Abstract
Previous studies have demonstrated that the use of integrated information from multi-modalities could significantly improve diagnosis of Alzheimer’s Disease (AD). However, feature selection, which is one of the most important steps in classification, is typically performed separately for each modality, which ignores the potential strong inter-modality relationship within each subject. Recent emergence of multi-task learning approach makes the joint feature selection from different modalities possible. However, joint feature selection may unfortunately overlook different yet complementary information conveyed by different modalities. We propose a novel multi-task feature selection method to preserve the complementary inter-modality information. Specifically, we treat feature selection from each modality as a separate task and further impose a constraint for preserving the inter-modality relationship, besides separately enforcing the sparseness of the selected features from each modality. After feature selection, a multi-kernel Support Vector Machine (SVM) is further used to integrate the selected features from each modality for classification. Our method is evaluated using the baseline PET and MRI images of subjects obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. Our method achieves a good performance, with an accuracy of 94.37% and an Area Under the ROC Curve (AUC) of 0.9724 for AD identification, and also an accuracy of 78.80% and an AUC of 0.8284 for Mild Cognitive Impairment (MCI) identification. Moreover, the proposed method achieves an accuracy of 67.83% and an AUC of 0.6957 for separating between MCI converters and MCI non-converters (to AD). These performances demonstrate the superiority of the proposed method over the state-of-the-art classification methods.
Keywords: Alzheimer’s Disease, Mild cognitive impairment, Multi-kernel support vector machine, Multi-task feature selection, inter-modality relationship
1. Introduction
Alzheimer’s Disease (AD) is a genetically complex and irreversible neurodegenerative disorder which is clinically characterized by progressive dementia and neuropsychiatric symptoms (Blennow et al., 2006). The number of subjects with AD has been predicted to quadruple by 2050 (Brookmeyer et al., 2007). Although numerous efforts have been made in the past decades to develop new treatment strategies, there is no effective treatment or effective diagnostic instrument until now. This causes substantial financial burden to the society, as well as psychological and emotional burden to patients and their families. Mild Cognitive Impairment (MCI), an intermediate stage between normal cognition and dementia, has a high risk of progressing to AD (Petersen et al., 1999). While the annual incidence rate of healthy subjects to develop AD is 1% to 2% (Bischkopf et al., 2002), the conversion rate from MCI to AD is reported to be approximately 10% to 15% per year (Grundman et al., 2004). Thus, it is of great interest to identify MCI and also predict its risk of progressing to AD.
Accumulating evidence demonstrates that individuals with AD have both functional and structural changes in the brain, such as loss of grey matter volume (Karas et al., 2003) and metabolic abnormalities (Matsuda, 2001). However, these findings are mainly obtained based on group-level statistical comparison, and thus are of limited value for individual-based disease diagnosis. To overcome this limitation, pattern classification methods have been used in recent years, and have shown great potential in neuroimaging studies (Fan et al., 2007; Wee et al., 2013). Unlike group-based comparison approaches, pattern classification methods are able to detect the fine-grained spatial discriminative patterns, which are critical for individual-based disease diagnosis. Moreover, some studies have shown that the combination of complementary information from different imaging modalities can improve the accuracy in diagnosis of AD and MCI. For example, Dai et al. (2012b) used both structural Magnetic Resonance Imaging (MRI) and resting-state functional MRI to classify 16 AD patients from 22 healthy subjects, and achieved a classification accuracy of 89.47%, which is an increase of 2.63% from the single-modality based method. Wee et al. (Wee et al., 2012) used both Diffusion Tensor Imaging (DTI) and resting-state functional MRI to identify 10 individuals with MCI from 17 matched Normal Controls (NC), and obtained a very promising classification accuracy of 96.3%, which is an increase of 7.4% from the single-modality based method. Although high classification accuracies were achieved, the small sample and large feature size problem in these studies may still lead to data overfitting. Since the original feature space of neuroimaging data is relatively high compared to the sample size, feature selection is one of the most important steps in neuroimaging classification. However, in the literature, feature selection in multimodal classification studies is often performed separately for each imaging modality without considering the potential strong relationship among different modalities, thus possibly affecting the final classification results. Hence, it is reasonable to consider preserving the inter-modality relationship during the feature selection for final improvement of classification.
