Abstract
Blast-related traumatic brain injury (TBI) has been one of the “signature injuries” of the wars in Iraq and Afghanistan. However, neuroimaging studies in concussive ‘mild’ blast-related TBI have been challenging due to the absence of abnormalities in computed tomography or conventional magnetic resonance imaging (MRI) and the heterogeneity of the blast-related injury mechanisms. The goal of this study was to address these challenges utilizing single-subject, module-based graph theoretic analysis of resting-state functional MRI (fMRI) data. We acquired 20 minutes of resting-state fMRI in 63 U.S. military personnel clinically diagnosed with concussive blast-related TBI and 21 U.S. military controls who had blast exposures but no diagnosis of TBI. All subjects underwent an initial scan within 90 days post-injury and 65 subjects underwent a follow-up scan 6 to 12 months later. A second independent cohort of 40 U.S. military personnel with concussive blast-related TBI patients served as a validation dataset. The second independent cohort underwent an initial scan within 30 days post-injury. 75% of scans were of good quality, with exclusions primarily due to excessive subject motion. Network analysis of the subset of these subjects in the first cohort with good quality scans revealed spatially localized reductions in participation coefficient, a measure of between-module connectivity, in the TBI patients relative to the controls at the time of the initial scan. These group differences were less prominent on the follow-up scans. The 15 brain areas with the most prominent reductions in participation coefficient were next used as regions of interest (ROIs) for single-subject analyses. In the first TBI cohort, more subjects than would be expected by chance (27/47 versus 2/47 expected, p < 0.0001) had 3 or more brain regions with abnormally low between-module connectivity relative to the controls on the initial scans. On the follow-up scans, more subjects than expected by chance (5/37, p = 0.044) but fewer subjects than on the initial scans had 3 or more brain regions with abnormally low between-module connectivity. Analysis of the second TBI cohort validation dataset with no free parameters provided a partial replication; again more subjects than expected by chance (8/31, p = 0.006) had 3 or more brain regions with abnormally low between-module connectivity on the initial scans, but the numbers were not significant (2/27, p = 0.276) on the follow-up scans. A single-subject, multivariate analysis by probabilistic principal component analysis of the between-module connectivity in the 15 identified ROIs, showed that 31/47 subjects in the first TBI cohort were found to be abnormal relative to the controls on the initial scans. In the second TBI cohort, 9/31 patients were found to be abnormal in identical multivariate analysis with no free parameters. Again, there were not substantial differences on the follow-up scans. Taken together, these results indicate that single-subject, module-based graph theoretic analysis of resting-state fMRI provides potentially useful information for concussive blast-related TBI if high quality scans can be obtained. The underlying biological mechanisms and consequences of disrupted between-module connectivity are unknown, thus further studies are required.
Keywords: functional connectivity, traumatic brain injury, graph theory, modularity, functional magnetic resonance imaging (fMRI), blast injury
INTRODUCTION
Traumatic brain injury (TBI) has been called a “signature injury” in the wars of Iraq and Afghanistan (Okie, 2006). As of the first quarter of 2012, the total incidence of TBI in U.S. military personnel since 2000 is 244,217 with 76.8% of these incidents are concussive or ‘mild’ TBI (Defense Medical Surveillance System and Theater Medical Data Store, http://www.health.mil/Libraries/TBI-Numbers-Current-Reports/dod-tbi-worldwide-2000-2012Q1-as-of-120516.pdf). Concussive or ‘mild’ TBI is characterized by loss of consciousness up to 30 minutes, altered consciousness and mental state up to 24 hours, post-traumatic amnesia up to 24 hours and the absence of abnormalities in computed tomography or conventional magnetic resonance imaging (MRI) (Casscells, 2007). However, utilizing advanced neuroimaging techniques such as functional magnetic resonance imaging (fMRI), diffusion tensor imaging (DTI), magnetoencephalography and electroencephalography, reports have described abnormalities in concussive TBI subjects (e.g., fMRI: Scheibel et al. (2012), Shumskaya et al. (2012), Slobounov et al. (2011), Tang et al. (2011); DTI: Levin et al. (2010), Mac Donald et al. (2011), Niogi et al. (2008, 2010), Shenton et al. (2012); fMRI and DTI: Mayer et al. (2011); magnetoencephalography: Castellanos et al. (2010, 2011); electroencephalography and DTI: Sponheim et al. (2011)).
Most of these previous functional neuroimaging studies in TBI have focused on group comparisons and have adopted hypothesis-driven approaches with predefined regions of interest, seed, or networks of interests. However, high individual variability of functional topology (van Essen and Dierker, 2007) is major source of variability in group analysis in healthy normal subjects. In TBI populations, the heterogeneity of injury types and locations (Doppenberg and Bullock, 1997; Saatman et al., 2008) further increases between-subject variability. In blast-related TBI (bTBI), the heterogeneity is further increased by the variety of blast-related injury mechanisms. Blast-related injuries may occur by (1) blast overpressure inducing mechanical damage to the brain, (2) having the head struck by debris or other objects set in motion by the blast (3) being thrown to the ground or against another stationary object or (4) inhaling toxic fumes, smoke or dust (Finkel, 2006; Warden, 2006). Different combinations of these injury types and other variables such as direction, distance and open field versus enclosed space associated with the blast exposures may make group analysis insufficient for the assessment of bTBI. The aforementioned heterogeneity of concussive bTBI also increases the chance for hypothesis-driven approaches with predefined regions or networks of interest to miss regions or networks with alterations of functional connectivity in concussive bTBI patients. Thus, single-subject based, data-driven approaches would be more meaningful in these heterogeneous concussive bTBI populations.
Recently, graph theory has become increasingly popular in neuroimaging research (see Rubinov and Sporns (2010) and Bullmore and Sporns (2009) for review), offering new insights into the understanding of the brain as a complex network. Several studies (Achard et al., 2006; He et al., 2007; Salvador et al., 2005; van den Heuvel et al., 2008) have found that the brain network has economical ‘small world’ properties having high levels of clustering and short path length for efficient global and local communications (Latora and Marchiori, 2001; Watts and Strogatz, 1998). Early studies of graph theoretic analysis in clinical populations have demonstrated disrupted ‘small world’ properties in patients with dementia of the Alzheimer’s type (Stam et al., 2006), schizophrenia (Micheloyannis et al., 2006) and epilepsy (Ponten et al., 2007). Taking advantage of the ‘small world’ properties of the brain network, subsequent studies (Chen et al., 2008; Hagmann et al., 2008; He et al., 2009; Power et al., 2011; Valencia et al., 2009; Yeo et al., 2011b) have identified modular or community structure of the normal, healthy human brain. With regard to clinical populations, Valencia et al. (2009) raised the possibility that characterizing the modular structure of the brain may be important to understand the brain organization during different pathological or cognitive states. Indeed, graph theoretic analysis of magnetoencephalography data has revealed disrupted modular structure in patients with dementia of the Alzheimer’s type (de Haan et al., 2012).
Another advantage of graph theoretic analyses over simple network approaches is that they do not require assumptions regarding hypothesized (thus predefined) seed regions or networks of interest. Thus, in this regard, graph theoretic analyses are useful in heterogeneous populations. With this advantage in heterogeneous populations over simple network approaches, recent studies (Caeyenberghs et al., 2012; Castellanos et al., 2011; Nakamura et al., 2009; Pandit et al., 2013) have utilized graph theoretic analyses to provide more comprehensive understanding of abnormal functional connectivity in TBI patients. In particular, Nakamura et al. (2009) demonstrated disrupted ‘small worldness’, defined as the level of clustering relative to path length, of functional networks in patients with moderate to severe TBI. To our knowledge, there are no previous studies that have investigated modular structure in resting-state functional connectivity MRI in patients with bTBI or any other concussive ‘mild’ TBI populations (though Pandit et al. (2013) included a wide range of injury severities).
In this study, we posited that module-based connectivity in patients with concussive bTBI may be disrupted. In our previous report (Mac Donald et al., 2011), we demonstrated DTI ‘abnormalities’ in white matter integrity of active duty U.S. military personnel with concussive bTBI relative to controls who had blast exposure but no diagnosis of TBI. At the time of the DTI and structural MRI collections in each of these subjects, resting-state blood oxygenation level dependent (BOLD) fMRI scans were also acquired. Here, we assessed modular organization of these active duty U.S. military personnel with concussive bTBI, utilizing whole brain, module-based graph theoretic analysis of these resting-state BOLD fMRI scans. Because of the heterogeneity of the concussive bTBI patients, we investigated module-based resting-state network properties at both the group and single-subject levels.
MATERIALS AND METHODS
2.1 Subjects
Three groups (controls and two TBI cohorts) of active duty U.S. military personnel deployed to the wars in Iraq and Afghanistan participated in this study. All of them had been exposed to blasts in a combat environment. The two TBI cohorts had sustained clinically diagnosed bTBI. The 21 controls (20 males; 19–49 years old with median = 29; 11–17.5 years of education with median = 12.5) had other injuries but screened negative for TBI (Dempsey et al., 2009). The first TBI cohort (TBI I cohort) consisted of a subset of the subjects about which we have reported previously (Mac Donald et al., 2011). Screening, enrollment, and initial scans were performed at the Landstuhl Regional Medical Center (LRMC), a U.S. Military hospital in Landstuhl, Germany. 63 TBI patients (all males; 19–57 years old with median = 25; 8–17 years of education with median = 12) were diagnosed with mild, uncomplicated traumatic brain injury based on the criteria from the Department of Defense (Casscells, 2007), marked by less than 30 minutes of loss of consciousness and the absence of abnormalities in conventional MRI and CT. Post blast exposure time on the initial scans at LRMC were within 90 days (median = 14). After 6–12 months from their initial scans, 65 out of these subjects traveled to Washington University in St. Louis for follow-up scans. More details and demographics of this cohort are in Mac Donald et al. (2011).
The same screening criteria as on the TBI I cohort and controls allowed the second TBI cohort (TBI II cohort) to comprise 40 additional concussive bTBI patients (37 males; 19–44 years old with median = 23; 9–16 years of education with median = 12). The TBI II cohort underwent the initial scans within 30 days (median = 7) after the blast exposure. After 6–12 months from their initial scans, 32 out of these subjects underwent follow-up scans at Washington University in St. Louis. The first cohort underwent initial scans in 2008–2009 whereas the second cohort was scanned in 2010–2011.
All subjects participated in this study after obtaining written informed consent and this study was approved by the Human Research Protection Office at Washington University in St. Louis, the Institutional Review Board for LRMC at Brooke Army Medical Center, and the Clinical Investigation Regulatory and Human Research Protection Offices of the U.S. Army Medical Research and Materiel Command. This study was also registered at clinicaltrials.gov (NCT00785304).
2.2 MRI data acquisition
Both initial scans at LRMC and follow-up scans in St. Louis were acquired using Siemens Magnetom Avanto 1.5 Tesla MRI scanners (Siemens, Germany) with identical imaging protocols. In each imaging session, three 412.5-second runs (total 1237.5 seconds) of resting-state BOLD fMRI were acquired using a 12-channel phase-arrayed head coil supplied by the manufacturer with T2*-weighted blipped EPI sequence (TR/TE = 2500/50 msec; flip angle (FA) = 90°; field of view (FOV) = 25.6 × 25.6 cm; matrix = 64 × 64) to obtain 165 images of each of 30 axial slices (4.0 mm thick) of the whole cerebrum. During resting-state fMRI acquisition, the subjects were asked to remain still during the scan, but no specific requests were made regarding eyes open versus eyes closed and no specific attempts were made to keep subjects awake. In the setting of acute injury, this was not feasible as some subjects had orbital injuries and extracranial injuries and analgesic medications after enrollment. See the discussion for the relevant limitations of the study findings due to these constraints.
For surface reconstruction and alignment to resting-state BOLD fMRI of each subject, the same head coil was used with one high resolution T1-weighted sagittal magnetization prepared rapid acquisition gradient echo (MPRAGE) image of the whole brain (TR/TE = 2000/2.92 msec; FA = 8°; FOV = 25.6 × 25.6 cm; matrix = 256 × 256; 176 slices, 1.0 mm thick).
2.3 MRI preprocessing
Briefly, our analyses consisted of cortical surface reconstruction of structural MRI, preprocessing of resting-state BOLD fMRI, projection of BOLD fMRI onto the reconstructed cortical surface, network construction and finally graph theoretic analysis (see Fig. 1). We used Freesurfer (Dale et al., 1999; Fischl et al., 1999a, 2002) for cortical surface reconstruction of structural MRI, AFNI (Cox, 1996) for fMRI preprocessing and SUMA (Saad et al., 2004) for surface mapping and surface-based analysis of fMRI time series. fMRI data were preprocessed in the three dimensional subject-native space of each participant.
Figure 1. An illustration of the analysis procedure.
For each subject, with volumetric structural MRI data (a), cortical surface (b) was reconstructed. Subsequently, the surface underwent the inter-subject alignment and spatial resampling close to the spatial resolution of resting-state BOLD fMRI (c) to allow surface-based, node-by-node cross-subject analyses. The preprocessed resting-state BOLD fMRI data (d) were converted to surface-based BOLD signal data (f) aligned to the individual cortical surface (e). BOLD fluctuation correlation coefficients between every pair of nodes in the brain (e.g., the gray square from red and cyan nodes in (e ) ) were obtained to yield a correlation matrix (g). A connectivity matrix (h) was derived by thresholding the correlation matrix, and a brain network (i) was constructed. In this illustration, yellow lines indicate connection between nodes. With the identified modules (three modules delineated by dashed lines in this example) in (j), modularity, within-module degree z-score (e.g., five magenta lines for the red-colored node in (k)) and participation coefficient (e.g., the distribution of magenta, cyan and olive lines for the red-colored node in (l)) were obtained for each node.
2.3.1 Surface reconstruction of structural imaging
Cortical surface reconstruction (Fig. 1(a) to (b)) was performed with the Freesurfer image analysis suite (version 5.1.0), online documented and freely available for download (http://surfer.nmr.mgh.harvard.edu/). The technical details of these procedures are described in previous publications (Dale et al., 1999; Dale and Sereno, 1993; Fischl et al., 1999a, 1999b, 2001b, 2002b, 2004a, 2004b; Jovicich et al., 2006; Segonne et al., 2004). Cortical surface reconstruction results for each image of the subjects were visually inspected to ensure the accuracy of skull stripping, Talairach transformation, gray/white matter boundary (white surface), gray matter/cerebrospinal fluid boundary (pial surface) and cerebral cortex label. When necessary, manual intervention was performed in order for Freesurfer to correctly reconstruct the cortical surface. The surface reconstruction was performed unblinded to group membership. See the discussion for the limitation of this study related to manual intervention and unblindness to group membership.
