rearrangements during speciation.
Conclusion
The linkage map based on ILP and SSR markers was constructed and used for QTL analysis of seed quality traits in yellow mustard. The markers tightly linked with the genes for different glucosinolate components will be used for marker-assisted selection and map-based cloning. The ILP markers and linkage map provide useful molecular tools for yellow mustard breeding.
Keywords: Yellow mustard, ILP marker, Linkage map, QTL analysis, Glucosinolate
Background
Yellow mustard (Sinapis alba L; genome SS, 2n = 24) is an obligate out-crossing crop due to its self-incompatibilty reproduction system. It is more heat and drought tolerant, and more resistant to pod shattering and diseases such as blackleg than Brassica napus and B. rapa[1-3]. Yellow mustard is well adapted to the semi-arid areas of western Canada and has been cultivated as a condiment crop in the Prairies since 1936 [4]. Condiment yellow mustard varieties contain a desirable high 4-hydroxybenzyl (HBEN) glucosinolate (GSL) component in the seed [3], which hydrolyses to produce the spicy "heat" sensation in the mouth. In addition, yellow mustard contains 3-indolylmethyl (IND), 4-hydroxy-3-indolylmethyl (HIND) and 2-hydroxy-3-butenyl (HBUT) GSL components in the seed.
Genetic linkage mapping has proven to be very useful for analyzing quantitative trait loci (QTL), tagging and cloning genes controlling desirable agronomic traits and studying genome organization and evolution. Construction of genetic linkage maps based on various molecular markers has revealed the occurrence of large-scale duplication as well as extensive chromosomal rearrangement in B. rapa[5,6] and B. oleracea[7], B. napus[8,9] and B. juncea[10,11]. QTL analysis identified two major QTLs for erucic acid content in B. napus and B. juncea[12,13]. Five QTLs for total glucosinolate content were mapped on chromosomes A2, A9, C2, C7, and C9 in B. napus[14,15]. Mahmood et al. [16] identified five QTLs explaining approximately 30 to 45% of the total aliphatic glucosinolate content variation in B. juncea.
Yellow mustard has lagged far behind oilseed Brassica species in molecular marker development and application. This could be due to the following reasons. Firstly, yellow mustard has a sporophytic self-incompatibility reproductive system which makes it difficult to develop homozygote parental and recombinant inbred lines required for linkage mapping. Secondly, yellow mustard is not a major food crop and therefore doesn’t receive much attention and funding for genomic research. So far, only one linkage map based on restriction fragment length polymorphism (RFLP) markers was constructed using populations derived from heterozygous parental lines in yellow mustard [17]. However, application of RFLP marker technology in genetic research and breeding is limited due to the laborious procedures and high cost. Single nucleotide polymorphism (SNP) and simple sequence repeat (SSR) markers have proven to be very useful for construction of high density maps in B. napus and B. juncea. However, these markers have not been developed in yellow mustard yet.
Intron length polymorphism (ILP) markers are highly polymorphic, co-dominant, cost-effective and cross-species applicable [9,18]. ILP primers in B. napus, B. rapa and A. thaliana have been developed [10,18]. Yellow mustard, B. napus and B. rapa belong in the Subtribe Brassicinae [19], and might have evolved from the same ancestor species as A. thaliana[10,20-22]. Therefore, the ILP primers from Brassica species and Arabidopsis could be used for genetic linkage mapping in yellow mustard. Doubled-haploid (DH) and inbred lines have lately been produced in yellow mustard at Agriculture and Agri-Food Canada-Saskatoon Research Centre (AAFC-SRC) [23,24]. Molecular markers for the fatty acid elongase 1 (FAE1) and self-(in) compatibility genes have also been developed in our lab [25]. The objectives of the present study were 1) to construct a genetic linkage map based on ILP and SSR markers using the F2 population derived from homozygote parental lines, 2) to identify QTLs for erucic acid content and GSL components, and 3) to assign the FAE1 and self-(in) compatibility genes to the respective linkage groups in yellow mustard.
Results
Polymorphism between the parental lines Y514 and Y517
A total of 1726 ILP primers – 383 from A. thaliana[10], 1093 from B. napus and 250 from B. rapa available in the Potential Intron Polymorphism (PIP) database [18] and 222 SSR markers with 73 from B. napus and 149 from B. juncea were used to screen the parental lines Y514 and Y517 for polymorphic markers. Of the 1726 ILP primers, 230 (13.3%) generated clear and scorable polymorphic bands between the parental lines varying in size from 100 bp to 1300 bp. Amongst the 230 polymorphic primers, 211 (91.7%) each amplified DNA fragments from one locus and 18 primers (7.8%) each revealed two loci while the remaining one (0.5%) revealed three polymorphic loci. To sum up, a total of 250 polymorphic loci were scored including 141 co-dominant and 109 dominant markers. In addition to the polymorphic loci, 253 monomorphic bands were amplified by 146 primers. Taking into account both polymorphic and monomorphic bands, a total of 503 loci were detected by the 230 ILP primers with an average of 2.2 loci/ILP primer. Only 14 (6.3%) out of the 222 SSR primers amplified polymorphic fragments between the parental lines, which comprised 5 (35.7%) co-dominant and 9 (64.3%) dominant loci. The 250 ILP and 14 SSR polymorphic markers were used to construct the linkage map with the F2 population of Y514 × Y517 in yellow mustard.