Recently, multi-task learning approach has attracted the increasing attention in machine learning, computer vision, and artificial intelligence (Evgeniou and Pontil, 2007; Zhou et al., 2011). The main goal of this approach is to capture the intrinsic relationship among different tasks with the assumption that these tasks are related to each other. Learning multiple related tasks simultaneously has been shown to often perform better than learning each task separately (Evgeniou and Pontil, 2007). Specially, recent emergence of multi-task learning method enables joint feature selection via group sparsity (i.e. using L2,1 norm) (Liu et al., 2009). However, this constraint may be too strong, since it forces common features to be selected for different tasks, without considering that different tasks may need different features.
In this paper, a novel multi-task learning based feature selection method is proposed to better preserve the complementary information conveyed by different modalities. More specifically, selection of features from different modalities is treated as different tasks. To better capture the complementary information among different modalities, it is also important to preserve the relationship between the feature vectors derived from different modalities, especially after their projection onto the lower dimensional feature space. To this end, a new constraint is imposed to preserve the inter-modality relationship, besides enforcing the sparseness of the selected features from each modality as popularly used in the literature. A multi-kernel Support Vector Machine (SVM) is finally used to combine the selected features from each modality for predicting the classification labels.
The remainder of this paper is organized as follows. Materials and Methods section furnishes information on the image dataset, preprocessing pipeline, and details of the proposed framework. Experimental results of the proposed method and some state-of-the-art methods on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset are summarized in the Experiment and Results section. The findings of proposed framework are extensively discussed in the Discussion section, which is followed by the Conclusion section.
2. Materials and Methods
2.1. Subjects
The data were taken from the ADNI dataset (www.adni.loni.ucla.edu/ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and nonprofit organizations, as a $60 million, 5-year public–private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Dr. Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years.
All the patients met the following inclusion criteria: 1) diagnosis of AD was made if the subject had a Mini-Mental State Examination (MMSE) score 20–26, a Clinical Dementia Rating (CDR) of 0.5 or 1.0, and meets the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS/ADRDA) criteria for probable AD. 2) Individuals were categorized as amnestic MCI if they had an MMSE score 24–30, a CDR of 0.5, a memory complaint, objective memory loss measured by education adjusted scores on Wechsler Memory Scale Logical Memory II, absence of significant levels of impairment in other cognitive domains, while essentially preserved activities of daily living, and an absence of dementia. All NC individuals met the following criteria: an MMSE score 24–30, a CDR of 0, nondepressed, non-MCI, and nondemented. The research protocol was approved by each local institutional review board, and written informed consent was obtained from each subject at the time of enrollment for imaging and genetic sample collection.
Two hundreds and two subjects from ADNI dataset: 51 patients with AD, 99 patients with MCI (43 MCI converters who had converted to AD within 18 months and 56 MCI non-converters who had not converted to AD within 18 months), and 52 NC are analyzed in this study. Table 1 presents a summary of the demographic characteristics of the used subjects.
Table 1.