2.3.2 fMRI preprocessing
Volumetric BOLD fMRI data were preprocessed (Fig. 1(c) to (d)) with standard methods using a modified version of a shell script generated by afni_proc.py (http://afni.nimh.nih.gov/pub/dist/doc/program_help/afni_proc.py.html) from AFNI (Cox, 1996). Each subject’s whole-brain structural images were first skull-stripped and coregistered (affine transform with 12 parameters) to the fifth time point of the first fMRI run. For each fMRI run, the initial four time points were discarded to allow T1 magnetization saturation. Standard preprocessing methods were then used, including despiking, slice timing correction, motion correction, normalization to whole brain mode of 1000, linear regression and band-pass filtering (0.009 < f < 0.08 Hz). At the motion correction stage, the 6 rigid body motion profiles were obtained for the linear regression. After the motion correction, subject masks indicating voxels that have fMRI signal were obtained for each of the subjects. In the linear regression, several sources of signal fluctuation unlikely to be of neuronal origin were regressed out as the nuisance variables: (1) six parameters for the rigid body head motion acquired from the motion correction (Johnstone et al., 2006), (2) the signal averaged over the lateral ventricles, (3) the signal averaged over a region centered in the deep cerebral white matter, (4) the signal averaged over the whole brain (Fox et al., 2005; see the control analyses and their results for the effects of global signal regression on graph theoretic analysis) and (5) the first temporal derivatives of aforementioned parameters. After the band-pass filtering, motion ‘scrubbing’ (Power et al., 2012) was performed with frame-to-frame head movement rate of 0.12 mm/s and standardized DVARS (http://www2.warwick.ac.uk/fac/sci/statistics/staff/academic-research/nichols/scripts/fsl/DVARS.sh) of 1.49 to prevent potential motion artifacts (Power et al., 2012; Satterthwaite et al., 2012; van Dijk et al., 2012). To prevent the introduction of artificial correlations in the fMRI signal between voxels (1) adjacent to each other in space distant in terms of cortical surface topology (e.g., voxels on opposite sides of the midline) or (2) that were located near the boundary of functional subdivisions unrelated to each other in functional connectivity (e.g., primary motor versus primary somatosensory cortex), spatial smoothing was not applied at this preprocessing step (van den Heuvel et al., 2008).
2.3.3 Inter-subject alignment and surface mapping of volumetric fMRI
The total number of mesh nodes in the reconstructed cortical surface by Freesurfer varies across subjects. To allow for cross-subject analysis while preserving geometry of sulcal and gyral patterns in the original surface of each individual and minimizing unnecessary interpolation artifacts (Argall et al., 2006), we used SUMA (Saad et al., 2004) to standardize surface meshes (i.e., coordinates) of each individual (i.e., the total number of mesh nodes is same across subjects and each mesh node corresponds to same anatomical location in each surface of the subjects). When calculating new coordinates, SUMA allowed us to set the total number of mesh nodes in the standard mesh surface of each subject (Fig. 1(e)) to be 11,524 so that the average distance between two nodes (3.7 mm) is close to the spatial resolution of original volumetric fMRI data (4 mm isotropic) while maintaining topology shown in the original high resolution (1 mm isotropic) structural MRI.
Volumetric functional time series were then projected onto these standard mesh surfaces of each subject by interpolating the time series located along the line between two matching nodes of the white and pial surfaces. For each mesh node, five equally-spaced coordinates were sampled between corresponding white and pial surfaces. At each time point, functional data were projected by averaging across the unique 3D voxels belonging to these coordinates. Consequently, surface-based functional time series (Fig. 1(f)) contained signal only from voxels within the cortical gray matter. In the same way, the voxel-based subject masks were converted to surface-based subject masks. For more technical details of the surface mapping procedure, readers are referred to Saad et al. (2004). Due to susceptibility artifacts (Ojemann et al., 1997) and inclusion of only cortical areas of surface (i.e., exclusion of the surface areas of the amygdala, putamen, hippocampus, caudate, ventricles and corpus callosum), not all nodes had fMRI signal, and surface-based subject masks indicating existence of fMRI signal on mesh nodes were different across the subjects. Thus, to make a comparison across subjects, further analyses on network measures considered only the mesh nodes (8,977 nodes) having fMRI signal across all subjects. This was performed by obtaining a subject-intersection mask and subsequently applying the intersection mask to surface-based functional time series of each of the subjects.
2.3.4 Quality assurance
We restricted our analysis to the subjects whose data quality was reliable within tolerable range. In the cortical surface reconstruction step, the quality of T1 images was visually inspected to determine if surface reconstruction was feasible. In fMRI preprocessing, the quality of preprocessed data was visually inspected at each step. After visual inspection, a subset of subjects’ data were excluded for following reasons: (1) a superior part of the functional images did not fall within the prescribed FOV due to substantial run-to-run change of head position, (2) intensity variation artifacts of low spatial frequency presumably due to constant oscillating head movement in a certain direction, (3) substantial susceptibility artifacts (Ojemann et al., 1997) in inferior frontal and inferior temporal regions, (4) motion correction failure due to large amount of abrupt motion and (5) lack of fMRI frames due to subject’s refusal to stay in the scanner. After motion ‘scrubbing’, additional subjects’ data were excluded if total length of remaining volumes after the ‘scrubbing’ was less than 4 minutes, the minimum length required to reliably estimate functional connectivity (van Dijk et al., 2010). See Table I for the details of the number of datasets excluded by this quality assurance procedure.
Table I.
Excluded datasets after the quality assurance procedure.
| Criteria | Ctrl | TBI I | TBI II | Total | |||
|---|---|---|---|---|---|---|---|
| Initial | Follow-up | Initial | Follow-up | Initial | Follow-up | ||
| Poor T1 image quality | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Brain outside FOV | 1 | 0 | 1 | 0 | 3 | 1 | 6 |
| Motion correction failure | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| Lack of frames | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Intensity distortion | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Susceptibility artifacts | 1 | 1 | 0 | 1 | 0 | 0 | 3 |
| <4 mins of data after motion scrubbing | 5 | 4 | 7 | 9 | 4 | 3 | 32 |
| Unidentifiable major modules | 1 | 1 | 4 | 0 | 2 | 1 | 9 |
| Total | 9 | 6 | 16 | 10 | 9 | 5 | 55 |
Note: FOV, field of view; Ctrl, Control; TBI I, TBI I cohort; TBI II, TBI II cohort.
Visual inspection on each module of the subjects identified by the Louvain algorithm allowed us to verify that, in 1 control, 4 TBI I and 2 TBI II subjects on the initial scans and 1 control and 1 TBI II subjects on the follow-up scans, the total number of modules was too few (less than 3) or module assignments were severely scattered yielding failure to identify major modules shown in group module assignment maps (Fig. 2). Thus, we conservatively excluded datasets with unidentifiable major modules in subsequent analyses as we could not be sure whether these module assignments were results of subjects’ condition or merely failures of the module identification algorithm (Table I).
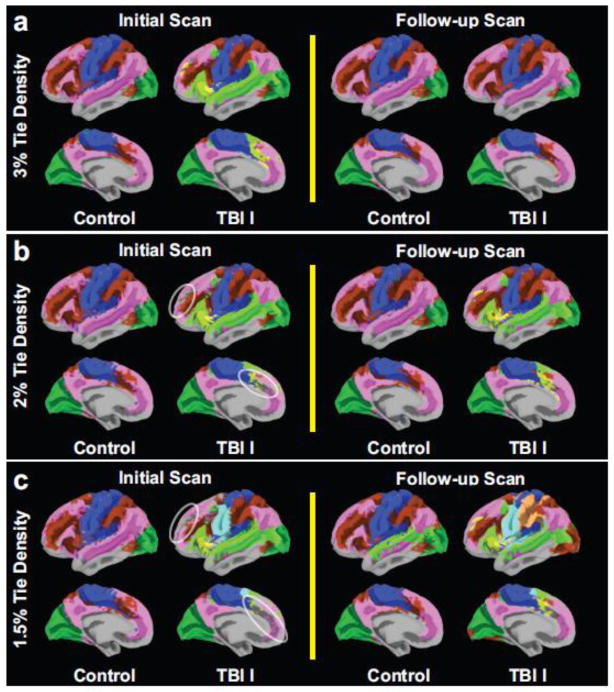
Figure 2. Group module assignments of each of the controls and the first TBI (TBI I) cohort.
The identified modules from group averaged correlation matrices were color-coded as a function of tie densities (densities of the retained strongest correlations): 3% (a), 2% (b) and 1.5% (c). Only modules of size greater than 1% of total number of nodes were displayed.
After quality assurance exclusion, we analyzed functional images of 12/21 control subjects on the initial scans, 12/18 controls subjects on the follow-up scans, 47/63 TBI I subjects on the initial scans, 37/47 TBI I subjects on the follow-up scans, 31/40 TBI II subjects on the initial scans and 27/32 TBI II subjects on the follow-up scans for the subsequent network analyses. Thus, 75% of all subjects’ data acquired as described above were of sufficient quality for further analyses. Note that we included subjects’ data if the data passed the quality assurance procedure for each scan separately. In other words, after the quality assurance procedure, included subjects on the follow-up scan analyses were no longer an exact subset of subjects included on the initial scan analyses.
2.4 Network analysis
2.4.1 Network construction
Weighted and undirected networks were constructed (Fig. 1(g)–(i)) for module-based graph theoretic analysis. For the network analysis, a node, a basic element of graph theoretic analysis, was defined as a mesh node in the cortical surface. An edge of the graph was defined from correlation matrix (Fig. 1(g)) whose components are Pearson correlation coefficients of time series at each pair of the mesh nodes in the brain. In other words, the weights of the edges were the correlation coefficients. An alternative to Pearson correlation coefficients would be the partial correlation coefficients. The partial correlation coefficients control for the influence of correlations from the other nodes on a correlation between two nodes of interest, which could replace global signal regression. Unfortunately, it was not feasible to use partial correlation coefficients in this study since the inversion of the covariance matrix to obtain the partial correlations was numerically unstable. More specifically, the total number of frames has to be greater than total number of nodes to ensure numerically stable estimation of the inverse covariance matrix. However, total number of frames (at most 495 frames) was far less than total number of nodes (8,977 nodes) in our case. Thus, we proceeded with the use of Pearson product moment correlations as edge weights.
Correlation coefficients between time series at short-distance nodes (20 mm in Euclidean distance), presumably associated with non-biological origins such as increased correlation by preprocessing and subject motion, were excluded in selecting edges of the graph (Power et al., 2011). The remaining correlation coefficients were thresholded at 3% tie density, i.e., density of the retained strongest correlations, to define edges of the graph (a colored dot and a yellow line in Fig. 1(h) and (i), respectively) for most analyses. Sparse tie densities were selected since module identification algorithms perform reliably when graphs are sparse (Fortunato, 2010). Further, inclusion of weak correlation coefficients (i.e., high tie density) could yield less reliable module-based graph measures due to artificial correlations between noise time courses. In addition, we explored the effects of 2% and 1.5% tie densities. For less than 1.5% tie densities, the TBI patients had excessive numbers of ‘trivial’ modules making it difficult to fairly compareal the remaining major modules of the TBI groups with those of the controls. These ‘trivial’ modules were defined as <1% of brain nodes; most generally had a single node without connection to other nodes. In these thresholding procedures, only positive correlation coefficients were considered for the network connections, as there is ongoing debate about the meaning of negative correlations assessed after global signal regression (Anderson et al., 2011; Chai et al., 2012; Chang and Glover, 2009; Fox et al., 2009; Murphy et al., 2009; Saad et al., 2012).
2.4.2 Module identification and module-based network properties
With the constructed binary and undirected brain networks, module-based graph theoretic analysis was performed using brain connectivity toolbox in MATLAB (Rubinov and Sporns, 2010) freely available online (http://www.brain-connectivity-toolbox.net) after applying the previously described subject-intersection mask for the nodes having functional times series across all subjects. First, the modules were identified (Fig. 1(j)). After module identification, global and node-specific module-based network properties were obtained.
Identification of modules is a complex and computationally demanding problem. For the module identification, modularity of a weighted and undirected network, , was defined:
where M is the number of modules, ls is the sum of the weights of all within-module connections in the module s, L is the total sum of all weights in the network, ds is the sum of the strength at each node in the module s and strength of a node is sum of the weights of all edges associated with the given node (Newman, 2004; Guimera and Amaral, 2005). In theory, is bounded between 0 and 1 (Newman, 2004; Guimera and Amaral, 2005). when nodes are randomly partitioned or all nodes belong to the same module. Thus, higher modularity means deviations from random networks with no community structure. In practice, modularity of typical networks with strong modular structure ranges from 0.3 to 0.7, and higher values are rare (Newman and Girvan, 2004). Assuming the brain network has modular structure (i.e., many within-module connections whereas few between-module connections), a module identification algorithm optimizes the total number of modules and the associated module membership of nodes for maximum modularity. For the implementation of our analysis, we used the Louvain algorithm (Blondel et al., 2008), a fast and relatively accurate algorithm, suitable for large networks. Due to “heuristic” nature of this algorithm (i.e., a ‘good enough’ approximation of the exact solution is implemented, resulting in faster execution time), the module identification algorithm was executed ten times. Then, we selected single module identification result from among the 10 executions that yielded the highest modularity to report modularity and assess subsequent module-based network measures. Overall, the variation of modularity over the executions was negligible, as in Blondel et al. (2008). For comparison, we additionally identified modules using the Infomap algorithm (Rosvall and Bergstrom, 2008), another module identification algorithm (See Supplemental Figs. S2, S3).
Given the identified modules, weighted within-module degree z-score (Fig. 1(k); Guimera and Amaral, 2005) and weighted participation coefficient (Fig. 1(l); Guimera and Amaral, 2005) were measured at each node of the individual brain network. In calculating within-module degree z-scores and participation coefficients, we excluded trivial modules whose size was less than 1% of the total number of nodes.