Construction of a genetic linkage map
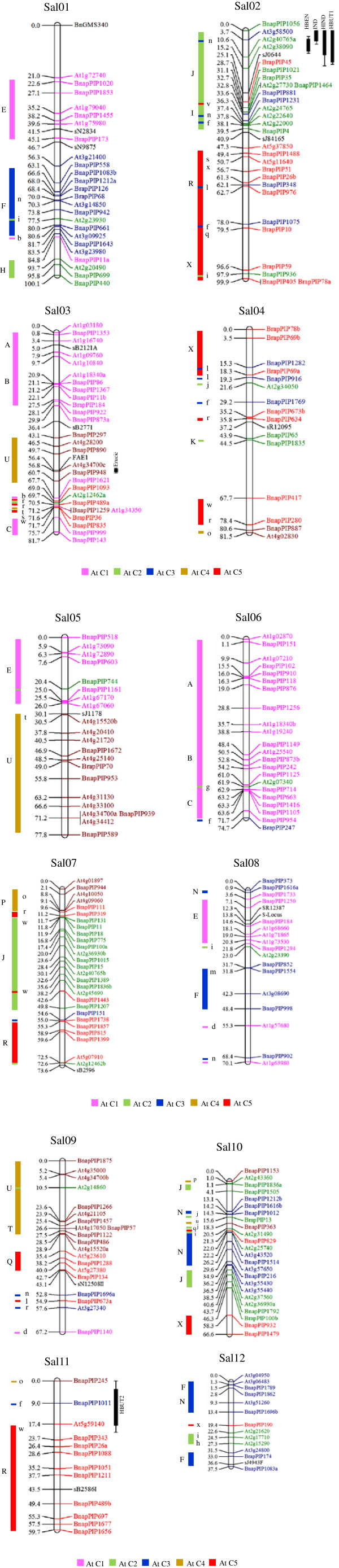
The 264 polymorphic loci between Y514 and Y517 were mapped on 12 linkage groups and covered a genetic length of 890.4 CentiMorgans (cM) (Table 1, Figure 1). The map length of the 12 linkage groups ranged from 37.5 to 100.1 cM with an average marker interval of 3.3 cM. They were designated as Sal01 to Sal12 in descending order of the genetic length. Sal01 and Sal02 had a similar long map length and average marker interval. However, Sal01 had one unmapped island (21.0 cM gap) located between the two markers BnGMS340 and At1g72740. Sal04 was similar with Sal03 in map length, but had the largest average marker interval (5.1 cM) and one unmapped island (23.2 cM gap) located between the two markers BnapPIP1835 and BnapPIP417. Sal05, Sal06, Sal07 and Sal08 had similar map length ranging from 77.8 cM to 70.1 cM. Sal07 had the smallest average marker interval of 2.6 cM. Sal09 and Sal10 were similar in both map length and number of markers. Sal11 had a map length of 59.7 cM with an average marker interval of 4.6 cM. The shortest linkage group Sal12 had 13 ILP and 1 SSR markers and a small average marker interval of 2.7 cM. The FAE1 gene was located adjacent to the marker At4g34700c on Sal03 with a genetic distance of 0.2 cM. The self-(in)compatibility gene was located close to the marker BnapPIP184 on Sal08 with a genetic distance of 0.8 cM. The ILP markers were evenly distributed on the 12 linkage groups which likely represented the 12 chromosomes in yellow mustard.
Table 1.
Characterization of the 12 linkage groups in yellow mustard
| Linkage group | Map length (cM) |
Marker interval (cM) |
No. of markers |
||||
|---|---|---|---|---|---|---|---|
| Average | Max distance (cM) | Min distance (cM) | ILP | SSR | Total | ||
| Sal01 |
100.1 |
3.7 |
21.0 |
0.3 |
24 |
3 |
27 |
| Sal02 |
99.9 |
3.3 |
17.1 |
0.0 |
28 |
2 |
30 |
| Sal03 |
81.7 |
2.8 |
11.2 |
0.0 |
27 |
2 |
29 |
| Sal04 |
81.5 |
5.1 |
23.2 |
0.6 |
15 |
1 |
16 |
| Sal05 |
77.8 |
3.5 |
12.8 |
0.1 |
21 |
1 |
22 |
| Sal06 |
74.7 |
3.4 |
9.8 |
0.1 |
22 |
0 |
22 |
| Sal07 |
73.6 |
2.6 |
12.9 |
0.1 |
27 |
1 |
28 |
| Sal08 |
70.1 |
3.9 |
13.1 |
0.1 |
17 |
1 |
18 |
| Sal09 |
67.2 |
3.2 |
13.1 |
0.0 |
20 |
1 |
21 |
| Sal10 |
66.6 |
2.8 |
12.0 |
0.1 |
24 |
0 |
24 |
| Sal11 |
59.7 |
4.6 |
8.4 |
2.2 |
12 |
1 |
13 |
| Sal12 | 37.5 | 2.7 | 6.5 | 0.2 | 13 | 1 | 14 |
Figure 1.
The 12 linkage groups (Sal01-12) in yellow mustard. For each linkage group, the ILP and SSR markers were shown on the right side, and the marker position in centiMorgan on the left side. The font colours of the markers and the colours of the highlighted conserved blocks indicated their chromosomal origin in A. thaliana. For each conserved block, the name was indicated on the left side and scattered markers were on the right side. The QTL for erucic acid content (ERU) co-localized with the FAE1 gene on Sal03. The QTLs for HBEN, IND, HIND and HBUT were mapped to Sal02. The other QTL for HBUT was mapped to Sal11. 1-LOD and 2-LOD supporting intervals of each QTL were marked by thick and thin bars, respectively.