Characteristics of the subjects used in this study.
| Characteristics | AD (n = 51) | MCI (n = 99) | NC (n = 52) |
|---|---|---|---|
| Gender (M/F) | 33/18 | 67/32 | 34/18 |
| Age (mean ± SD) | 75.2 ± 7.4 | 75.3 ± 7.0 | 75.3 ± 5.2 |
| Education (mean ± SD) | 14.7 ± 3.6 | 15.9 ± 2.9 | 15.8 ± 3.2 |
| MMSE (mean ± SD) | 23.8 ± 2.0 | 27.1 ± 1.7 | 29.0 ± 1.2 |
2.2. Data acquisition and preprocessing
All structural MRI scans used were acquired using 1.5T scanners. MRI acquisitions were performed according to the ADNI acquisition protocol (Jack et al., 2008). For the image preprocessing, we first performed Anterior Commissure - Posterior Commissure (AC-PC) correction on all images, and then used N3 algorithm (Sled et al., 1998) to correct intensity inhomogeneity. Skull-stripping (Wang et al., 2011) was then performed, followed by the registration-based cerebellum removal. We used FAST in FSL (Zhang et al., 2001) to segment brain into three different tissue types: Grey Matter (GM), White Matter (WM) and Cerebrospinal Fluid (CSF). We registered the template into subject specific space using HAMMER (Shen and Davatzikos, 2002) to preserve the absolute image volume of each subjects. Subsequently, we parcellated the brain in 93 regions-of-interest, based on the Jacob template (Kabani, 1998). GM volume of each ROI was extracted as the feature for the MRI modality. PET images were acquired 30–60 min post-injection, averaged, spatially aligned, interpolated to a standard voxel size, intensity normalized, and smoothed to a common resolution of 8-mm full width at half maximum. For each subject, we aligned the preprocessed PET image to its respective MRI image using affine registration. Then we calculated the average intensity of each regions-of-interest and treated it as the feature for the PET modality. Thus, we finally have 93 features from the MRI image and 93 features from the PET image, for each subject.
2.3. Overview of our method
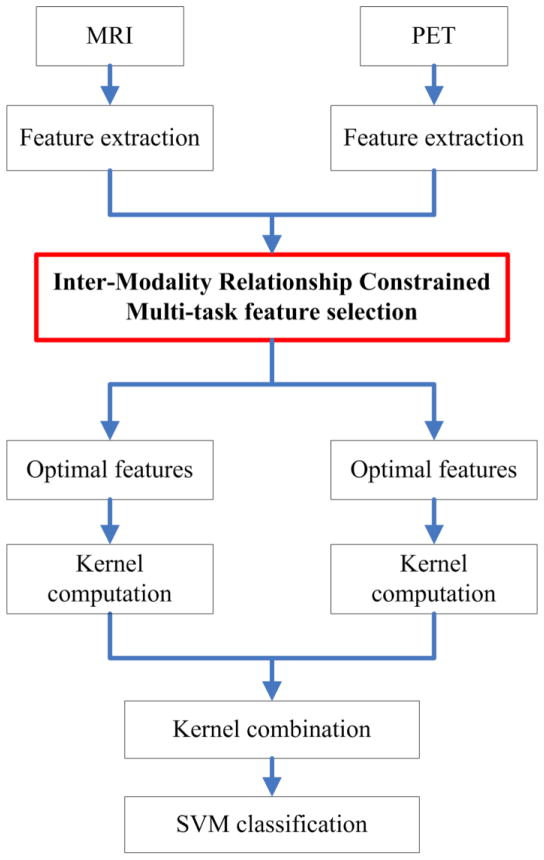
An overview of the proposed classification pipeline was illustrated in Fig. 1. From the preprocessed PET and MRI images, we first extracted the respective regional features as motioned above. Based on these features, inter-modality relationship constrained multi-task feature selection was applied to select the important features from each modality. Based on the selected features, a kernel matrix was constructed for each modality. Then, a fused kernel matrix was further constructed, based on the individual matrix from each modality, for classification by using SVM.
Fig. 1.
Schematic diagram illustrating the proposed classification framework.
Although we used regional features, the number of features was relatively large compared to the sample size, and most importantly, some of them were irrelevant or redundant for classification. Thus, an effective feature selection could not only speed up computation, but also improve the classification performance. Unlike the traditional single-task feature selection approaches which treat each task independently, such as Least Absolute Shrinkage and Selection Operator (LASSO) (Tibshirani, 1996), multi-task feature selection approaches can often significantly improve the classification performance by learning multiple tasks simultaneously (Evgeniou and Pontil, 2007).