Briefly, weighted within-module degree z-score of node i, , measures normalized strength of connections from a node within the corresponding module s. zi can be written as:
where si is the module containing node i, , within-module strength, is the total sum of weights of edges connecting node i and all other nodes within si, and σkw(si) are the respective mean and standard deviation of the for all nodes j ∈ si. So, high weighted within-module degree z-score of a node means the node has a larger than expected strength within its own module.
Weighted participation coefficient of node i, , is defined as:
where is the total sum of weights of edges connecting node i and all other nodes in module s and , strength of node i, is the total sum of weights of edges connecting between node i and all other nodes in the entire network. Weighted participation coefficient shows how well a node communicates with other modules. Weighted participation coefficient is close to 1 if the distribution of connections at a node across modules is uniform. Weighted participation coefficient becomes 0 if there is no inter-module connection. A high value of weighted participation coefficient means nodes’ inter-module connections are ‘well-distributed’ over multiple modules, thus are likely to span more modules.
Each node-specific measure was then spatially smoothed (10 mm full-width-at-half-maximum (FWHM)) on the cortical surface of each individual to increase signal-to-noise ratio as in van den Heuvel et al. (2008). To identify ‘abnormal’ regions in the TBI patients, counted thewe number of patients whose network measures were outside two standard deviations from the mean of the controls.
2.5 Region of interest analysis
In the region of interest (ROI) analysis, the TBI I cohort served as an exploratory dataset to identify functional ROIs exhibiting noticeable difference in node-specific network measures between the controls and TBI patients from the TBI I cohort. The TBI II cohort served as a validation dataset with no free parameters with regard to ROI selection. Surface-based ROIs were selected on the standard mesh template in reference to the Destrieux surface atlas (Destrieux et al., 2010) using SUMA (Saad et al., 2004) to define the center of each ROI. Similar to the method described in Hagler et al. (2006), we slid a threshold level between puncorr = 0.05 and 0.01 from the group comparison map for participation coefficients to identify functional ROIs. We first identified ROI candidates with cluster area (white matter surface) greater than 150 mm2 at puncorr = 0.05. Then we subdivided large clusters in reference to the Destrieux atlas (Destrieux et al., 2010) and slid the threshold level up to puncorr = 0.01. With peaks that survived at puncorr = 0.01, we selected ROIs comprising nodes within 15 mm geodesic distance (along the white matter surface) from the peaks and whose puncorr < 0.05. If nodes within 15 mm geodesic distance from the peaks of these preliminary ROIs were part of 2 neighboring ROIs such that there was overlap, the boundaries of these ROIs were determined by sliding the threshold down from puncorr = 0.01 towards 0.05 allowing the clusters to grow until the ROIs reached the edge of the neighboring clusters. With these identified ROIs from the first dataset (i.e., the controls versus TBI I cohort), we performed ROI analysis on the second dataset (i.e., the controls versus TBI II cohort). For each ROI, we defined that a TBI patient had an ‘abnormal’ network measure relative to the controls in the ROI if the average network measure of the patient in the ROI was outside the mean plus or minus two-standard deviation band of the controls group. This procedure to identify TBI patients with ‘abnormal’ network measure in ROIs was carried out after the normality test of the controls’ ROI-specific network measures.
2.6 Multivariate region of interest analysis
A multivariate approach was then used to decide which TBI patients had ‘abnormal’ measures relative to the controls over all ROIs by aggregating average node-specific measures within each of the ROIs. After confirming that controls’ network measure at each ROI passed the normality test, multivariate Gaussian distribution of the network measures for the controls was estimated. Since the sample size of the controls after the module identification was small compared to the number of ROIs, estimating covariance structure was challenging. In order to circumvent this ‘curse of dimensionality’ issue (Duda et al., 2001), the vector dimension was reduced using probabilistic principal component analysis (PPCA; Minka, 2000). PPCA automatically estimates the number of reduced components preserving the variability of the original high dimensional vector while eliminating spurious and noisy components. Based on reduced components of ROI-based network measures via PPCA, we defined relatively ‘abnormal’ TBI patients whose components were located within the lower and upper tails (less than the 2.5th percentile and greater than the 97.5th percentile) of the estimated multivariate normal distribution from the controls.
2.7 Statistical analyses
All statistical analyses were assessed in MATLAB. First, we performed the Shapiro-Wilk test at α = 0.05 to assess the normality of distributions of each group’s demographics (age, years of education and post-injury time at the initial scan) and each network measure. The aforementioned demographics did not pass the Shapiro-Wilk normality test. Thus, the Mann-Whitney U test was used to compare the demographics between each pair of groups: (1) the controls versus TBI I cohort, (2) control versus TBI II cohort and (3) TBI I cohort versus TBI II cohort. Chi-square tests were used to compare the gender distributions between each pair of groups.
All network measures for the control group passed the Shapiro-Wilk normality test, but some measures for the TBI groups did not. Thus, for group comparison of the network measures, two sided hypothesis tests were taken using permutation tests (10,000 permutations; Nichols and Holmes, 2001) on group means of each measure by permuting group membership. For global and region-specific network properties, permutation tests were performed on the t-statistics.
We performed the one-sided z-test (TBI > control) to compare the distributions of the number of TBI patients with more than two relatively ‘abnormal’ regions versus those expected. To calculate the expected number of TBI patients with relatively ‘abnormal’ regions, the binomial distribution was used with the probability that a region is relatively ‘abnormal.’ This probability was calculated from both upper and lower tails (i.e., two-standard deviations ± mean) of the normal distribution. More specifically, the estimated number of TBI patients with m ‘abnormal’ regions out of total n ROIs was:
where |S| is total number of TBI patients, is n choose m and p is the probability that one region is ‘abnormal’ by chance. The number of ‘abnormal’ regions expected to occur by chance in each TBI patient was estimated based on the assumption that the ROIs are statistically independent (see Mac Donald et al., (2011) for the details of this statistical test procedure).
To minimize potential bias from possible non-independence across ROIs in the single-subject analyses, we additionally performed one-sided z-tests (TBI > control) on the distributions of the number of TBI patients as a function of the proportion of ‘abnormal’ nodes versus those expected. The expected distributions were obtained by permuting group memberships (10,000 permutations) and redefining abnormality based on permuted ‘controls’.
2.8 Control analyses
To assess effects of motions on module-based graph theoretic measures obtained from our cohorts, each subject’s average frame-to-frame head movement after censoring was first calculated and compared between cohorts. To compare the average frame-to-frame head movement after motion censoring between cohorts, subjects whose images passed all quality assurance procedure up to the motion censoring criterion were included (see Table I). To assess effects of motion on module-based graph theoretic measures, i.e., average participation coefficients within each cohort and the number of ‘abnormal’ ROIs in each TBI cohort, subjects whose images passed all quality assurance procedure were included. We also assessed the effect of thresholds on our findings by applying two different threshold levels at 2% and 1.5% tie densities to the correlation matrices and subsequently performing group and ROI analyses of participation coefficients. To assess the effect of global signal regression on our results, we additionally preprocessed our data without global signal regression and constructed a correlation and connectivity matrix. ROI analyses after group comparisons were then performed for participation coefficients. Lastly, we selected a different version of ROIs comprising nodes within 20 mm geodesic distance from the peaks, and we repeated the ROI analyses to verify effects of different size of ROIs on our results.
RESULTS
3.1 Demographics comparisons between the groups
There were no statistically significant differences in age, education, gender, or post-injury time between the subjects whose scans were included in graph theoretical analyses and their respective whole cohorts (Table II). The whole cohorts did not show statistically significant differences in age, education and post-injury time comparing the control vs. TBI I, control vs. TBI II, and TBI I vs. TBI II cohorts. There were differences in gender that were significant only between the two TBI cohorts (p = 0.03; chi-square test) as only the TBI II cohort had females (Table II). For the subjects whose scans (initial or follow-up scans) were included in graph theoretical analyses, there were no statistically significant differences in the demographics between each cohort except gender between the two TBI cohorts (p = 0.04; chi-square test).
Table II.
Demographics of the controls, TBI I cohort and TBI II cohort.
| Demographicsa | Ctrl, all (N=21) | Ctrl, subsetb (N=14) | TBI I, all (N=63) | TBI I, subsetb (N=54) | TBI II, all (N=40) | TBI II, subsetb (N=38) |
|---|---|---|---|---|---|---|
| Age (years)c | 19–49, 29 | 20–49, 29 | 19–57, 25 | 19–44, 24 | 19–44, 23 | 19–44, 23 |
| Gender (males, females) | 20,1 | 14,0 | 63,0 | 54,0 | 37,3e | 35,3f |
| Education (years)c | 11–17.5, 12.5 | 11–16, 12 | 8–17, 12 | 8–17, 12 | 9–16, 12 | 9–16, 12 |
| Post-injury time (days)c,d | N/A | N/A | 0–90, 14 | 0–90, 14 | 0–30, 7 | 0–30, 7 |
Demographics of the controls and TBI I cohort were reproduced from Table I in Mac Donald et al. (2011).
Subsets of subjects that were included in graph theoretical analyses of either the initial scans, the follow-up scans, or both.
Range and median values were reported.
Post-injury time on the day of the initial scan.
p < 0.05 (chi-square) vs. TBI I, all.
p < 0.05 (chi-square) vs. TBI I, included subset.
3.2. Seed-based approach results for the default mode network
To ensure that the preprocessed resting-state BOLD fMRI had acceptable data quality for the further network analyses, we obtained the seed-based correlation maps from the left posterior cingulate cortex (−7, −55, 27) per each subject dataset that passed the quality assurance procedure prior to checking module assignment results. Group statistic maps (Fig. S1) for the seed-based correlation maps of each control and TBI I group on both scans showed the default mode network (See Fig. S1 legend for the detailed methods for the seed-based approach we used).
3.3. Module identification results
Color maps for identified modules from group averaged correlation matrix (Fig. 2) allowed us to compare major modules (comprising more than 1% of the total number of nodes) between groups at each tie density. In the control group, there were typically 4 major modules identified (except at 1.5% tie density on the follow-up scans, where there were 5). The identified 4 major modules in the control group corresponded to the default mode (Greicius et al., 2002; Raichle et al., 2001), executive control (Seeley et al., 2007; Vincent et al., 2008), visual (Lowe et al., 1998) and somatosensory-motor (Biswal et al., 1995) networks. In contrast, the TBI I cohort generally had more than 4 modules (except at 3% tie density on the follow-up scans). These module identification results were consistent with modular organization maps from young healthy subjects reported elsewhere (Liang et al., 2013) using the same module identification algorithm. At 3% tie density, subdivided modules (light green and yellow modules) in the TBI I cohort on the initial scans reorganized (merged to other modules) on the follow-up scans and their module assignments became similar to modular structures of the control groups. At 2% and 1.5% tie densities, many subjects in the TBI I cohort had substantial numbers of trivial modules in lateral prefrontal cortex and anterior cingulate cortex (white circles) on the initial scans, whereas fewer trivial modules were observed on the follow-up scans (Fig. 2b–c). Further assessment revealed that these trivial modules were disconnected from major modules and most of them consisted of single node, i.e., a node with no connections to other nodes.
An alternative module identification algorithm (Infomap) also demonstrated a large number of trivial modules and increased total number of major modules in the TBI I cohort on the initial scans (Fig. S3). For the controls, module assignments were largely similar to the results of Power et al. (2011), but direct comparisons at the same tie densities were not feasible as other parameters were different. Specifically, definition of nodes (i.e., modified voxel or large functional areas versus vertex of surface) and brain regions that involved the analyses (i.e., whole-brain including subcortical versus cortex only) were different. Note that we did not further compare Louvain algorithm results with those obtained by the Infomap algorithm as the Infomap algorithm did not reliably identify modules in some of our cohorts at the single-subject level (e.g., see Fig. S3).
3.4 Global network properties
Group differences in modularity (Fig. 3) were statistically significant at α = 0.05 on the initial scans, but not on the follow-up scans. The TBI I cohort had higher modularity on the initial scans than the controls. Modularity in both groups passed the Shapiro-Wilk normality test. Modularity of the controls ranged from 0.3 to 0.55 (Fig. 3). This range is slightly low, especially on the initial scans, but the range is comparable to the modularity range (0.4 to 0.6) previously described in healthy normal subjects (Meunier et al., 2009a, 2009b; Valencia et al., 2009). At the single subject level, 14 TBI patients on the initial scans had ‘abnormally’ high modularity and 2 had ‘abnormally’ low modularity (Fig. 3). Note that by chance only 2 subjects out of 47 (4.8%) would be expected to be outside of the 2-standard deviation range; 1 above and 1 below. With regard to trivial modules observed in the TBI patients (Fig. 2), the effect of excluding trivial modules on the modularity value was negligible (< 10−6) since most of the trivial modules comprised of single nodes (see the modularity equation in the methods section).
Figure 3. Whole brain modularity of the controls and the first TBI (TBI I) cohort.
Each symbol represents a single individual’s modularity. The I bars indicate the means and standard deviation of the control, the dotted horizontal bar is two standard deviations from the mean of the control and the solid horizontal bar in the TBI I cohort is the mean of the TBI I cohort. Filled triangles represent TBI patients with relatively ‘abnormal’ modularity, located outside of the dotted horizontal bars. The number of relatively ‘abnormal’ TBI patients for modularity was labeled in parentheses, and the p-values were obtained from permutation tests (10,000 permutations) on group mean difference. Because the quality control procedures for inclusion or exclusion were performed on each scan individually, the subjects whose modularity data is shown for the follow-up scans were not an exact subset of subjects whose data is shown for the initial scans.
Higher modularity in the TBI cohort on the initial scans was associated with lower average participation coefficient, a measure of between-module connectivity, in these subjects. Modularity and average participation coefficient were highly inversely correlated on both scans (Fig. S4a, b), and average participation coefficient was lower in the TBI I cohort than in the controls on the initial scans (Fig S4c).