The majority (83.1%) of the 264 mapped markers segregated with the expected 1:2:1 or 3:1 Mendelian ratios. However, 16.9% markers deviated from the expected segregation ratio of 1:2:1 or 3:1 (0.01 ≤ P ≤ 0.05). These distorted markers were unevenly distributed on linkage groups Sal03, Sal06, Sal08 and Sal10. Among the distorted marker loci, 29 (64.4%) loci skewed towards the homozygous Y517 genotype whereas 11 (24.4%) loci skewed towards the homozygous Y514 genotype. The remaining 5 loci (11.1%) skewed towards the heterozygous genotype. Interestingly, DNA fragments not observed in the parental lines were generated in the F1 hybrid plants by 59 ILP primers. The new bands observed in the F1 plants appeared in the heterozygote F2 plants.
Inheritance of HBEN, IND, HIND and HBUT GSL contents
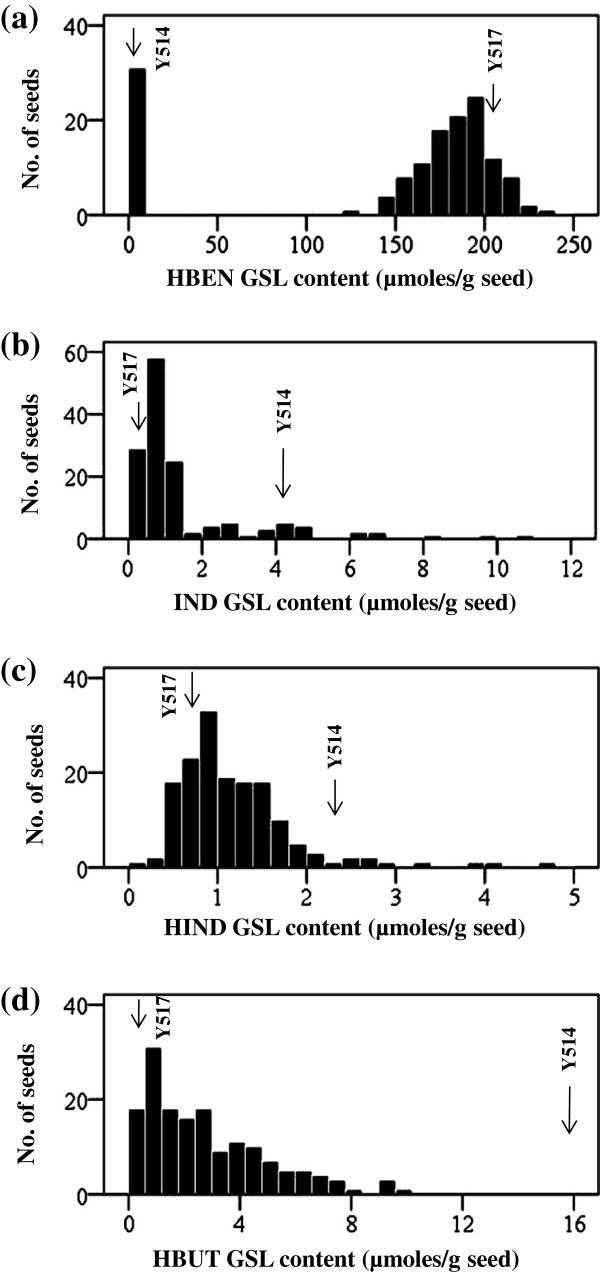
The F1 seed had similar HBEN, IND, HIND and HBUT GSL contents as the selfed female parental seed (Table 2), indicating that they were controlled by the maternal genotype. The F3 seeds borne on F2 plants were classified into two groups: seeds with zero (<0.3 μmoles/g seed), and seeds with medium to high (124.8-237.5 μmoles/g seed) HBEN contents, fitting well with a phenotypic ratio of 1 (zero):3 (medium to high) (χ2 = 1.0, P = 0.31) (Figure 2a). This result suggested that HBEN GSL was controlled by one gene locus with the dominance of high over the low content. The segregation of IND GSL content fitted with a phenotypic ratio of 3 (seeds with low content (0.2-1.6 μmoles/g seed)):1 (seeds with medium to high content (2.0-10.6 μmoles/g seed)) (Figure 2b), and was therefore under monogenic control with the dominance of low over the high content. HIND and HBUT contents of the F3 seeds exhibited continuous distribution and could not be classified into discrete groups (Figure 2c-d).
Table 2.
Erucic acid content, HBEN, IND, HIND and HBUT glucosinolate contents of parental lines Y514, Y517 and their reciprocal crosses F1 seeds
| Genotype | Erucic acida (%) |
Glucosinolate (μmoles/g seed) |
|||
|---|---|---|---|---|---|
| HBEN | IND | HIND | HBUT | ||
| Parenal lines |
|
|
|
|
|
| Y514 |
0.1 ± 0.0 |
0.1 |
4.3 |
2.3 |
16.3 |
| Y517 |
52.9 ± 2.6 |
210.4 |
0.5 |
0.8 |
1.0 |
| F1 |
|
|
|
|
|
| Y514 × Y517 |
34.6 ± 1.4 |
0.1 |
4.4 |
3.3 |
8.2 |
| Y517 × Y514 | 217.0 | 0.5 | 0.7 | 0.6 | |
aErucic acid expressed as mean value ± standard deviation.
Figure 2.
Frequency distribution of HBEN (2a), IND (2b), HIND (2c) and HBUT (2d) glucosinolate contents of F2 plants of Y514 × Y517. The arrows indicate the contents of the parental lines.