2.4. Multi-task feature selection
Let be a n × d matrix that represents d features of n training samples for modality j, j = 1, …, m, where m is the total number of modalities. Let be a n dimensional corresponding target vector (with classification labels as values of +1 or −1 in this study) for modality j. Linear regression model used for prediction can be defined as follows (Zhou et al., 2011):
| (1) |
where wj ∈ Rd×1 and ŷj denote, respectively, the regression coefficient vector and the predicted label vector of the j-th modality. To estimate all m regression coefficient vectors from all m modalities such as W = [w1, …, wj, …, wm], one of the popular approaches is to minimize the following objective function:
| (2) |
where λ1 > 0 is a regularization parameter which controls the sparsity of the model, with a higher value leading to a sparser model, i.e., more elements in W are zero. ||W||1 is the L1 norm of W, which is defined as . This regression approach is known as LASSO.
The limitation of above-mentioned model is that each task is treated independently. Although we can use the recent emergence of multi-task feature learning method, under framework of group sparsity (i.e., L2,1 norm (Liu et al., 2009)), to guide selection of common features from different modalities, the complementary information conveyed by different modalities might be discarded after this too strong group sparsity constraint and thus affect the final classification results.
To address this problem, we propose to preserve the relative distance between the feature vectors extracted from different modalities of the same subject (also called as inter-modality relationship), before and after feature projection, by imposing the following constraint:
| (3) |
where and denote the feature vectors of the j-th and k-th modalities in the i-th subject, respectively. is the relative distance between the feature vectors and before feature projection, while is the respective distance after feature projection (or the distance between the corresponding predictions). Basically, if is small, is also required to be small after projection. The goal of providing this additional constraint or guidance as described in Eq. (3) is to better preserve the manifold of relative distributions among different modalities. Although using the sparse regression (based on L1 norm) can effectively select discriminative features from each modality separately, it ignores the relationship among different modalities, which could make the final selected features over-fitting to the data and thus affect the final performance. Therefore, the use of Eq. (3) is mainly for keeping the joint relationship among different modalities, when each modality selects its own discriminative features. Note that, in this way, the relationships among different modalities can be preserved via this constraint after projection onto the low dimensional feature space.
By using the constraint given in Eq. (3), the objective function of the proposed multi-task feature selection model can be further defined as
| (4) |
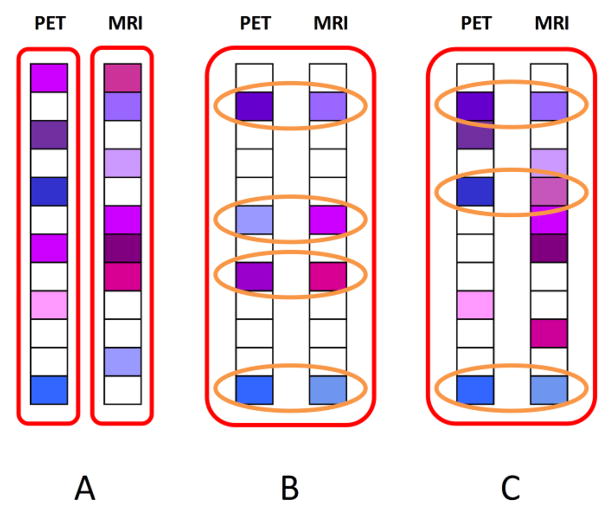
where λ1 > 0 and λ2 > 0 are the regularization parameters controlling, respectively, the sparseness and the degree of preserving the inter-modality relationship. The optimization of objective function in Eq. (4) can be solved using the Accelerated Proximal Gradient (APG) method (Nesterov, 2003). In our application, the target vectors are the classification labels, and we just have two modalities (MRI and PET) with the same target vector, i.e., m = 2 and y1 = y2. The models constructed by different approaches are compared in Fig. 2.