When the modularities were compared across the initial and follow-up scans at each group, the modularities were apparently changed in the controls whereas they appeared to remain stable on average in the TBI I cohort (Fig. 3). However, the interpretation of this result is not straightforward for two reasons: 1) we acquired MRIs at a different site for the follow-up scans and 2) the subjects on the follow-up scans included in the analyses were not an exact subset of subjects on the initial scans due to the quality assurance procedures. In other words, longitudinal comparisons between each group should not be made until unknown amounts of (1) effects of different sites on the module-based network properties and (2) inter-subject variability of the module-based network properties are clarified. Therefore, to further investigate if the modularities of TBI patients remained stable while those of controls changed over the two scans at each subject, we performed additional, direct comparisons between the initial and follow-up scans with subjects who underwent both scans at the single-subject level (See Fig. S5). At the group level, the overall result was unchanged: there was increased modularity in the control group over time and stable modularity in the TBI I group (Fig. S5, 1st, 2nd columns). However, single subject analyses demonstrated a wide variety of changes (both increases and decreases) in modularity over time in both controls and the TBI I cohort (Fig. S5, 3rd column). See discussion for the limitations of this study relevant to this observation. Further, notable group differences in longitudinal changes in these measures were mainly due to TBI patients with ‘abnormal’ modularities (Fig. S4, 4th column). More specifically, it was the ‘abnormal ’ TBI patients on the initial scans that shifted the group average of longitudinal changes toward zero. Without these ‘abnormal’ TBI patients, average longitudinal changes in the TBI I (dotted horizontal bar in Fig. S4, 4th column) were increased as well, making group differences in longitudinal changes no longer statistically significant at α = 0.05. Thus, the most likely explanation is that there were systematic differences between the two scanners as well as changes in the most abnormal subjects over time.
3.5 Node-specific network properties
Node-specific analysis allowed us to identify localized patterns of module-based network measures that differed between TBI and control subjects (Figs. 4, 5). Overall, the spatial pattern of group comparisons (Fig. 4) and the number of TBI patients with ‘abnormal’ network measures relative to the controls (Fig. 5) were clear on the initial scans but less prominent on the follow-up scans. On the initial scans, the TBI I cohort had small areas of increased and decreased within-module connectivity relative to the controls (Fig. 4a) On the contrary, the TBI subjects had extensive and more markedly decreased participation coefficient (Fig. 4b) compared with the controls. Maps for count of the TBI patients with ‘abnormal’ node-specific measures relative to the controls were similar to the corresponding group comparison maps (Fig. 5).
Figure 4. Node-specific network properties of the controls and TBI I cohort.
Group mean comparison maps (puncorr < 0.05) of within-module degree z-score (a) and participation coefficient (b), respectively. All color maps were superimposed on the averaged cortical surface from all participants in the controls and TBI I cohort. R G_precentral: right precentral gyrus, R G_pariet_inf-Supramar: right supramarginal gyrus, R G_front_inf-Opercular: right opercular part of the inferior frontal gyrus, R G_front_sup-medial : right medial superior frontal gyrus, R G_front_sup-dorsomedial: right dorsomedial superior frontal gyrus.
Figure 5. Count of the TBI patients from the TBI I cohort with ‘abnormal’ node-specific network properties relative to the controls.
Color maps of the number of TBI patients whose measures (within-module degree z-score and participation coefficient, respectively) were outside two standard deviations from the mean of the control. All color maps were superimposed on the averaged cortical surface from all participants in the controls and TBI I cohort.
3.5.1 Group comparisons
The spatial pattern of group differences in within-module connectivity changed over time. At the time of the initial scan, within-module connectivity in the TBI I cohort was slightly elevated, on average, compared with the controls in the right precentral gyrus, right medial superior frontal gyrus, and right dorsomedial superior frontal gyrus (R G_precentral, R G_front_sup-medial and R G_front_sup-dorsomedial in Fig. 4a left panel). Subtle decreases in within-module connectivity of the TBI patients compared with the controls were also observed in the right supramarginal gyrus and right opercular part of the inferior frontal gyrus (R G_pariet_inf-Supramar and R G_front_inf-Opercular in Fig. 4a left panel). Though there were trends in group differences on the initial scans, none of the group differences were statistically significant at qFDR < 0.05 (FDR: false discovery rate (Genovese et al., 2002)). At the time of the follow-up scan, such disturbances in within-module connectivity became less prominent (Fig. 4a, right panel).
In contrast to the scattered subtle increases and decreases in the within-module connectivity, the group comparison maps for participation coefficient (Fig. 4b left panel) exhibited more widespread decreases in the TBI I cohort compared with the controls. At the time of the initial scan, such patterns were localized over the central sulcus, left anterior transverse temporal gyrus, right long insular gyrus and central sulcus of the insula, superior frontal gyrus and sulcus, anterior part of the cingulate gyrus and sulcus, right superior part of the precentral sulcus, right superior temporal sulcus, right orbital gyri, posterior-ventral part of the cingulate gyrus near the calcarine sulcus, lingual gyrus, right parieto-occipital sulcus and left cuneus. These differences on the initial scans were significant based on uncorrected p-values. However, such group differences on the initial scans did not survive after correction for multiple comparisons at qFDR < 0.05. At the time of the second scan, the widespread group differences in participation coefficient mostly became less prominent. (Fig. 4b, right panel)
3.5.2 Counts of the numbers of TBI patients with relatively ‘abnormal’ node-specific network measures
Color maps for the number of TBI patients with ‘abnormal’ network measures relative to the controls in each of the nodes (Fig. 5) allowed us to identify regions where functional connectivity appeared most vulnerable to bTBI. Here, ‘abnormal’ was defined if a network measure of a patient was outside two standard deviations from the mean of the controls. Though abnormalities revealed on group comparison maps did not survive at qFDR < 0.05, the ‘abnormality’ maps demonstrated that substantial portions of the TBI patients (up to 25%) had ‘abnormally’ low participation coefficient relative to the controls (Fig. 5c). None had ‘abnormally’ high participation coefficient. These findings are notable in that by chance 2.4% of the TBI I cohort would be expected to be lower than the mean of the controls minus the 2-standard deviations.
3.6 ROI analysis results
Since participation coefficient was the most prominent among the module-based network measures in the group-wide node-specific analysis, we focused on participation coefficient in further analyses. From the group comparison map in participation coefficient (Fig. 4b), we identified 15 regions (Fig. 6) for ROI analysis using the Destrieux atlas (Destrieux et al., 2010), a standard surface-based atlas available from Freesurfer. See the discussion section for explanation of utilizing structural parcellations as opposed to functional parcellations in this ROI analysis. The identified ROIs were (1) central sulcus (S_central), (2) left anterior transverse temporal gyrus (L G_temp_sup-G_T_transv), (3) right long insular gyrus and central sulcus of the insula (R G_Ins_lg_and_S_cent_ins), (4) superior frontal sulcus (S_front_sup), (5) the medial portion of the superior frontal gyrus (G_front_sup-medial), (6) the anterior portion of the superior frontal gyrus (G_front_sup-anterior), (7) deep anterior part of the cingulate gyrus and sulcus (G_and_S_cingul-Ant-deep), (8) superficial anterior part of the cingulate gyrus and sulcus (G_and_S_cingul-Ant-superficial), (9) right superior part of the precentral sulcus (R S_precentral-sup-part), (10) right superior temporal sulcus (R S_temporal_sup), (11) right orbital gyri (R G_orbital), (12) posterior-ventral part of the cingulate gyrus (G_cingul-Post-ventral) near the calcarine sulcus, (13) lingual gyrus (G_oc-temp_med-Lingual), (14) right parieto-occipital sulcus (R S_parieto_occipital) and (15) left cuneus (L G_cuneus). For detailed locations and surface area of these ROIs, see Fig. 6 and Table III.
Figure 6. Regions of interest (ROIs) for participation coefficient.
15 ROIs are colored and labeled (1: central sulcus, 2: left anterior transverse temporal gyrus, 3: right long insular gyrus and central sulcus of the insula, 4: superior frontal sulcus, 5: medial superior frontal gyrus, 6: anterior superior frontal gyrus, 7: deep anterior part of the cingulate gyrus and sulcus, 8: superficial anterior part of the cingulate gyrus and sulcus, 9: right superior part of the precentral sulcus, 10: right superior temporal sulcus, 11: right orbital gyri, 12: posterior-ventral part of the cingulate gyrus, 13: lingual gyrus, 14: right parieto-occipital sulcus and 15: left cuneus). See Table III for the coordinates and areas of these ROIs.
Table III.
Regions-of-interest (ROI) for participation coefficient analyses.
| Indexa | ROI name | MNI coordinates (x,y,z) of center b | Surface area (mm2)c | ||
|---|---|---|---|---|---|
| Left | Right | Left | Right | ||
| 1 (45) | S_central | (−51, −11, 28) | (51, −11, 30) | 531.5 | 616.5 |
| 2 (33) | L G_temp_sup-G_T_transv | (−41, −20, 4) | 419.0 | ||
| 3 (17) | R G_Ins_lg_and_S_cent_ins | (39, −15, 10) | 560.4 | ||
| 4 (54) | S_front_sup | (−21, 25, 46) | (28, 27, 41) | 281.0 | 310.5 |
| 5 (16) | G_front_sup-medial | (−5, 39, 31) | (9, 41, 22) | 409.3 | 411.5 |
| 6 (16) | G_front_sup-anterior | (−8, 63, 25) | (13, 57, 29) | 490.9 | 473.2 |
| 7 (6) | G_and_S_cingul-Ant-deep | (−4, 37, −8) | (7, 38, −7) | 262.3 | 356.0 |
| 8 (6) | G_and_S_cingul-Ant-superficial | (−8, 56, 3) | (9, 55, 2) | 203.8 | 338.7 |
| 9 (69) | R S_precentral-sup-part | (23, −13, 61) | 260.7 | ||
| 10 (73) | R S_temporal_sup | (55, −20, −12) | 291.8 | ||
| 11 (24) | R G_orbital | (41, 28, −16) | 237.3 | ||
| 12 (10) | G_cingul-Post-ventral | (−8, −52, 3) | (12, −53, 5) | 300.0 | 199.5 |
| 13 (22) | G_oc-temp_med-Lingual | (−14, −60, 1) | (29, −41, −11) | 441.9 | 381.2 |
| 14 (65) | R S_parieto_occipital | (18, −81, 38) | 302.2 | ||
| 15 (11) | L G_cuneus | (−2, −85, 14) | 465.1 | ||
Initial index numbers indicate regions of interest labeled in Fig. 6. Numbers in parentheses refer to the corresponding index numbers in Destrieux et al. (2010).
MNI coordinates correspond to a mid point between pial surface and white matter surface.
Surface area of white matter surface
Note: S_central, central sulcus (Rolando’s fissure); L G_temp_sup-G_T_transv, left anterior transverse temporal gyrus (of Heschl); R G_Ins_lg_and_S_cent_ins, right long insular gyrus and central sulcus of the insula, S_front_sup, superior frontal sulcus; G_front_sup-medial, medial superior frontal gyrus (F1); G_front_sup-anterior, anterior superior frontal gyrus (F1); G_and_S_cingul-Ant-de, deep anterior part of the cingulate gyrus and sulcus; G_and_S_cingul-Ant-su perficial, superficial anterior part of the cingulate gyrus and sulcus; R S_precentral-sup-part, right superior part of the precentral sulcus; R S_temporal_sup, right superior temporal sulcus (parallel sulcus); R G_orbital, right orbital gyri; G_cingul-Post-ventral, posterior-ventral part of the cingulate gyrus (vPCC, isthmus of the cingulate gyrus); G_oc-temp_med-Lingual, lingual gyrus, ligual part of the medial occipito-temporal gyrus (O5); R S_parieto_occipital, right parieto-occipital sulcus (or fissure); L G_cuneus, left cuneus (O6); MNI, Montreal Neurological Institute (Evans et al., 1993)
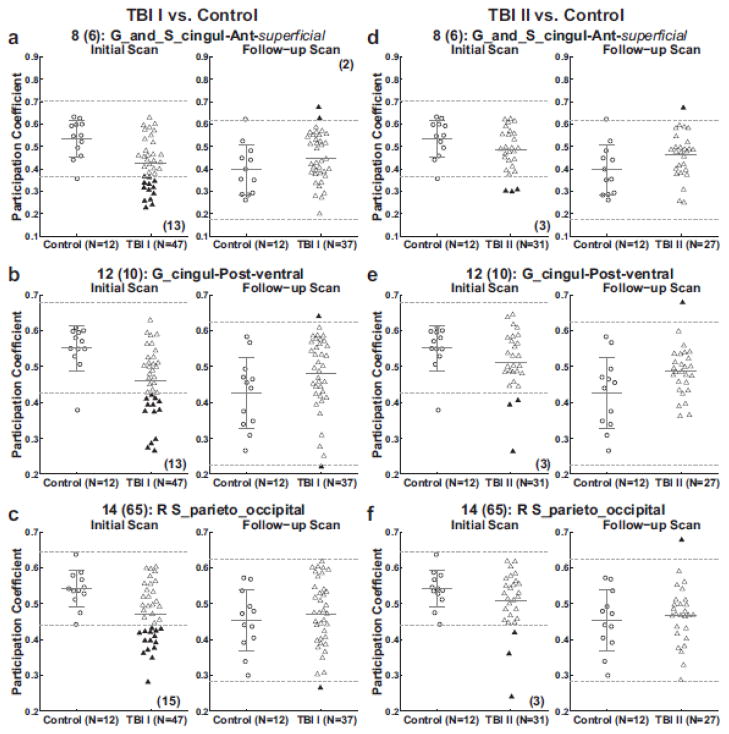
Scatter plots were made (Fig. 7) to examine the distributions of average participation coefficient of each subject in three ROIs (G_and_S_cingul-Ant-superficial (left: (-8, 56, 3), right: (9, 55, 2)), G_cingul-Post-ventral (left: (−8, −52, 3), right: (12, −53, 5)) and R S_parieto_occipital (18, −81, 38)). These three ROIs were representative of all 15 ROIs analyzed (see table IV for the complete list of the ROI analysis results). Similar to the trends observed in global and node-specific network measures, the TBI I cohort, on average, had lower participation coefficients than the controls on the initial scans, yielding up to 15 (36.6%) relatively ‘abnormal’ TBI patients. (Fig. 7a–c left panel). Such group differences that were marked by many relatively ‘abnormal’ TBI patients became less prominent at the time of the follow-up scans (Fig. 7a–c right panel).
Figure 7. ROI analysis of participation coefficient of the controls and patients with TBI.