QTL analysis of erucic acid content, HBEN, IND, HIND and HBUT GSL contents
QTL analysis was performed for erucic acid content, HBEN, IND, HIND and HBUT contents (Table 3). One QTL (LOD = 83.5), accounting for 92.3% of erucic content variation, was detected and co-localized with the FAE1 gene on Sal03 (Figure 1). One QTL (LOD = 83.1), explaining 93.1% of the HBEN content variation was assigned to Sal02. It was located between the two markers At3g58500 and At2g40765a (Figure 1). One QTL (LOD = 36.1) explained 68.8% of the phenotypic variation of IND and was mapped in the region between the two markers BnapPIP1056 and At3g58500 on Sal02. One QTL (LOD = 13.4) for HIND was detected and located in the vicinity of the marker At3g58500 on Sal02, which accounted for 35.1% of the total variation. Two QTLs for HBUT content were detected. The first QTL (LOD = 7.1) accounted for 20.4% of the total variation and was mapped to the region near the marker At2g40765a on Sal02. The second QTL (LOD = 6.5) for HBUT explained 19.2% of the total variation and was located adjacent to the marker BnapPIP1011on Sal11. The QTLs for HBEN, IND and HIND GSL contents as well as one of the two QTLs for HBUT content were mapped to a terminal region on the same linkage group Sal02 (Figure 1).
Table 3.
Summary of QTLs for erucic content, HBEN, IND, HIND and HBUT glucosinolate contents in the F2 population derived from Y514 × Y517
| Trait | Linkage group | Marker or interval | Position cM | LOD score | LOD threshold | R2 a |
|---|---|---|---|---|---|---|
| Erucic acid |
Sal03 |
FAE1 |
56.4 |
83.5 |
3.8 |
92.3 |
| HBEN |
Sal02 |
At3g58500-At2g40765a |
5.7 |
83.1 |
3.6 |
93.1 |
| IND |
Sal02 |
BnapPIP1056 |
0.0 |
36.1 |
3.6 |
68.8 |
| HIND |
Sal02 |
At3g58500 |
3.7 |
13.4 |
3.5 |
35.1 |
| HBUT |
Sal02 |
At2g40765a |
10.6 |
7.1 |
3.5 |
20.4 |
| Sal11 | BnapPIP1011 | 9.0 | 6.5 | 3.5 | 19.2 |
aPercentage of the total phenotypic variation explained by QTL.
Synteny relationships between yellow mustard and A. thaliana
ILP markers were used for comparative analysis of the linkage maps of S. alba and A. thaliana (Figure 1, Table 4). All linkage groups of yellow mustard were chimeric in relation to A. thaliana chromosomes. Sal01 and Sal08 had markers of A. thaliana chromosome (AtC) 1, AtC2 and AtC3, which corresponded to three different conserved synteny blocks. Sal02 had four conserved blocks with two from AtC2 and another two from AtC5. Sal03 comprised 4 conserved blocks with three from AtC1 and one from AtC4. Sal04, Sal07 and Sal10 contained markers of AtC2, C3, C4 and C5, which were grouped into two conserved blocks in Sal04, and three conserved blocks in Sal07 and Sal10. Sal05 contained a large conserved block of AtC4 with 12 markers covering a genetic length of 40 cM (51.4% of the total map length), and one conserved block of AtC1 (Figure 1). Sal06 mainly contained marker loci of AtC1 whereas Sal11 had majority of markers of AtC5. Sal09 was the most chimeric linkage group with marker loci from all the five A. thaliana chromosomes (Figure 1). Sal12 carried three conserved blocks of AtC3. In addition to the conserved blocks, it was also observed that some markers of one At chromosome were scattered in a conserved block of a different At chromosome in the linkage groups of yellow mustard.
Table 4.
Summary of conserved blocks of A. thaliana in yellow mustard genome
| Linkage group | AKBa | No. of loci | Length of block (cM) | A. thaliana chromosome |
|---|---|---|---|---|
| Sal01 |
E |
6 |
24.1 |
1 |
| Sal01 |
F |
12 |
27.2 |
3 |
| Sal01 |
H |
3 |
6.4 |
2 |
| Sal02 |
J |
7 |
32.8 |
2 |
| Sal02 |
I |
4 |
2.1 |
2 |
| Sal02 |
R |
4 |
12.0 |
5 |
| Sal02 |
X |
3 |
3.3 |
5 |
| Sal03 |
A |
8 |
21.2 |
1 |
| Sal03 |
B |
4 |
7.8 |
1 |
| Sal03 |
U |
5 |
17.6 |
4 |
| Sal03 |
C |
2 |
6.0 |
1 |
| Sal04 |
X |
3 |
18.3 |
5 |
| Sal04 |
K |
2 |
0.6 |
2 |
| Sal05 |
E |
6 |
26.0 |
1 |
| Sal05 |
U |
12 |
40.0 |
4 |
| Sal06 |
A |
10 |
38.8 |
1 |
| Sal06 |
B |
5 |
12.6 |
1 |
| Sal06 |
C |
4 |
8.8 |
1 |
| Sal07 |
P |
2 |
0.5 |
4 |
| Sal07 |
J |
11 |
38.1 |
2 |
| Sal07 |
R |
5 |
17.5 |
5 |
| Sal08 |
N |
2 |
0.9 |
3 |
| Sal08 |
E |
7 |
18.2 |
1 |
| Sal08 |
F |
2 |
6.1 |
3 |
| Sal09 |
U |
8 |
22.3 |
4 |
| Sal09 |
T |
2 |
0.4 |
4 |
| Sal09 |
Q |
3 |
4.6 |
5 |
| Sal10 |
J |
3 |
3.1 |
2 |
| Sal10 |
N |
3 |
1.2 |
3 |
| Sal10 |
N |
5 |
10.3 |
3 |
| Sal10 |
J |
4 |
7.8 |
2 |
| Sal10 |
X |
2 |
8.3 |
5 |
| Sal11 |
R |
9 |
36.0 |
5 |
| Sal12 |
F |
4 |
2.8 |
3 |
| Sal12 |
N |
2 |
4.1 |
3 |
| Sal12 | F | 3 | 6.0 | 3 |
aAncestral karyotype blocks proposed by Schranz et al. [46].