Fig. 2.
Comparison of different feature selection models. Fig. 2A shows the model constructed by the L1 norm, where feature selection is performed independently on each individual modality. Fig. 2B shows the model constructed by the L2,1 norm, where a common set of features is selected from all modalities. Fig. 2C shows the proposed feature selection model, where, in additon to the common features selected across all modalities, the complementary information conveyed by the modality-specific features is also effectively preserved.
Of note, all the procedures of feature selection are always performed on the training set, without using the information from the test set. Also, each feature in the training set is transformed into z score via the following transformation:
| (5) |
where and σr are the mean and standard deviation of the r-th feature across all the training subjects. The and σr are also used to normalize the r-th feature fr in the test subject.
It is also worth noting that, for feature selection, we just keep those features with non-zero regression coefficients for the subsequent classification with SVM. For simplicity, in the following, we still use to denote a vector of the selected features for the j-th modality in the i-th subject.
2.5. Multi-kernel SVM
Let represents a feature vector of the i-th subject with m modalities and yi ∈ {1, −1} denotes the corresponding class label. The primal optimization problem of the traditional single-kernel SVM is given as:
| (6) |
where ξi denotes non-negative slack variable which measures the degree of misclassification of the data, C denotes the penalty parameter which controls the amount of constraint violations introduced by ξi, b denotes the bias term, q denotes the normal vector of hyperplane, and Ø denotes the kernel-induced mapping function.
The dual form of single kernel SVM can be represented as below:
| (7) |
where α is the Lagrange multiplier and k(xi, xp) = Ø(xi)TØ(xp) is the kernel function for training samples xi and xp.
Given a new test sample v = {v1, …, vj, …, vm}, the decision function of single-kernel SVM for predicted label is
| (8) |
Previous study demonstrated that multi-kernel SVM can effectively integrate the features from different modalities compared to single-kernel SVM (Dai et al., 2012a; Zhang et al., 2011). The primal optimization problem of a multi-kernel SVM is given as:
| (9) |
where βj ≥ 0 denotes the weighting factor on the j-th modality and with the constraint of . Similarly as in the single-kernel SVM, the dual form of multi-kernel SVM is given as:
| (10) |
where denotes the kernel matrix for training samples and for the j-th modality.
Given a new test sample v = {v1, …, vj, …, vm}, the decision function of multi-kernel SVM for predicted label is
| (11) |
The multi-kernel SVM can be embedded into the single-kernel SVM by interpreting as a mixed kernel between the multi-modality training samples xi and xp, and as a mixed kernel between the multi-modality training sample xi and the test sample v.
2.6. Validation
SVM classifier was implemented by using the LIBSVM toolbox (Chang and Lin, 2011). A ten-fold cross-validation strategy was used to evaluate the classification performance. Specifically, the whole samples were randomly partitioned into ten subsets and then nine subsets were chose for training and the remaining one was used for testing, and this procedure was repeated ten times to avoid any bias introduced by randomly partitioning in the cross-validation. To determine the optimal values for the parameters, i.e., the regularization parameters λ1, λ2 and the above-mentioned kernel combination weight, we performed another ten-fold cross-validation on the training samples. SVM model with the best performance in this nested cross-validation was then used to classify the unseen test samples. ACCuracy (ACC), SENsitivity (SEN) and SPEcificity (SPE) were calculated to quantify the performance of all compared methods, which were defined respectively as
| (12) |
where TP is the number of true positives (number of patients correctly classified), TN is the number of true negatives (number of NC correctly classified), FP is the number of false positives (number of NC classified as patients), and FN is the number of false negatives (number of patients classified as NC).