Scatter plots for averaged participation coefficients within each of three selected ROIs. See Fig. 3 for the details of the scatter plots, Table III for abbreviations and Table IV for results from all ROIs. Again, because the quality control procedures for inclusion or exclusion were performed on each scan individually, the subjects whose modularity data is shown for the follow-up scans were not an exact subset of subjects whose data is shown for the initial scans.
Table IV.
ROI analysis results of participation coefficients.
| Index | ROI name | ‘Abnormal’ TBIa | |||
|---|---|---|---|---|---|
| TBI I vs. Control | TBI II vs. Control | ||||
| Initial | Follow-up | Initial | Follow-up | ||
| 1 (45) | S_central | 13/0 | 3/0 | 1/0 | 1/1 |
| 2 (33) | L G_temp_sup-G_T_transv | 12/1 | 4/0 | 2/1 | 4/0 |
| 3 (17) | R G_Ins_lg_and_S_cent_ins | 10/1 | 4/3 | 1/1 | 2/1 |
| 4 (54) | S_front_sup | 10/0 | 2/1 | 1/0 | 0/1 |
| 5 (16) | G_front_sup-medial | 11/0 | 1/1 | 0/0 | 0/1 |
| 6 (16) | G_front_sup-anterior | 14/0 | 1/3 | 2/0 | 0/1 |
| 7 (6) | G_and_S_cingul-Ant-deep | 12/0 | 1/2 | 3/0 | 0/1 |
| 8 (6) | G_and_S_cingul-Ant-superficial | 13/0 | 0/2 | 3/0 | 0/1 |
| 9 (69) | R S_precentral-sup-part | 11/0 | 1/0 | 1/0 | 1/1 |
| 10 (73) | R S_temporal_sup | 13/0 | 2/0 | 3/0 | 1/0 |
| 11 (24) | R G_orbital | 15/0 | 2/1 | 1/0 | 0/0 |
| 12 (10) | G_cingul-Post-ventral | 13/0 | 1/1 | 3/0 | 0/1 |
| 13 (22) | G_oc-temp_med-Lingual | 15/0 | 2/0 | 3/1 | 0/1 |
| 14 (65) | R S_parieto_occipital | 15/0 | 1/0 | 3/0 | 0/1 |
| 15 (11) | L G_cuneus | 8/0 | 2/0 | 0/0 | 0/1 |
The number of TBI patients with ‘abnormally’ low participation coefficients / the number of TBI patients with ‘abnormally’ high participation coefficients.
Note: See Table III for the abbreviations of ROI names.
Subsequently, without free parameters with regard to ROI selection, we used the TBI II cohort as a validation dataset to test the hypotheses generated with the TBI I cohort. On the initial scans, in the three ROIs, relative ‘abnormality’ patterns in the TBI II cohort were less striking than those in the TBI I cohort, yielding up to 3 (9.6%) TBI patients with relatively ‘abnormal’ participation coefficients (Fig. 7d–f left panels). Table IV indicates that the number of TBI patients from the TBI II cohort with relatively ‘abnormal’ ROI-specific participation coefficients on the initial scans was greater than the expected number by chance (0 above and 0 below). Relatively abnormal participation coefficient on the initial scans in TBI patients from the TBI II cohort were identified in all ROIs except in the G_front_sup-medial and L G_cuneus. However, there were fewer TBI patients with relatively abnormal participation coefficient over the ROIs in the TBI II cohort compared with the TBI I cohort (Fig. 7 and Table IV).
Turning to multiple ROI single-subject analyses, we counted the number of relatively ‘abnormal’ ROIs in each individual TBI patient and tested if this observed distribution of ‘abnormal’ ROIs differed from the distribution expected by chance (Fig. 8). On the initial scans, the observed numbers of TBI patients with more than two ‘abnormal’ ROIs were markedly different from those expected by chance, and these differences were statistically significant at α = 0.05 in both datasets (27/47 observed versus 2/47 expected in the TBI I cohort, p < 0.0001, one-sided z-test; 8/31 observed versus 1/31 expected in the TBI II cohort, p = 0.006, one-sided z-test). As expected considering the nature of these analyses, the proportion of TBI patients from the TBI II cohort with more than two relatively ‘abnormal’ ROIs was lower than those from the TBI I cohort: 44.9% reduction from the TBI I cohort to TBI II cohort on the initial scans (i.e., 57.4% = 27/47 versus 25.8% = 8/31).
Figure 8. Bar graphs for observed and expected ‘abnormal’ ROIs in the patients with TBI relative to the controls.
The distribution of expected relatively ‘abnormal’ ROIs was calculated from the binomial distribution with the probability that one region deems ‘abnormal’ by chance (0.0455; the probability that participation coefficient for a TBI patient falls outside two standard deviations from the mean of the controls). The p-values were obtained by the one-sided z-test (TBI I > controls in the number of ‘abnormal’ ROIs).
On the follow-up scans, the proportion of patients in the TBI I cohort with more than two ‘abnormal’ ROIs was different than expected by chance with marginal statistical significance (p = 0.044). The proportion of patients in the TBI II cohort with more than two ‘abnormal’ ROIs on follow-up scans was not statistically significant (p = 0.276).
Multivariate analysis aggregating all 15 ROIs after dimensionality reduction via probabilistic principal component analysis (Fig. 9) demonstrated a similar reduction in the proportion of the TBI patients with relatively ‘abnormal’ ROIs from the TBI I cohort to the TBI II cohort. Specifically, 31/47 (66.0%) of the patients from the TBI I cohort (Fig. 9a) and 9/31 (29.0%) of the patients from the TBI II cohort (Fig. 9b) were deemed to be relatively ‘abnormal’ across the 15 ROIs on the initial scans. On the follow-up scans, 4/37 (10.8%) of the patients from the TBI I cohort (Fig. 9c) and 1/27 (3.7%) of the patients from the TBI II cohort (Fig. 9d) were found to be ‘abnormal’ using this multivariate analysis.
Figure 9. Scatter plots for multivariate analysis of participation coefficients within the ROIs after dimensionality reduction by probabilistic principal component analysis.
Solid triangles represent relatively ‘abnormal’ TBI patients whose values are locating within the lower and upper tail, accounting for 2.5% in each tail, of the estimated multivariate normal distribution from the control. The label in each panel indicates the number of relatively ‘abnormal’ TBI patients and the number of TBI patients on the corresponding scan.
3.7 Permutation-based omnibus abnormality analysis results
We next asked whether the above results could have been biased by non-independence of the ROIs. Therefore, we counted the proportion of ‘abnormal’ nodes in each individual TBI patient and compared the observed distributions in TBI patients with those expected by chance using permutation of group membership (Fig. 10). Again, ‘abnormal’ was defined as participation coefficient outside of 2 standard deviations of the mean of the controls. Overall, these omnibus nodal abnormality analysis results were consistent with the ROI analysis results shown in Fig. 8. Specifically, on the initial scans, the differences between the observed number of TBI patients with more than 15% ‘abnormal’ nodes in the TBI I cohort or more than 10% in the TBI II cohort vs. those expected by chance were statistically significant at α = 0.05 in both datasets (19/47 observed versus 1/47 expected in the TBI I cohort, p < 0.0001, one-sided z-test; 7/31 observed versus 1/31 expected in the TBI II cohort, p = 0.012, one-sided z-test). Again, the trends on the follow-up scans shown in Fig. 10 were similar to those in Fig. 8. The observed proportion of patients in the TBI I cohort was statistically significant (p = 0.0008), but not in the TBI II (p = 0.080).
Figure 10. Bar graphs for observed and expected proportion of ‘abnormal’ nodes in the patients with TBI relative to the controls without ROI selection.
The distributions of expected relatively ‘abnormal’ ROIs were calculated from the permutation of group memberships (10,000 permutations). The p-values were obtained by the one-sided z-test (TBI I > controls in the proportion of ‘a bnormal ’ nodes).
3.8 Control analysis results
3.8.1 Assessment of the effects of subtle head motion
Recently, the effects of subtle motion have been found to substantially influence resting-state functional connectivity MRI findings (Power et al., 2012; Satterthwaite et al., 2012; van Dijk et al., 2012). Our motion analysis (Fig. S6) revealed that there were not statistically significant group differences in average frame-to-frame movement in the analyzed data, after censoring and exclusion of scans with excessive motion (Table I). There were no associations between average participation coefficient and head motion (Fig. S7) in any of the groups. Furthermore, there was no relationship between the number of ‘abnormal’ ROIs and head motion (Fig. S8).
3.8.2 Assessment of the effects of network connectivity thresholds
The differences between TBI subjects and controls in the number of regions on interest with ‘abnormal’ participation coefficient were replicated at 2 additional threshold levels for connectivity matrices (Figs. S9, S10 for 2% and 1.5% tie densities, respectively).
3.8.3 Effects of global signal regression
Analyses of the number of ROIs with abnormal participation coefficient in TBI patients vs. controls based on connectivity matrices created without global signal regression (Fig. S11) essentially replicated the results with global signal regression (Fig. 8). The only exception was that the number of TBI I patients with relatively ‘abnormal’ ROI values on the follow-up scans was not quite statistically significant at α = 0.05.
3.8.4 Effects of region of interest size
To explore the effect of ROI size, we re-analyzed the data using all nodes within 20 mm rather than 15 mm geodesic distance of the local peaks of participation coefficient difference between groups as the ROIs (Fig. S12). The analysis of the number of ROIs with abnormal participation coefficient in TBI patients versus controls using 20 mm geodesic distance ROIs (Fig. S13) again essentially replicate the results with 15 mm geodesic distance ROIs (Fig. 8).
DISCUSSION
In summary, we found disrupted community organization of resting-state functional connectivity in a subset of U.S. military personnel following concussive bTBI using module-based graph theoretic analysis. Of the module-based graph theoretical measures studied, participation coefficient, a measure of between-module connectivity, showed the most pronounced disruptions in the TBI patients relative to the controls. There were spatially localized ‘abnormalities’ over multiple brain regions in the TBI patients. Importantly, these abnormalities were detected in comparison with U.S. military controls that had blast-exposures but no clinical diagnosis of TBI. At the time of the initial scans, the distribution of the number of the ‘abnormal’ regions was different from the expected distribution by chance. Multivariate analysis aggregating the 15 ROI values of between-module connectivity consistently demonstrated the substantial portion of TBI patients had relatively ‘abnormal’ between-module connectivity. The number of TBI patients with relatively ‘abnormal’ between-module connectivity was less prominent at the time of the follow-up scans. In an independent group of concussive bTBI patients, we were able to partially replicate these results with no free parameters.
4.1 Technological innovations effectively analyze heterogeneous concussive bTBI patients at the single subject level
The single-subject analyses performed in this study were useful to effectively analyze a heterogeneous group of bTBI patients in this report. Scatter plots (Figs. 3, 7) illustrate skewed distribution of the network measures in the TBI patients. The distributions of the TBI patients with multiple ‘abnormal’ regions or nodes (Fig. 8, 10) were consistently different from those expected by chance. The multivariate analysis (Fig. 9) also consistently revealed the TBI patients with relatively ‘abnormal’ regions over the two independent cohorts. Thus, we suggest that single-subject analysis should be considered along with group analysis for the identification of ‘abnormalities’ in heterogeneous subject populations such as concussive bTBI patients. Obviously, single-subject analyses have greater potential clinical applicability than group analyses.
We adopted a surface-based approach to reduce between-subject variability arising from brain anatomy. An advantage of the surface-based approach is an increase in sensitivity by (1) matching sulcus-to-sulcus and gyrus-to-gyrus of cortical surface across subjects to circumvent the issues of improper registration and (2) utilizing spatial smoothing on the cortical surface rather than the voxel space (Jo et al., 2007, 2008; Tucholka et al., 2012). On the other hand, a disadvantage of the surface-based approach is that it does not represent the whole brain. The surface based approach does not assess cerebellum and subcortical regions such as basal ganglia and thalamus.
Complex network analysis using the graph theory is advantageous in bTBI populations with heterogeneous injury mechanisms since it does not make assumptions regarding networks or regions of interest. Thus, our data-driven approach may provide more ‘comprehensive’ view than hypothesis-driven approaches do in the studies of heterogeneous TBI populations. Disrupted between-module connectivity in the TBI patients over multiple regions (Figs. 4b, 5c, 7–10, Table IV) indicates that multiple regions or networks should be included to detect ‘abnormalities’ in TBI patients if hypothesis-driven approaches are adopted. Like complex network analysis using the graph theory, independent component analysis (ICA) is another data driven approach with potential to identify abnormalities in heterogeneous TBI populations. Indeed, Shumskaya et al. (2012) identified abnormalities of resting-state networks in concussive TBI patients utilizing the whole-brain ICA. However, Shumskaya et al. (2012) applied the ICA to a relatively more homogenous concussive TBI group including only patients with fronto-occipital injuries. Thus, the utility of whole brain ICA in heterogeneous concussive bTBI populations warrants further investigations.
In contrast to other graph theoretic analyses on TBI populations (Castellanos et al., 2011; Nakamura et al., 2009), we exploited the spatial resolution of fMRI by defining each vertex of the cortical surface as a node. Network analyses with higher spatial resolution of nodes are beneficial over region-based network analyses in identifying network properties of interests with greater sensitivity and specificity and visualizing spatially localized network properties of interest (Hayasaka and Laurienti, 2010). Thus, we attempted to use a vertex as a node considering that the spatial extent of ‘abnormalities’ in concussive bTBI populations measured by the graph theoretic analysis in fMRI was unknown. Indeed, the utilization of high spatial resolution of fMRI allowed us to identify localized ‘abnormalities’ in the TBI patients in greater detail, which would be missed if a node was defined from coarse parcellations. However, a vertex itself is arguably less biologically meaningful than a functional brain region in the context of graph theoretic analysis (Wig et al., 2011). Unfortunately our current understanding of the functional subdivisions of the human brain is not yet sufficient for optimal prespecified ROI analyses. For the same reason, we selected the 15 ROIs in reference to the Destrieux atlas, a structural parcellation, as oppose to a functional parcellation.