Discussion
Doubled haploid and highly inbred lines of yellow mustard were successfully produced in our lab. This allowed us to construct a genetic linkage map by using an F2 population derived from homozygote parents in this out-crossing crop species for the first time. ILP primers were designed based on the conserved exon sequence flanking the introns to exploit intron polymorphism. Therefore, each ILP marker locus revealed by any particular primer would likely represent a gene copy in the genome. Taking both polymorphic and monomophic loci into account, 50.4% of the 230 ILP primers in the present study revealed duplicated or triplicated loci, thereby suggesting that yellow mustard is a secondary polyploidy species. This is in agreement with the previous RFLP mapping results [26]. The twelve linkage groups most likely represented the twelve chromosomes in yellow mustard. The presence of the unmapped islands in linkage groups Sal01 and Sal04 could be due to the low polymorphism in the two regions between the parental lines. The possible reasons for the occurrence of distorted ILP loci could be related to the linkage of the markers with self-incompatibility and also to natural selection for the heterozygous genotypes with higher vigour than the homozygotes. Distorted segregation ratio of markers were reported in linkage mapping of B. napus[27] and B. carinata[28]. In the F1 hybrid, some ILP primer pairs generated new DNA fragments that were absent in the parents, suggesting the occurrence of extensive divergence of either side of the flanking exon sequences in the parental lines. Each of the two parental lines might only have flanking exon sequence complementary to either the forward or reverse primer, thereby leading to no amplification. However, the F1 plants derived from such two parental lines would contain flanking exon sites that could anneal with both forward and reverse primers and therefore produced a new band.
One QTL was detected for erucic content, which was in agreement with the monogenic control of this trait in previous reports [29,30]. The biosynthesis of erucic acid is controlled by the FAE1 gene [31]. As expected, the QTL for erucic content co-localized with the FAE1 gene on Sal03. The biosynthesis of glucosinolates occurs in the silique wall (maternal tissue) and is then transferred to the developing seeds [32]. Therefore, the HBEN, IND, HIND and HBUT contents were controlled by the maternal genotype. Genetic studies and QTL mapping indicated that HBEN content was controlled by one gene locus with dominance of the high over low content as reported by Drost et al. [33]. The two markers At3g58500 and At2g40765a were tightly linked with the HBEN content and could be used for marker-assisted selection and map-based cloning. The allele for high HBEN content was linked with the allele for low IND content with a recombination frequency of 5.7%. Of the two genes for HBUT, one had a recombination frequency of 4.9% with the gene for HBEN on Sal02, and the other one was located on the linkage group Sal11. These results indicate that it is possible to develop new germplasm with different GSL profile in yellow mustard.
HBEN is an aromatic GSL produced from tyrosine. IND and HIND are indole GSLs derived from tryptophan, and HBUT is an aliphatic GSL with methionine as the precursor [34]. In A. thaliana, the QTL controlling indolic GSL was not overlapped with QTL for aliphatic GSLs, but the major QTL controlling the accumulation of seed benzyl GSL is linked to the GS-Elong locus that controls total leaf aliphatic glucosinolates [35]. The present study revealed that the QTLs for HBEN, IND, HIND and one QTL of HBUT were overlapped with each other at 1-LOD confidence interval in Sal02. It remains to be investigated whether the four GSL QTLs located in the same region are controlled by the same gene or due to linkage of different genes for the various GSL components in yellow mustard.
Comparative synteny analysis revealed that the linkage groups of yellow mustard shared many conserved blocks with that of A. thaliana. In particular, Sal06 and Sal11 mainly had common marker loci with AtC1 and AtC5, respectively. This suggested that Sal06 and AtC1 could be derived from the same ancestor chromosome while Sal11 and AtC5 from different one. However, it was also observed that most linkage groups of yellow mustard shared markers with over three chromosomes of A. thaliana, implying that extensive structural rearrangements such as translocations involving various chromosomes of the ancestor species and inversions had occurred during the speciation of yellow mustard. This is in agreement with the previous mapping results based on RFLP markers in yellow mustard [36]. Comparative mapping of Brassica species and A. thaliana also revealed the occurrence of extensive segmental rearrangement in B. nigra[20], B. napus[37], B. juncea[10] and B. oleracea[38]. The occurrence of conserved blocks between S. alba and A. thaliana would enable the genomics knowledge transfer from this model species to yellow mustard.
The current cultivars in yellow mustard are heterogeneous population varieties. With the objective to develop high yielding synthetic varieties, elite inbred lines tolerant to inbreeding have been produced by purging the deleterious alleles in each inbred generation [24]. Characterization of the genetic diversity of different inbred lines is essential for selection of synthetic component lines with high heterotic potential. The ILP markers and constructed genetic linkage map in this study will greatly facilitate molecular assisted breeding in our yellow mustard program.
Methods
Plant material and production of mapping population
The quality profiles of the two parental lines Y514 and Y517 were shown in Table 2. Y514 was the DH line SaMD3 produced at AAFC-SRC [23]. It had a zero erucic content (average: 0.1%) and a zero HBEN content (average: 0.1 μmoles/g seed). Y517 was produced by seven generations of inbreeding of the F1 plant between the variety Sabre and the Svalöf high oil line (T. Olson, personal communication, 2010). It had a high erucic acid content (average: 52.9%) and a high HBEN content (average: 210.4 μmoles/g seed). In addition, Y514 and Y517 differed in IND, HIND and HBUT GSL contents (Table 2). Reciprocal crosses were made between Y514 and Y517 to produce the F1 seed. The F1 plants were self-pollinated to produce F2 seeds. One hundred fifty F2 plants from one F1 plant (Y514 × Y517) were used for constructing the genetic linkage map. The parental lines, F1 plants and F2 population were grown in the greenhouse at AAFC-SRC.