3. Experiment and Results
3.1. Experiment settings
The proposed approach was compared with three different classification approaches, i.e., Single modality approach using Single-Task feature selection (i.e., LASSO) and conventional Single-kernel SVM (SSTS), Multi-modality approach using Single-Task feature selection (i.e., LASSO) on each modality and Multi-kernel SVM (MSTM), and Multi-modality approach using Joint feature Selection (i.e., L2,1 norm) and Multi-kernel SVM (MJSM). It is worth noting that the same training and test data were used in all methods for fair comparison. Performance of each comparison method was evaluated through two different classification tasks: AD vs. NC and MCI vs. NC. The MCI dataset used was the combination of all MCI converters and MCI non-converters.
3.2. Comparison of Results
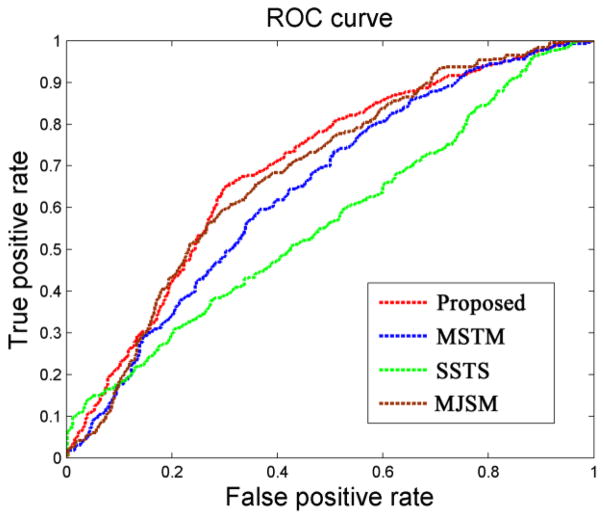
As can be seen from Table 2, our proposed method consistently obtains better performance than any of other three methods in AD/MCI classification. Fig. 3 further plots the Receiver Operating Characteristic (ROC) curves of different methods for AD classification and MCI classification, respectively. Specifically, for classifying AD from NC, the proposed method achieves a classification accuracy of 94.37%, sensitivity of 94.71%, specificity of 94.04%, and the Area Under the ROC Curve (AUC) of 0.9724, indicating excellent diagnostic power. In contrast, the best performance is only 88.25% by using individual modality (i.e., SSTS, when using PET), 91.02% by MSTM, and 91.10% by MJSM. On the other hand, for classifying MCI from NC, the proposed method achieved a classification accuracy of 78.80%, sensitivity of 84.85%, specificity of 67.06%, and the AUC of 0.8284, while the best performance is only 71.41% using the individual modality (i.e. SSTS, when using PET), 72.08% by MSTM, and 73.54% by MJSM. We also performed paired t-tests on the classification accuracy between the proposed method and the other three comparison methods, and the p values were provided in Table 2.
Table 2.
Classification performance of all comparison methods.
| Method | AD vs. NC | MCI vs. NC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p-value | ACC (%) | SEN (%) | SPE (%) | AUC | p-value | ACC (%) | SEN (%) | SPE (%) | AUC | |
| SSTS | <0.001 | 88.25 | 84.91 | 91.54 | 0.9004 | <0.001 | 71.41 | 77.78 | 59.23 | 0.7575 |
| MSTM | <0.001 | 91.02 | 89.02 | 92.88 | 0.9655 | <0.001 | 72.08 | 75.56 | 65.38 | 0.7826 |
| MJSM | <0.001 | 91.10 | 91.57 | 90.58 | 0.9584 | <0.001 | 73.54 | 81.01 | 59.23 | 0.7706 |
| Proposed | - | 94.37 | 94.71 | 94.04 | 0.9724 | - | 78.80 | 84.85 | 67.06 | 0.8284 |
Fig. 3.