4.2 Findings in relation to prior literature
4.2.1 Multiple regions with disrupted between-module connectivity as potential candidates for hypothesis-driven network-specific approaches
Most of the previous functional connectivity studies in fMRI on civilian TBI patients (Arenivas et al., 2012; Bonnelle et al.,2011; Caeyenberghs et al., 2012; Kasahara et al., 2010; Marquez de la Plata et al., 2011; Mayer et al., 2011; Sharp et al., 2011; Slobounov et al., 2011; Tang et al., 2011) used hypothesis-driven approaches within the default mode (Greicius et al., 2002; Raichle et al.,2001), executive control (Seeley et al., 2007; Vincent et al., 2008), motor (Biswal et al., 1995), thalamic (Zhang et al., 2008) and hippocampal (Rombouts et al., 2003) networks. However, to our knowledge, there are no current fMRI studies of resting-state functional connectivity in concussive bTBI patients using these networks. In this regard, future work based on the identified ‘abnormal’ regions in Figs. 4–7 could involve assessment of within-network and between-network connectivity in these specific resting-state networks.
4.2.2 Transient change of module-based organizations following concussive bTBI
A prevailing view on the plasticity of TBI patients based on functional neuroimaging studies (Castellanos et al., 2010, 2011; Mayer et al., 2011; Nakamura et al., 2009; Sponheim et al., 2011) is that the recovery of functional network at the chronic stage is incomplete. Even after 7 to 12 months of active rehabilitation treatments, it has been reported that many TBI patients had incomplete recovery of the graph-theory measures, associated with the incomplete reestablishment of cognitive function (Castellanos et al., 2011). Furthermore, concussive bTBI patients with 32 months of mean post-injury time demonstrated relatively reduced inter-hemispheric connectivity in lateral frontal lobe despite no observed deficits in neuropsychological measures (Sponheim et al., 2011).
In contrast, in our study at the global, region-and node-specific levels (Figs. 3–5, 7–10), group differences in module-based connectivity observed on the initial scans were less prominent at the follow-up scan 6–12 months later. This transient change in module-based connectivity supports the utility of early scanning (if feasible) on bTBI patients to identify localized modular disruptions in brain networks. However, less prominent group differences in between-module connectivity at the follow-ups do not necessarily mean complete restoration of bTBI patients’ network architectures to baseline.
4.2.3 Marginally perturbed within-module connectivity, but markedly unbalanced between-module connectivity with increased modularity
Node-specific analysis of the module-based network measures (Figs. 4, 5) demonstrates that the TBI patients had disrupted between-module connectivity with marginal change in within-module connectivity relative to the controls. Disrupted between-module connectivity in the TBI patients indicate that the blast-related injuries may interfere with integration across functional brain networks, as between-module connectivity may be necessary for complex tasks spanning multiple modules. In healthy normal subjects, modules in the spontaneous brain network are closely associated with auditory, visual, somatosensory/motor, attention, sub-cortical and default mode networks (He et al., 2009). Though we did not directly assess in this study, traumatic axonal injuries in long range fibers mediating connections with multiple modules may lead to disruptions in between-module connectivity. In TBI patients, disrupted white matter integrity in the splenium of the corpus callosum has been correlated with PCC functional connectivity in the default mode network (Sharp et al., 2011). Our previous DTI study showed disrupted white matter integrity in the TBI I cohort relative to the controls (Mac Donald et al., 2011). However, it is too early to interpret the association of altered white matter integrity reported in Mac Donald et al. (2011) with disrupted between-module functional connectivity following bTBI without direct investigation of structural white matter connectivity at a subject by subject level. Since strong functional connectivity between regions is often maintained without monosynaptic connections between these regions (Honey et al. 2009), direct comparisons of functional and structural connectivity are not straightforward. Therefore, further studies on the association between structural and functional connectivity on these concussive bTBI patients are needed to understand the underlying mechanisms of disrupted between-module connectivity following bTBI. This will require further investigations that are beyond the scope of the current manuscript.
4.3 Technical Limitations
The present study has several limitations due to data quality. First, our analysis results were from a subset (75% of all subjects’ data; 62%, 76%, 81% of the controls, TBI I cohort and TBI II cohort, respectively) of the subjects’ data to ensure data quality. Our study results may be more representative if more subjects’ data had passed the quality assurance procedure. As such, the quality assurance procedure yielded relatively small sample size (N=12 for each scan) of controls. Small sample size could be an alternative explanation for the apparent resolution of the module-based graph measures in the TBI patients on follow-up scans. In other words, decreases in the numbers of relatively abnormal TBI patients could have been partially explained by the increased standard deviations of the module-based graph measures for the controls on the follow-up scans. The exclusion of a part of the subjects’ data for data quality assurance may not be surprising to neuroimaging researchers, particularly in fMRI, considering difference in subjects’ conditions between clinical populations and healthy subjects. However, this is an important consideration clinically as it may not be feasible to collect reliable data from every patient to be used as early indicators of injury, disease, or for non-invasive monitoring of rehabilitation progress.
Second, moderate data quality of the T1-weight MRI due to subject motion was not sufficient for Freesurfer to reconstruct cortical surfaces in fully automated fashion. Thus, following the well-documented instructions, a substantial amount of time was required for manual intervention during cortical surface reconstruction to compensate MRI data quality of our study. Cortical surface reconstruction would be more reliable (and require less manual intervention) if we increased signal-to-noise ratio of the T1-weighted MRI by acquiring two or more images as the Freesurfer instructions suggest. However, a practical limitation of studies of relatively acutely injured subjects is that time in the scanner must be kept relatively short for patient comfort and safety.
Third, we were not able to include much of the orbitofrontal cortex and inferior temporal cortex in our analysis due to signal loss by fMRI susceptibility artifacts (Ojemann et al., 1997) and low signal to noise due to distance from the MR receiver coils. Particularly, orbitofrontal cortex was of interest based on a previous simulation study on the effect of blasts on the brain (Taylor and Ford, 2009) and our previous DTI study (Mac Donald et al., 2011) involving the TBI I cohort. An optimized pulse sequence to account for susceptibility artifacts (Domsch et al., 2012) would be beneficial in this regard for future studies.
Fourth, we did not include negative correlations for the network constructions as the meaning of negative correlations after global signal regression is still unclear (Anderson et al., 2011; Chai et al., 2012; Chang and Glover, 2009; Fox et al., 2009; Murphy et al., 2009; Saad et al., 2012). Assessment of anticorrelations in bTBI populations would be of interest if the meaning of negative correlations is clarified by future studies.
Finally, we did not make direct comparison of the initial and follow-up scans even though we acquired MRI data at two time points. Instead, we performed only a cross-sectional study at two time points and observed change of modular organization of the TBI patients relative to the controls at each time point. Lack of quantification on the effects of the two different MRI scanners, medications between the time of the initial and follow-up scans and sleep deprivation at the time of the initial scans prevented us from directly analyzing our data in a subject-by-subject longitudinal fashion.
4.4 Interpretations
The underlying biological mechanisms and consequences of the findings in this study are not yet clear. Possible explanations for the relatively disrupted between-module functional connectivity in the TBI cohorts on initial scans may be related to white matter injury. This hypothesis is based on previous combined DTI and fMRI connectivity studies on TBI populations (Bonnelle et al., 2012; Mayer et al., 2011; Sharp et al., 2011). Several relevant investigations into the underlying biological mechanisms of concussive TBI in the context of white matter injury have been performed recently. In magnetic resonance spectroscopy (MRS) studies of concussive TBI (Gasparovic et al., 2009; Johnson et al., 2012, Yeo et al., 2011a), three main findings have been reported: (1) reduced N-acetylaspartate (NAA) in the white matter, reflective of disruptions in neuronal integrity, (2) increased creatine in the white matter, reflective of alterations in cellular energy metabolism, and (3) increased choline in the white matter, associated with injury and repair of myelin and/or inflammation. In particular, Johnson et al. (2012) demonstrated that altered metabolic integrity measured by NAA/choline in the splenium of the concussive TBI patients was associated with reduced functional inter-hemispheric connectivity following TBI.
Another possibility is unmeasured pathophysiology following TBI in the gray matter. Previous metabolic imaging studies in concussive TBI reported metabolic changes in the gray matter following TBI that might lead to changes in BOLD signals (see Lin et al. (2012) for review). For example, single photon emission computed tomography (SPECT) identified reduced regional cerebral blood flow (rCBF) in concussive TBI patients (Jacobs et al., 1996). Positron emission tomography (PET) revealed decreased cerebral glucose metabolic rate (CMRglu) in veterans with repetitive concussive bTBI (Peskind et al., 2011). MRS studies (Gasparovic et al., 2009; Yeo et al., 2011a) demonstrated decreased glutamate-glutamine (Glx) in the gray matter of the concussive TBI group and reduced BOLD response following TBI. Taken together, future studies involving DTI, MRS, SPECT, PET and other methods sensitive to structural and metabolic derangements will be required to assess the underlying pathological mechanisms of findings in this study.
The biological mechanisms for apparent resolution of between-module functional connectivity in the TBI patients on the follow-up scans are not known. One possibility is that the TBI patients could have been subjected to more sleep deprivation at the time of initial scans; sleep deprivation has been shown to affect BOLD signal (Gujar et al., 2010). Medications used by TBI patients at the acute stage and during rehabilitation could also be partially accountable.
There are several alternative explanations for our findings:
It is possible that some of these results may have occurred by chance as group differences at the node- and region-specific level were not statistically significant after correction for multiple comparisons (qFDR < 0.05). We do not think that this is likely to be the case for all of our results since the analysis of the TBI II cohort was performed with no free parameters after we obtained the analysis results of the TBI I cohort. In other words, the TBI II cohort served as a validation dataset. Thus, the single-subject analyses of the initial scan (Figs. 7–9) should be regarded as the most solid.
Unmeasured systematic differences in the time spent in the scanner with eyes open versus closed or awake versus asleep could also be confounding factors. It has been reported that functional connectivity strength in the default mode network and attention network with eyes closed is decreased over network connectivity with eyes opened (van Dijk et al., 2010). Functional connectivity in the default mode, attention, executive control, motor, visual and auditory network is retained (Larson-Prior et al., 2009) during light sleep, but functional connectivity in the default mode network is partially changed during deep sleep (Horovitz et al., 2009). We were not able to control the extent of eyes-open vs. eyes closed time, nor assess sleep in the scanner due to the logistical challenges of obtaining scans in acutely injured U.S. military personnel.
The effects of analgesic medications could be another confounding factor. To our knowledge, immediate effects of analgesics on resting-state functional connectivity in fMRI are unknown, but alterations in regional synchrony of resting-state functional connectivity in fMRI in the right anterior cingulate cortex and left precentral frontal gyrus in chronic ketamine users has been reported (Liao et al., 2012).
Other, unmeasured differences in pre- and post-injury characteristics among three groups could also provide alternative explanations for our findings. The initial scan data of the controls and TBI I cohort were acquired in 2008–2009 while the TBI II cohort in 2010–2011. During this period, the pace of the wars was changed and rehabilitation strategies and treatments may have improved (See van Wingen et al. (2012) for the effects of combat stress on functional connectivity). All these characteristics were unmeasured. Thus, it is unclear whether reduction in the proportion of TBI patients with relatively ‘abnormal’ participation coefficient across ROIs (Figs. 7–9) from the TBI I cohort to the TBI II cohort was solely due to different detection performance of the multivariate analysis over the two datasets or due to intrinsic difference between the subjects in the 2 TBI cohorts.
Biases due to incomplete blinding during the manual portions of the Freesurfer analysis could also affect our results though the associated effects are not likely to be substantial considering the difference in spatial resolutions: 43 mm3 in fMRI versus 1 mm3 in structural MRI.
Test-retest reliability of our findings is still unknown. Two previous reports, one indicating moderately good reliability (Braun et al., 2012) and another indicating relatively low test-retest reliability for many graph theoretical measures (Wang et al., 2011) used coarser parcellation and less aggressive motion scrubbing than the current work.
4.4 Future Directions
The most important future direction for this line of investigation is an assessment of the relationship between these graph-theoretically derived neuroimaging variables and clinical outcomes in these cohorts. This will be the topic of future communications. Further studies will be required to address the technical concerns discussed above and assess the mechanisms underlying these observed network properties.
CONCLUSIONS
In conclusion, we demonstrated disrupted modular organization of resting-state cortical functional network in U.S. military personnel with concussive bTBI. Module-based graph theoretic analysis revealed altered between-module connectivity in the TBI patients relative to the controls who had blast exposures without a diagnosis of TBI. Single-subject multivariate analysis fairly consistently detected the TBI patients with relatively ‘abnormal’ ROIs over two independent cohorts. Our single-subject analysis approach may be useful for heterogeneous populations and potentially complement hypothesis-driven approaches for these populations in future resting-state fMRI studies. Further studies are required to explain the underlying mechanisms and consequences of these phenomena.
Supplementary Material
With each subject’ data that passed the quality assurance procedure before checking the module assignment results, the seed was placed at the left posterior cingulate cortex (PCC; −7, −55, 27, red spot) including nodes within 6 mm Euclidean distance from the PCC. The colors represent statistical significance of the group means of Fisher-Z transformed correlation coefficients. After ensuring that the Fisher-Z transformed correlations across subjects within each group passed the Shapiro-Wilk test at α = 0.05, the t-test on the group means were performed, thresholded (puncorr < 0.001) and converted to z-score.
See Fig. 2 for the details of labels.
The ρ-values in panels a–b represent the Spearman rank correlation coefficients. See Fig. 3 for the details of scatter plots in panel c. On the initial scans, network measures of the TBI patients within the gray oval were distant from the inverse linear trends (a). On the follow-up scans, only one subject was an outlier from the trends (b). As expected, the TBI I cohort had lower average participation coefficient, than the controls on the initial scans (Fig. S4c, left panel). As for modulatity, group differences in average participation coefficient were not observed on the follow-up scans (Fig. S4c, right panel).
The first and second columns are scatter plots for the modularity of subjects who had acceptable data quality for both the initial and follow-up scans. Changes in this modularity at each subject are on the third column. Scatter plots on the fourth column are symmetrized percent change, i.e., (follow-up scans – initial scans)/average of initial and follow-up scans. Filled triangles on the third and fourth columns are relatively ‘abnormal’ TBI patients on the initial scans. Dotted magenta horizontal bars on the TBI I in the fourth column represent the group average values of the TBI patients without these ‘abnormal’ TBI patients. See Fig. 3 for the other details of the scatter plots.
The p-values were obtained from permutation tests (10,000 permutations) on group mean difference. See Fig. 3 for the details of the scatter plots. Note that subjects whose images passed all quality assurance procedure up to the motion censoring criteria were included for this analysis (See Table I).
The ρ-values are the Spearman correlation coefficients.