DNA extraction and polymerase chain reaction (PCR)
Genomic DNA was extracted from young leaves of the parental, F1 and F2 plants using the modified sodium dodecyl sulfate method [39]. PCR for ILP markers was carried out according to Javidfar and Cheng [30]. For SSR marker analysis, the PCR mixture contained 50 ng genomic DNA, 200 μM of each dNTP, 1x PCR buffer, 200 nM of each primer and 1 U of Taq polymerase (New England Biolabs) in a final volume of 20 μl. The PCR was performed with an initial 5 min denaturation at 94°C; 20 cycles of 30 s at 94°C, 30 s at 56°C, 45 s at 72°C; 20 cycles of 30 s at 94°C, 30 s at 47°C, 45 s at 72°C; and a final 6 min extension at 72°C. Fragments were amplified with T7 fluorochrome- labeled primers (FAM, VIC, NED and PET) (Life Technologies). PCR products were combined and analyzed on a 3130xl DNA Analyzer with 600LIZ size standard (Life Technologies), and scored using GeneMapper 4 software (Life Technologies).
Construction of genetic linkage map and QTL analysis
The genetic linkage map was constructed by using JoinMap version 4.0 at LOD scores ≥ 4.0 [40]. Recombination frequencies were converted to map distances in cM using the Kosambi mapping function [41] and the genetic map was drawn with MapChart [42]. Chi-square test for goodness-of-fit was performed to determine if marker segregation deviated from the expected ratio. The threshold of P < 0.01 was used to exclude the distorted markers from the map construction. An interval mapping analysis [43,44] was conducted using the MapQTL 6.0 software [45] to detect QTLs for erucic acid content, HBEN, IND, HIND and HBUT GSL contents. Permutation test (1,000 replications) was used to determine the significance level for LOD with a genome-wide probability of P < 0.05. An island was defined as a region with a gap of greater than 20 cM between two adjacent markers [10].
Comparative synteny analysis
The constructed genetic linkage map in yellow mustard was compared with the established 24 genomic blocks (A-X) of A. thaliana[46]. A conserved block in the linkage map of yellow mustard was defined as a region with at least two closely linked ILP marker loci from the same block of A. thaliana.
Mapping of the FAE1 and self-(in)compatibility genes
Primer pair No 1 OF (ATGACGTCCGTTAACGTA) /PR (AAGACTTGTCGTCAGCTCCA) was designed based on the FAE1 gene sequence of Y517 and generated a dominant marker of 928 bp for FAE1 gene in Y517 (F. Zeng and B.F. Cheng, personal communication, 2012), while the primer pair No 2 Sal-SRK I (GATTATCTCGTGTCTGAATG/ GGTAATGTCGAATCTCTCCT) was designed based on the class I S haplotype of Y514 and produced a dominant marker of 640 bp for the self-(in) compatibility gene in Y514 [25]. The FAE1 and self-(in)compatibility genes were mapped to their respective linkage groups based on the segregation of the two markers in the F2 population of Y514 × Y517. The PCR reaction mixture (20 μl) contained 1x PCR buffer, 1.5 mM MgCl2, 200 μM of each dNTP, 0.1 μM of each forward and reverse primer, 1 U of Taq polymerase (New England Biolabs) and 50 ng of genomic DNA. Polymerase chain reaction was performed with an initial denaturation at 94°C for 5 min followed by 30 cycles of 45 s at 94°C, 45 s at annealing temperature and 1 min at 72°C. A final extension cycle of 72°C for 5 min was conducted. All PCR products were analyzed by electrophoresis in 2% agarose gels in 1x TAE buffer. Gels were visualized by staining in 0.5 mg/L ethidium bromide and photographed on a digital gel documentation system.
Erucic acid content and GSL profile assay
The erucic acid contents of the parental lines, F1 and F2 seeds were determined using the half-seed technique [47] and the gas chromatographic method of Thies [48], except that gas chromatography of the methyl esters was performed with a HP-INNOWax fused silica capillary column (0.25 mm × 7.5 m and 0.5 μm) (Agilent Technologies) at 250°C using hydrogen as the carrier gas. At least 20 seeds of each of the parental lines and 20 F1 seeds were half-seed analyzed for erucic acid content. Bulk samples of 30 self-pollinated seeds from each of the parental lines, 10 F1 seeds and 10 F3 seeds from each F2 plant were assayed for GSL profile using the method described by Raney et al. [49]. Benzyl GSL was isolated from the nasturtium seed in the chemistry lab at AAFC-SRC and used as standard.
Conclusions
We have constructed a genetic linkage map with ILP and SSR markers and used it for QTL analysis of erucic acid content and glucosinolate components in yellow mustard. The markers tightly linked with the genes controlling different glucosinolate components will be used for marker-assisted selection and map-based cloning. The ILP markers and linkage map in this study provide useful molecular tools for yellow mustard breeding.
Abbreviations
AAFC-SRC: Agriculture and Agri-Food Canada-Saskatoon Research Centre; AtC: Arabidopsis thaliana chromosome; cM: CentiMorgans; DH: Doubled-haploid; FAE1: Fatty acid elongase 1; GSL: Glucosinolate; HBEN: 4-hydroxybenzyl; HBUT: 2-hydroxy-3-butenyl; HIND: 4-hydroxy-3-indolylmethyl; ILP: Intron length polymorphism; IND: 3-indolylmethyl; PCR: Polymerase chain reaction; PIP: Potential intron polymorphism; QTL: Quantitative trait loci; RFLP: Restriction fragment length polymorphism; SNP: Single nucleotide polymorphism; SSR: Simple sequence repeat.