See Fig. 8 for the details of the bar graphs.
See Fig. 8 for the details of the bar graphs.
See Fig. 8 for the details of the bar graphs.
Regions are nodes within 20 mm geodesic distance from the local peaks. See Fig. 6 for the details of labels.
See Fig. 8 for the details of the bar graphs.
Highlights.
Module-based graph theoretic analysis of resting-state fMRI in patients with concussive bTBI
Spatially localized reductions in between-module connectivity in the TBI patients on the initial scans
Less prominent group differences on the follow-up scans
Analysis of the second TBI cohort dataset with no free parameters provided a partial replication
Acknowledgments
First of all, we would like to thank all participants, their families, the commanding officers and the clinical care providers that supported this study. Our thanks also go to Dr. Nico U. Dosenbach, Dr. Steven E. Petersen and Dr. Abraham Z. Snyder for their helpful comments. This work has been supported by Department of Defense CDMRP grants PT075299 and PT090444 to DLB. We also would like to thank the facilities of the Washington University Center for High Performance Computing, which were partially provided through grant NCRR 1S10RR022984-01A1 for providing computational resource for conducting this study. Finally, we would like to thank the two anonymous reviewers for their comments which substantially improved this manuscript.
Footnotes
The views and opinions expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Army, Department of the Air Force, Department of Defense or United States Government.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore ET. A Resilient, Low-Frequency, Small-World Human Brain Functional Network with Highly Connected Association Cortical Hubs. J Neurosci. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Druzgal TJ, Lopez-Larson M, Jeong E, Desai K, Yurgelun-Todd D. Network Anticorrelations, Global Regression, and Phase-shifted Soft Tissue Correction. Hum Brain Mapp. 2011;32:919–934. doi: 10.1002/hbm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenivas A, Diaz-Arrastia R, Spence J, Cullum CM, Krishnan K, Bosworth C, Culver C, Kennard B, Marquez de la Plata C. Three Approaches to Investigating Functional Compromise to the Default Mode Network After Traumatic Axonal Injury. Brain Imaging Behav. 2012 doi: 10.1007/s11682-012-9191-2. in press. [DOI] [PubMed] [Google Scholar]
- Argall BD, Saad ZS, Beauchamp MS. Simplified Intersubject Averaging on the Cortical Surface Using SUMA. Hum Brain Mapp. 2006;27:14–27. doi: 10.1002/hbm.20158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional Connectivity in the Motor Cortex of Resting Human Brain Using Echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J, Lambiotte R, Lefebvre E. Fast Unfolding of Communities in Large Networks. J Stat Mech: Theory and Exp. 2008;2008:P10008. [Google Scholar]
- Bonnelle V, Leech R, Kinnunen KM, Ham TE, Beckmann CF, de Boissezon X, Greenwood RJ, Sharp DJ. Default Mode Network Connectivity Predicts Sustained Attention Deficits After Traumatic Brain Injury. J Neurosci. 2011;31:13442–13451. doi: 10.1523/JNEUROSCI.1163-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnelle V, Ham TE, Leech R, Kinnunen KM, Mehta MA, Greenwood RJ, Sharp DJ. Salience Network Integrity Predicts Default Mode Network Function After Traumatic Brain Injury. Proc Natl Acad Sci USA. 2012;109:4690–4695. doi: 10.1073/pnas.1113455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun Urs, Plichta MM, Esslinger C, Sauer C, Haddad L, Grimm O, Mier D, et al. Test–retest Reliability of Resting-state Connectivity Network Characteristics Using fMRI and Graph Theoretical Measures. Neuroimage. 2012;59:1404–1412. doi: 10.1016/j.neuroimage.2011.08.044. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Sporns O. Complex Brain Networks: Graph Theoretical Analysis of Structural and Functional Systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Caeyenberghs K, Leemans A, Heitger MH, Leunissen I, Dhollander T, Sunaert S, Dupont P, Swinnen SP. Graph Analysis of Functional Brain Networks for Cognitive Control of Action in Traumatic Brain Injury. Brain. 2012;135:1293–1307. doi: 10.1093/brain/aws048. [DOI] [PubMed] [Google Scholar]
- Casscells SW. Traumatic Brain Injury: Definition and Reporting. Washington, DC: Department of Defense; 2007. (memorandum) ( http://mhs.osd.mil/Content/docs/pdfs/policies/2007/07-030.pdf.) [Google Scholar]
- Castellanos NP, Paul N, Ordonez VE, Demuynck O, Bajo R, Campo P, Bilbao A, Ortiz T, del-Pozo F, Maestu F. Reorganization of Functional Connectivity as a Correlate of Cognitive Recovery in Acquired Brain Injury. Brain. 2010;133:2365–2381. doi: 10.1093/brain/awq174. [DOI] [PubMed] [Google Scholar]
- Castellanos NP, Leyva I, Buldu JM, Bajo R, Paul N, Cuesta P, Ordonez VE, et al. Principles of Recovery from Traumatic Brain Injury: Reorganization of Functional Networks. Neuroimage. 2011;55:1189–1199. doi: 10.1016/j.neuroimage.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Chai XJ, Castanon AN, Ongur D, Whitfield-Gabrieli S. Anticorrelations in Resting State Networks Without Global Signal Regression. Neuroimage. 2012;59:1420–1428. doi: 10.1016/j.neuroimage.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Effects of Model-based Physiological Noise Correction on Default Mode Network Anti-correlations and Correlations. Neuroimage. 2009;47:1448–1459. doi: 10.1016/j.neuroimage.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Germann J, Evans AC. Revealing Modular Architecture of Human Brain Structural Networks by Using Cortical Thickness from MRI. Cereb Cortex. 2008;18:2374–2381. doi: 10.1093/cercor/bhn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Dale AM, Sereno MI. Improved Localization of Cortical Activity by Combining EEG and MEG with MRI Cortical Surface Reconstruction: a Linear Approach. J Cogn Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-based Analysis. I. Segmentation and Surface Reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- de Haan W, van der Flier WM, Koene T, Smits LL, Scheltens P, Stam CJ. Disrupted Modular Brain Dynamics Reflect Cognitive Dysfunction in Alzheimer’s Disease. Neuroimage. 2012;59:3085–3093. doi: 10.1016/j.neuroimage.2011.11.055. [DOI] [PubMed] [Google Scholar]
- Dempsey KE, Dorlac WC, Martin K, Fang R, Fox C, Bennett B, Williams K, Flaherty S. Landstuhl Regional Medical Center: Traumatic Brain Injury Screening Program. J Trauma Nurs. 2009;16:6–12. doi: 10.1097/01.JTN.0000348063.41099.a7. [DOI] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale AM, Halgren E. Automatic Parcellation of Human Cortical Gyri and Sulci Using Standard Anatomical Nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsch S, Linke J, Heiler PM, Kroll A, Flor H, Wessa M, Schad LR. Increased BOLD Sensitivity in the Orbitofrontal Cortex Using Slice-dependent Echo Times at 3 T. Mag Reson Imaging. 2012;31:201–211. doi: 10.1016/j.mri.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Doppenberg EMR, Bullock R. Clinical Neuro-Protection Trials in Severe Traumatic Brain Injury: Lessons from Previous Studies. J Neurotrauma. 1997;14:71–80. doi: 10.1089/neu.1997.14.71. [DOI] [PubMed] [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern Classification. 2. Wiley; 2001. [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3D Statistical Neuroanatomical Models from 305 MRI Volumes. Proc IEEE Nucl Sci Symp Med Imaging Conf; IEEE. 1993. pp. 1813–1817. [Google Scholar]
- Finkel MF. The Neurological Consequences of Explosives. J Neurol Sci. 2006;249:63–67. doi: 10.1016/j.jns.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical Surface-Based Analysis: II: Inflation, Flattening, and a Surface-Based Coordinate System. Neuroimage. 1999a;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution Intersubject Averaging and a Coordinate System for the Cortical Surface. Hum Brain Mapp. 1999b;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated Manifold Surgery: Constructing Geometrically Accurate and Topologically Correct Models of the Human Cerebral Cortex. IEEE Trans Med Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe AJW, et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe AJW, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, et al. Automatically Parcellating the Human Cerebral Cortex. Cereb Cortex. 2004a;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Segonne F, Quinn BT, Dale AM. Sequence-independent Segmentation of Magnetic Resonance Images. Neuroimage. 2004b;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fortunato Santo. Community Detection in Graphs. Phys Rep. 2010;486:75–174. [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, van Essen DC, Raichle ME. From The Cover: The Human Brain Is Intrinsically Organized into Dynamic, Anticorrelated Functional Networks. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The Global Signal and Observed Anticorrelated Resting State Brain Networks. J Neurophysiol. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparovic Charles, Yeo Ronald, Mannell Maggie, Ling Josef, Elgie Robert, Phillips John, Doezema David, Mayer Andrew R. Neurometabolite Concentrations in Gray and White Matter in Mild Traumatic Brain Injury: An 1H–Magnetic Resonance Spectroscopy Study. J Neurotrauma. 2009;26:1635–1643. doi: 10.1089/neu.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols TE. Thresholding of Statistical Maps in Functional Neuroimaging Using the False Discovery Rate. NeuroImage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional Connectivity in the Resting Brain: A Network Analysis of the Default Mode Hypothesis. Proc Natl Acad Sci U S A. 2002;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera R, Amaral LAN. Functional Cartography of Complex Metabolic Networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujar N, Yoo S, Hu P, Walker MP. The Unrested Resting Brain: Sleep Deprivation Alters Activity Within the Default-mode Network. J Cog Neurosci. 2010;22:1637–1648. doi: 10.1162/jocn.2009.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler DJ, Saygin AP, Sereno MI. Smoothing and Cluster Thresholding for Cortical Surface-based Group Analysis of fMRI Data. Neuroimage. 2006;33:1093–1103. doi: 10.1016/j.neuroimage.2006.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the Structural Core of Human Cerebral Cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka S, Laurienti PJ. Comparison of Characteristics Between Region-and Voxel-based Network Analyses in Resting-state fMRI Data. Neuroimage. 2010;50:499–508. doi: 10.1016/j.neuroimage.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. Small-World Anatomical Networks in the Human Brain Revealed by Cortical Thickness from MRI. Cereb Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, et al. Uncovering Intrinsic Modular Organization of Spontaneous Brain Activity in Humans. PLoS ONE. 2009;4:e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting Human Resting-state Functional Connectivity from Structural Connectivity. Proc Natl Acad Sci USA. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Braun AR, Carr WS, Picchioni D, Balkin TJ, Fukunaga M, Duyn JH. Decoupling of the Brain’s Default Mode Network During Deep Sleep. Proc Natl Acad Sci USA. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A, Put E, Ingels M, Put T, Bossuyt A. One-Year Follow-up of Technetium-99m-HMPAO SPECT in Mild Head Injury. J Nuc Med. 1996;37:1605–1609. [PubMed] [Google Scholar]
- Jo HJ, Lee J, Kim J, Shin Y, Kim I, Kwon JS, Kim SI. Spatial Accuracy of fMRI Activation Influenced by Volume- and Surface-based Spatial Smoothing Techniques. Neuroimage. 2007;34:550–564. doi: 10.1016/j.neuroimage.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Jo HJ, Lee J, Kim J, Choi C, Gu B, Kang D, Ku J, Kwon JS, Kim SI. Artificial Shifting of fMRI Activation Localized by Volume- and Surface-based Analyses. Neuroimage. 2008;40:1077–1089. doi: 10.1016/j.neuroimage.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Johnson B, Zhang K, Gay M, Neuberger T, Horovitz S, Hallett M, Sebastianelli W, Slobounov S. Metabolic Alterations in Corpus Callosum May Compromise Brain Functional Connectivity in MTBI Patients: An 1H-MRS Study. Neurosci Lett. 2012;509:5–8. doi: 10.1016/j.neulet.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone T, Walsh KSO, Greischar LL, Alexander AL, Fox AS, Davidson RJ, Oakes TR. Motion Correction and the Use of Motion Covariates in Multiple-subject Fmri Analysis. Hum Brain Mapp. 2006;27:779–788. doi: 10.1002/hbm.20219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley Elizabeth, van der Kouwe AJW, Gollub R, Kennedy D, et al. Reliability in Multi-site Structural MRI Studies: Effects of Gradient Non-linearity Correction on Phantom and Human Data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Menon DK, Salmond CH, Outtrim JG, Taylor Tavares JV, Carpenter TA, Pickard JD, Sahakian BJ, Stamatakis EA. Altered Functional Connectivity in the Motor Network After Traumatic Brain Injury. Neurology. 2010;75:168–176. doi: 10.1212/WNL.0b013e3181e7ca58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior LJ, Zempel JM, Nolan TS, Prior FW, Snyder AZ, Raichle ME. Cortical Network Functional Connectivity in the Descent to Sleep. Proc Natl Acad Sci USA. 2009;106:4489–4494. doi: 10.1073/pnas.0900924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V, Marchiori M. Efficient Behavior of Small-World Networks. Phys Rev Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Levin HS, Wilde EA, Troyanskaya M, Petersen NJ, Scheibel R, Newsome M, Radaideh M, et al. Diffusion Tensor Imaging of Mild to Moderate Blast-Related Traumatic Brain Injury and Its Sequelae. J Neurotrauma. 2010;27:683–694. doi: 10.1089/neu.2009.1073. [DOI] [PubMed] [Google Scholar]
- Liang X, Zou Q, He Y, Yang Y. Coupling of Functional Connectivity and Regional Cerebral Blood Flow Reveals a Physiological Basis for Network Hubs of the Human Brain. Proc Natl Acad Sci. 2013;110:1929–1934. doi: 10.1073/pnas.1214900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Tang J, Fornito A, Liu T, Chen X, Chen H, Xiang X, Wang X, Hao W. Alterations in Regional Homogeneity of Resting-state Brain Activity in Ketamine Addicts. Neurosci Lett. 2012;522:36–40. doi: 10.1016/j.neulet.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Lin AP, Liao HJ, Merugumala SK, Prabhu SP, Meehan WP, Ross BD. Metabolic Imaging of Mild Traumatic Brain Injury. Brain Imaging Behav. 2012;6:208–223. doi: 10.1007/s11682-012-9181-4. [DOI] [PubMed] [Google Scholar]
- Lowe MJ, Mock BJ, Sorenson JA. Functional Connectivity in Single and Multislice Echoplanar Imaging Using Resting-State Fluctuations. Neuroimage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Mac Donald CL, Johnson AM, Cooper D, Nelson EC, Werner NJ, Shimony JS, Snyder AZ, et al. Detection of Blast-related Traumatic Brain Injury in U.S. Military Personnel. N Engl J Med. 2011;364:2091–2100. doi: 10.1056/NEJMoa1008069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez de la Plata CD, Garces J, Kojori ES, Grinnan J, Krishnan K, Pidikiti R, Spence J, et al. Deficits in Functional Connectivity of Hippocampal and Frontal Lobe Circuits After Traumatic Axonal Injury. Arch Neurol. 2011;68:74–84. doi: 10.1001/archneurol.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AR, Mannell MV, Ling J, Gasparovic C, Yeo RA. Functional Connectivity in Mild Traumatic Brain Injury. Hum Brain Mapp. 2011;32:1825–1835. doi: 10.1002/hbm.21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore ET. Age-related Changes in Modular Organization of Human Brain Functional Networks. Neuroimage. 2009a;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. Hierarchical Modularity in Human Brain Functional Networks. Front Neuroinform. 2009b;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheloyannis S, Pachou E, Stam CJ, Breakspear M, Bitsios P, Vourkas M, Erimaki S, Zervakis M. Small-world Networks and Disturbed Functional Connectivity in Schizophrenia. Schizophr Res. 2006;87:60–66. doi: 10.1016/j.schres.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Minka T. Tech Rep 514. Massachusetts Inst. Technol; Cambridge: 2000. Automatic Choice of Dimensionality for PCA. [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The Impact of Global Signal Regression on Resting State Correlations: Are Anti-correlated Networks Introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hillary FG, Biswal BB. Resting Network Plasticity Following Brain Injury. PLoS ONE. 2009;4:e8220. doi: 10.1371/journal.pone.0008220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Fast Algorithm for Detecting Community Structure in Networks. Phys Rev E. 2004;69:066133. doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- Newman M, Girvan M. Finding and Evaluating Community Structure in Networks. Phys Rev E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric Permutation Tests for Functional Neuroimaging: A Primer with Examples. Hum Brain Mapp. 2001;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niogi SN, Mukherjee P, Ghajar J, Johnson CE, Kolster R, Lee H, Suh M, Zimmerman RD, Manley GT, McCandliss BD. Structural Dissociation of Attentional Control and Memory in Adults with and without Mild Traumatic Brain Injury. Brain. 2008;131:3209–3221. doi: 10.1093/brain/awn247. [DOI] [PubMed] [Google Scholar]
- Niogi SN, Mukherjee P. Diffusion Tensor Imaging of Mild Traumatic Brain Injury. J Head Trauma Rehabil. 2010;25:241–255. doi: 10.1097/HTR.0b013e3181e52c2a. [DOI] [PubMed] [Google Scholar]
- Ojemann JG, Akbudak E, Snyder AZ, McKinstry RC, Raichle ME, Conturo TE. Anatomic Localization and Quantitative Analysis of Gradient Refocused Echo-planar fMRI Susceptibility Artifacts. Neuroimage. 1997;6:156–167. doi: 10.1006/nimg.1997.0289. [DOI] [PubMed] [Google Scholar]
- Okie Susan. Reconstructing Lives — A Tale of Two Soldiers. N Engl J Med. 2006;355:2609–2615. doi: 10.1056/NEJMp068235. [DOI] [PubMed] [Google Scholar]
- Pandit AS, Expert P, Lambiotte R, Bonnelle V, Leech R, Turkheimer FE, Sharp DJ. Traumatic Brain Injury Impairs Small-world Topology. Neurology. 2013;80:1826–1833. doi: 10.1212/WNL.0b013e3182929f38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskind ER, Petrie EC, Cross DJ, Pagulayan K, McCraw K, Hoff D, Hart K, et al. Cerebrocerebellar Hypometabolism Associated with Repetitive Blast Exposure Mild Traumatic Brain Injury in 12 Iraq War Veterans with Persistent Post-concussive Symptoms. Neuroimage. 2011;54:S76–S82. doi: 10.1016/j.neuroimage.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten SC, Bartolomei F, Stam CJ. Small-world Networks and Epilepsy: Graph Theoretical Analysis of Intracerebrally Recorded Mesial Temporal Lobe Seizures. Clin Neurophysiol. 2007;118:918–927. doi: 10.1016/j.clinph.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, et al. Functional Network Organization of the Human Brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but Systematic Correlations in Functional Connectivity MRI Networks Arise from Subject Motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A Default Mode of Brain Function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts SARB, Stam CJ, Kuijer JPA, Scheltens Ph, Barkhof F. Identifying Confounds to Increase Specificity During a ‘no Task Condition’: Evidence for hippocampal connectivity using fMRI. Neuroimage. 2003;20:1236–1245. doi: 10.1016/S1053-8119(03)00386-0. [DOI] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT. Maps of Random Walks on Complex Networks Reveal Community Structure. Proc Natl Acad Sci. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex Network Measures of Brain Connectivity: Uses and Interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Argall B, Japee S, Cox RW. SUMA: an interface for surface-based intra- and inter-subject analysis with AFNI. 2004 2nd IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2004;1 and 2:1510–1513. [Google Scholar]
- Saad ZS, Gotts SJ, Murphy K, Chen G, Jo HJ, Martin A, Cox RW. Trouble at Rest: How Correlation Patterns and Group Differences Become Distorted After Global Signal Regression. Brain Connect. 2012;2:25–32. doi: 10.1089/brain.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatman KE, Duhaime A, Bullock R, Mass AIR, Valadka A, Manley GT Workshop scientific team and advisory panel members. Classification of Traumatic Brain Injury for Targeted Therapies. J Neurotrauma. 2008;25:719–738. doi: 10.1089/neu.2008.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore ET. Neurophysiological Architecture of Functional Magnetic Resonance Images of Human Brain. Cereb Cortex. 2005;15:1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of In-scanner Head Motion on Multiple Measures of Functional Connectivity: Relevance for Studies of Neurodevelopment in Youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel RS, Newsome MR, Troyanskaya M, Lin X, Steinberg JL, Radaideh M, Levin HS. Altered Brain Activation in Military Personnel with One or More Traumatic Brain Injuries Following Blast. J Int Neuropsychol Soc. 2012;18:89–100. doi: 10.1017/S1355617711001433. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable Intrinsic Connectivity Networks for Salience Processing and Executive Control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonne F, Dale AM, Busa E, Glessner M, Salat DH, Hahn HK, Fischl B. A Hybrid Approach to the Skull Stripping Problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Beckmann CF, Greenwood R, Kinnunen KM, Bonnelle V, de Boissezon X, Powell JH, Counsell SJ, Patel MC, Leech R. Default Mode Network Functional and Structural Connectivity After Traumatic Brain Injury. Brain. 2011;134:2233–2247. doi: 10.1093/brain/awr175. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Hamoda HM, Schneiderman JS, Bouix S, Pasternak O, Rathi Y, Vu MA, et al. A Review of Magnetic Resonance Imaging and Diffusion Tensor Imaging Findings in Mild Traumatic Brain Injury. Brain Imaging Behav. 2012;6:137–192. doi: 10.1007/s11682-012-9156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumskaya E, Andriessen TMJC, Norris DG, Vos PE. Abnormal Whole-brain Functional Networks in Homogeneous Acute Mild Traumatic Brain Injury. Neurology. 2012;79:175–182. doi: 10.1212/WNL.0b013e31825f04fb. [DOI] [PubMed] [Google Scholar]
- Slobounov SM, Gay M, Zhang K, Johnson B, Pennell D, Sebastianelli W, Horovitz S, Hallett M. Alteration of Brain Functional Network at Rest and in Response to YMCA Physical Stress Test in Concussed Athletes: RsFMRI Study. Neuroimage. 2011;55:1716–1727. doi: 10.1016/j.neuroimage.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponheim SR, McGuire KA, Kang SS, Davenport ND, Aviyente S, Bernat EM, Lim KO. Evidence of Disrupted Functional Connectivity in the Brain After Combat-related Blast Injury. Neuroimage. 2011;54:S21–S29. doi: 10.1016/j.neuroimage.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Stam C, Jones B, Nolte G, Breakspear M, Scheltens P. Small-World Networks and Functional Connectivity in Alzheimer’s Disease. Cereb Cortex. 2006;17:92–99. doi: 10.1093/cercor/bhj127. [DOI] [PubMed] [Google Scholar]
- Tang L, Ge Y, Sodickson DK, Miles L, Zhou Y, Reaume J, Grossman RI. Thalamic Resting-State Functional Networks: Disruption in Patients with Mild Traumatic Brain Injury. Radiology. 2011;260:831–840. doi: 10.1148/radiol.11110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PA, Ford CC. Simulation of Blast-induced Early-time Intracranial Wave Physics Leading to Traumatic Brain Injury. J Biomech Eng. 2009;131:061007. doi: 10.1115/1.3118765. [DOI] [PubMed] [Google Scholar]
- Tucholka A, Fritsch V, Poline J, Thirion B. An Empirical Comparison of Surface-based and Volume-based Group Studies in Neuroimaging. Neuroimage. 2012;63:1443–1453. doi: 10.1016/j.neuroimage.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Valencia M, Pastor MA, Fernandez-Seara MA, Artieda J, Martinerie J, Chavez M. Complex Modular Structure of Large-scale Brain Networks. Chaos. 2009;19:023119. doi: 10.1063/1.3129783. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Boersma M, Hulshoff Pol HE. Small-world and Scale-free Organization of Voxel-based Resting-state Functional Connectivity in the Human Brain. Neuroimage. 2008;43:528–539. doi: 10.1016/j.neuroimage.2008.08.010. [DOI] [PubMed] [Google Scholar]
- van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic Functional Connectivity as a Tool for Human Connectomics: Theory, Properties, and Optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk KRA, Sabuncu MR, Buckner RL. The Influence of Head Motion on Intrinsic Functional Connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Essen DC, Dierker D. On Navigating the Human Cerebral Cortex: Response to ‘in Praise of Tedious Anatomy’. Neuroimage. 2007;37:1050–1054. doi: 10.1016/j.neuroimage.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wingen GA, Geuze E, Caan MWA, Kozicz T, Olabarriaga SD, Denys D, Vermetten E, Fernandez G. Persistent and Reversible Consequences of Combat Stress on the Mesofrontal Circuit and Cognition. Proc Natl Acad Sci USA. 2012;109:15508–15513. doi: 10.1073/pnas.1206330109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL. Evidence for a Frontoparietal Control System Revealed by Intrinsic Functional Connectivity. J Neurophysiol. 2008;100:3328–3342. doi: 10.1152/jn.90355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zuo X, Gohel S, Milham MP, Biswal BB, He Y. Graph Theoretical Analysis of Functional Brain Networks: Test-Retest Evaluation on Short- and Long-Term Resting-State Functional MRI Data. PLoS ONE. 2011;6:e21976. doi: 10.1371/journal.pone.0021976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden DL. Military TBI During the Iraq and Afghanistan Wars. J Head Trauma Rehabil. 2006;21:398–402. doi: 10.1097/00001199-200609000-00004. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective Dynamics of ‘small-world’ Networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wig GS, Schlaggar BL, Petersen SE. Concepts and Principles in the Analysis of Brain Networks. Ann NY Acad Sci. 2011;1224:126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- Yeo RA, Gasparovic C, Merideth F, Ruhl D, Doezema D, Mayer AR. A Longitudinal Proton Magnetic Resonance Spectroscopy Study of Mild Traumatic Brain Injury. J Neurotrauma. 2011a;28:1–11. doi: 10.1089/neu.2010.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, et al. The Organization of the Human Cerebral Cortex Estimated by Intrinsic Functional Connectivity. J of Neurophysiol. 2011b;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Snyder AZ, Fox MD, Sansbury MW, Shimony JS, Raichle ME. Intrinsic Functional Relations Between Human Cerebral Cortex and Thalamus. J Neurophysiol. 2008;100:1740–1748. doi: 10.1152/jn.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
With each subject’ data that passed the quality assurance procedure before checking the module assignment results, the seed was placed at the left posterior cingulate cortex (PCC; −7, −55, 27, red spot) including nodes within 6 mm Euclidean distance from the PCC. The colors represent statistical significance of the group means of Fisher-Z transformed correlation coefficients. After ensuring that the Fisher-Z transformed correlations across subjects within each group passed the Shapiro-Wilk test at α = 0.05, the t-test on the group means were performed, thresholded (puncorr < 0.001) and converted to z-score.
See Fig. 2 for the details of labels.
The ρ-values in panels a–b represent the Spearman rank correlation coefficients. See Fig. 3 for the details of scatter plots in panel c. On the initial scans, network measures of the TBI patients within the gray oval were distant from the inverse linear trends (a). On the follow-up scans, only one subject was an outlier from the trends (b). As expected, the TBI I cohort had lower average participation coefficient, than the controls on the initial scans (Fig. S4c, left panel). As for modulatity, group differences in average participation coefficient were not observed on the follow-up scans (Fig. S4c, right panel).
The first and second columns are scatter plots for the modularity of subjects who had acceptable data quality for both the initial and follow-up scans. Changes in this modularity at each subject are on the third column. Scatter plots on the fourth column are symmetrized percent change, i.e., (follow-up scans – initial scans)/average of initial and follow-up scans. Filled triangles on the third and fourth columns are relatively ‘abnormal’ TBI patients on the initial scans. Dotted magenta horizontal bars on the TBI I in the fourth column represent the group average values of the TBI patients without these ‘abnormal’ TBI patients. See Fig. 3 for the other details of the scatter plots.
The p-values were obtained from permutation tests (10,000 permutations) on group mean difference. See Fig. 3 for the details of the scatter plots. Note that subjects whose images passed all quality assurance procedure up to the motion censoring criteria were included for this analysis (See Table I).
The ρ-values are the Spearman correlation coefficients.
See Fig. 8 for the details of the bar graphs.
See Fig. 8 for the details of the bar graphs.
See Fig. 8 for the details of the bar graphs.
Regions are nodes within 20 mm geodesic distance from the local peaks. See Fig. 6 for the details of labels.
See Fig. 8 for the details of the bar graphs.