Competing interests
The authors declare that they do not have competing interests.
Authors’ contributions
BC conceived the project. FJ performed research and analyzed data. FJ and BC wrote the article. Both authors read and approved the final manuscript.
Contributor Information
Farzad Javidfar, Email: farzad.javidfar@agr.gc.ca.
Bifang Cheng, Email: bifang.cheng@agr.gc.ca.
Acknowledgements
The condiment yellow mustard breeding and research programs are supported by the Developing Innovative Agri-Products Initiative of the Growing Canadian Agri-Innovations Program (DIAP), Agri-Innovation Program, Canada, Mustard 21 Canada Inc. and the Agriculture Development Fund (ADF) of Saskatchewan, Canada.
References
- Brun H, Plessis J, Renard M. Resistance of some crucifers to Alternaria brassicae (Berk) Sacc. Poznan, Poland; 1987. pp. 1222–1225. (Proceedings of the 7th International Rapeseed Congress: 11–14 May 1987). [Google Scholar]
- Zhu JS, Spannier A. Resistance sources to Phoma lingam and Alternaria brassicae. Eucarpia Cruciferae Newslett. 1991;14/15:143. [Google Scholar]
- Hemingway JS. In: Brassica Oilseeds Production and Utilization. Kimber D, McGregor DI, editor. Wallingford: CAB International; 1995. The mustard species: condiment and food ingredient use and potential as oilseed crops; pp. 373–383. [Google Scholar]
- Downey RK, Rakow G. In: Harvest of Gold: The History of Field Crop Breeding in Canada. Slinkard AE, Knott DR, editor. Saskatchewan: University of Saskatchewan; 1995. Mustard. [Google Scholar]
- Choi S, Teakle G, Plaha P, Kim J, Allender C, Beynon E, Piao Z, Soengas P, Han T, King G. et al. The reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Theor Appl Genet. 2007;115(6):777–792. doi: 10.1007/s00122-007-0608-z. [DOI] [PubMed] [Google Scholar]
- Kim H, Choi SR, Bae J, Hong CP, Lee SY, Hossain MJ, Van Nguyen D, Jin M, Park B-S, Bang J-W. Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics. 2009;10(1):432. doi: 10.1186/1471-2164-10-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez-Luy F, Lukens L, Farnham M, Amasino R, Osborn T. Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. Theor Appl Genet. 2009;120(1):31–43. doi: 10.1007/s00122-009-1157-4. [DOI] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet. 2006;114(1):67–80. doi: 10.1007/s00122-006-0411-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Lydiate DJ, Parkin IAP, Falentin C, Delourme R, Carion PWC, King GJ. Integration of linkage maps for the Amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics. 2011;12(1):101. doi: 10.1186/1471-2164-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pradhan AK, Pental D. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics. 2008;9(1):113. doi: 10.1186/1471-2164-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchiary N, Padmaja KL, Sharma S, Gupta V, Sodhi YS, Mukhopadhyay A, Arumugam N, Pental D, Pradhan AK. Mapping of yield influencing QTL in Brassica juncea: implications for breeding of a major oilseed crop of dryland areas. Theor Appl Genet. 2007;115(6):807–817. doi: 10.1007/s00122-007-0610-5. [DOI] [PubMed] [Google Scholar]
- Mahmood T, Ekuere U, Yeh F, Good AG, Stringam GR. RFLP linkage analysis and mapping genes controlling the fatty acid profile of Brassica juncea using reciprocal DH populations. Theor Appl Genet. 2003;107(2):283–290. doi: 10.1007/s00122-003-1244-x. [DOI] [PubMed] [Google Scholar]
- Thormann CE, Romero J, Mantet J, Osborn TC. Mapping loci controlling the concentrations of erucic and linolenic acids in seed oil of Brassica napus L. Theor Appl Genet. 1996;93(1–2):282–286. doi: 10.1007/BF00225758. [DOI] [PubMed] [Google Scholar]
- Howell PM, Sharpe AG, Lydiate DJ. Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus) Genome. 2003;46(3):454–460. doi: 10.1139/g03-028. [DOI] [PubMed] [Google Scholar]
- Quijada PA, Udall JA, Lambert B, Osborn TC. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet. 2006;113(3):549–561. doi: 10.1007/s00122-006-0323-1. [DOI] [PubMed] [Google Scholar]
- Mahmood T, Ekuere U, Yeh F, Good AG, Stringam GR. Molecular mapping of seed aliphatic glucosinolates in Brassica juncea. Genome. 2003;46(5):753–760. doi: 10.1139/g03-051. [DOI] [PubMed] [Google Scholar]
- Nelson MN, Nixon J, Lydiate DJ. Genome-wide analysis of the frequency and distribution of crossovers at male and female meiosis in Sinapis alba L.(white mustard) Theor Appl Genet. 2005;111(1):31–43. doi: 10.1007/s00122-005-1961-4. [DOI] [PubMed] [Google Scholar]
- Yang L, Jin G, Zhao X, Zheng Y, Xu Z, Wu W. PIP: a database of potential intron polymorphism markers. Bioinformatics. 2007;23(16):2174–2177. doi: 10.1093/bioinformatics/btm296. [DOI] [PubMed] [Google Scholar]
- Gómez-Campo C. In: Brassica crops and wild allies Biology and breeding. Gómez-Campo C, Tsunoda S, Hinata K, editor. Tokyo: Japan scientific Societies Press; 1980. Morphology and morpho-taxonomy of the tribe Brassiceae; pp. 3–31. [Google Scholar]
- Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics. 1998;150(3):1217–1228. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Gao M, Yang B, Quiros CF. Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor Appl Genet. 2003;107(1):168–180. doi: 10.1007/s00122-003-1236-x. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171(2):765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundrock T. Doubled haploidy in yellow mustard (Sinapis alba L.). MS thesis. Saskatchewan, Saskatoon, Canada: University of Saskatchewan; 1998. [Google Scholar]
- Cheng B, Williams DJ, Zhang Y. Genetic Variation in Morphology, Seed Quality and Self-(in)Compatibility among the Inbred Lines Developed from a Population Variety in Outcrossing Yellow Mustard (Sinapis alba) Plants. 2012;1(1):16–26. doi: 10.5923/j.plant.20110101.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Cheng B. Self-(in)compatibility inheritance and allele-specific marker development in yellow mustard (Sinapis alba) Mol Breed. 2013. doi:10.1007/s11032-013-9943-8. [DOI] [PMC free article] [PubMed]
- Nelson MN, Lydiate DJ. New evidence from Sinapis alba L. for ancestral triplication in a crucifer genome. Genome. 2006;49(3):230–238. doi: 10.1139/G05-099. [DOI] [PubMed] [Google Scholar]
- Chen G, Geng J, Rahman M, Liu X, Tu J, Fu T, Li G, McVetty PE, Tahir M. Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus) Euphytica. 2010;175(2):161–174. doi: 10.1007/s10681-010-0144-9. [DOI] [Google Scholar]
- Guo S, Zou J, Li R, Long Y, Chen S, Meng J. A genetic linkage map of Brassica carinata constructed with a doubled haploid population. Theor Appl Genet. 2012;125(6):1113–1124. doi: 10.1007/s00122-012-1898-3. [DOI] [PubMed] [Google Scholar]
- Drost WJ, Rakow G, Raney P. Inheritance of erucic acid content in yellow mustard (Sinapis alba L.) Canberra, Australia: Proceedings of the 10th International Rapeseed Congress; 1999. [Google Scholar]
- Javidfar F, Cheng B. Single Locus, Multiallelic Inheritance of Erucic Acid Content and Linkage Mapping of FAE1 Gene in Yellow Mustard. Crop Sci. 2013;53(3):825–832. doi: 10.2135/cropsci2012.09.0552. [DOI] [Google Scholar]
- James DW, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. The Plant Cell Online. 1995;7(3):309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liangcheng D, Ann Halkier B. Biosynthesis of glucosinolates in the developing silique walls and seeds of Sinapis alba. Phytochemistry. 1998;48(7):1145–1150. doi: 10.1016/S0031-9422(97)00877-7. [DOI] [Google Scholar]
- Drost WJ, Rakow G, Raney P. Inheritance of glucosinolate content in yellow mustard (Sinapis alba L.) Canberra, Australia: Proceedings of the 10th International Rapeseed Congress; 1999. [Google Scholar]
- Halkier BA, Du L. The biosynthesis of glucosinolates. Trends Plant Sci. 1997;2(11):425–431. doi: 10.1016/S1360-1385(97)90026-1. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T. Comparative Quantitative Trait Loci Mapping of Aliphatic, Indolic and Benzylic Glucosinolate Production in Arabidopsis thaliana Leaves and Seeds. Genetics. 2001;159(1):359–370. doi: 10.1093/genetics/159.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MN, Parkin IAP, Lydiate DJ. The mosaic of ancestral karyotype blocks in the Sinapis alba L. genome. Genome. 2010;54(1):33–41. doi: 10.1139/G10-097. [DOI] [PubMed] [Google Scholar]
- Parkin IA, Sharpe AG, Lydiate DJ. Patterns of genome duplication within the Brassica napus genome. Genome. 2003;46(2):291–303. doi: 10.1139/g03-006. [DOI] [PubMed] [Google Scholar]
- Kaczmarek M, Koczyk G, Ziolkowski PA, Babula-Skowronska D, Sadowski J. Comparative analysis of the Brassica oleracea genetic map and the Arabidopsis thaliana genome. Genome. 2009;52(7):620–633. doi: 10.1139/G09-035. [DOI] [PubMed] [Google Scholar]
- Somers DJ, Friesen KRD, Rakow G. Identification of molecular markers associated with linoleic acid desaturation in Brassica napus. Theor Appl Genet. 1998;96(6–7):897–903. [Google Scholar]
- Van Ooijen JW. JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations Kyazma BV. Netherlands: Wageningen; 2006. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Voorrips RE. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121(1):185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet. 1992;84(7–8):803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. MapQTL 6. Wageningen, Netherlands: Software for the mapping of quantitative trait loci in experimental populations of diploid species Kyazma BV; 2009. [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in plant science. 2006;11(11):535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Downey RK, Harvey BL. Methods of breeding for oil quality in rape. Can J Plant Sci. 1963;43:271–275. doi: 10.4141/cjps63-054. [DOI] [Google Scholar]
- Thies W Schnelle und einfache Analysen der Fettsäurezusammensetzung in einzelnen Raps-Kotyledonen. 1. Gaschromatographische und papierchromatographische Methoden Z Pflanzenzüchtg 197165181–202.12287983 [Google Scholar]
- Raney P, Rakow G, Olson T. Development of low erucic, low glucosinolate Sinapis alba. Cambridge, UK: Proceedings of the 9th International Rapeseed Congress; 1995. pp. 416–418. [Google Scholar]