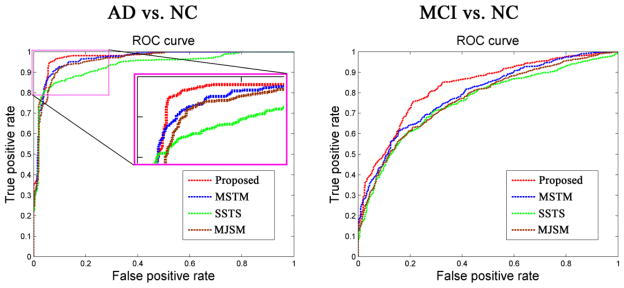
ROC curves of different methods for classification of AD (left) and MCI (right).
3.3. Comparison with other existing methods
Furthermore, we compare the classification results of the proposed method with some results reported in the literature, also mainly based on both PET and MRI data of the ADNI subjects. Specifically, we compare with three recent studies as briefly described next. Hinrichs and colleagues (2011) used 48 AD patients and 66 NC for AD diagnosis, and obtained an accuracy of 87.60% by using imaging modalities (PET+MRI) and an accuracy of 92.40% by using five modalities (MRI+PET+CSF+APOE+cognitive scores). No MCI vs. NC classification result was reported in this paper. In Gray et al. (2013), they used 37 AD patients, 75 MCI patients, and 35 NC for AD and MCI classifications. By using four modalities (CSF+MRI+PET+Genetic) as features, they reported an accuracy of 89.00% for AD classification and an accuracy of 74.60% for MCI classification. One of our previous studies (Zhang et al., 2011), which used the same dataset as the present study, reported an accuracy of 90.60% for AD classification by using MRI and PET and an accuracy of 93.20% for AD classification by using MRI, PET and CSF. Besides, they reported an accuracy of 76.40% for MCI classification by using three modalities, and obtained an accuracy of 73.79% if using only PET and MRI (which was obtained when we ran their algorithm). The results of these methods, along with our proposed method, as reported in Table 3, further validate the efficacy of our proposed method in both AD and MCI classifications.
Table 3.
Comparison with the classification accuracies reported in the literature.
| Method | Subjects | Modalities | AD vs. NC (%) | MCI vs. NC (%) |
|---|---|---|---|---|
| Zhang et al. (2011) | 51AD+99MCI+52HC | PET+MRI | 90.60 | - |
| Zhang et al. (2011) | 51AD+99MCI+52HC | PET+MRI+CSF | 93.20 | 76.40 |
| Hinrichs et al. (2011) | 48AD+66NC | PET+MRI | 87.60 | - |
| Hinrichs et al. (2011) | 48AD+66NC | MRI+PET+CSF+APOE+cognitive scores | 92.40 | - |
| Gray et al. (2013) | 37AD+75MCI+35NC | PET+MRI+CSF+Genetic | 89.00 | 74.60 |
| Proposed | 51AD+99MCI+52NC | PET+MRI | 94.37 | 78.80 |
3.4. Effective of feature selection procedure
We also compare the performances of the aforementioned classification tasks using our proposed feature selection method, or without using any feature selection (i.e., using all features) since the number of original features (186) is comparable to the number of subject samples. The same multi-kernel SVM framework is applied for both comparison methods. As can be seen in Table 4, the proposed feature selection method performs much better than the case without feature selection.
Table 4.
Classification performance with or without feature selection step.
| Method | Subjects | Modalities | AD vs. NC (%) | MCI vs. NC (%) | MCI subgroup (%) |
|---|---|---|---|---|---|
| Without | 51AD+99MCI+52NC | PET+MRI | 89.90 | 70.89 | 59.18 |
| With | 51AD+99MCI+52NC | PET+MRI | 94.37 | 78.80 | 67.83 |
3.5. Classification between MCI converters and MCI non-converters (to AD)
Due to the heterogeneity of MCI patients, it is important to predict whether a certain MCI patient will progress to AD within a certain period of time. Thus, a good AD/MCI classification framework should be able to differentiate between different MCI subgroups, i.e. MCI converters and MCI non-converters to AD. Based on Table 5 and Fig. 4, the proposed method outperforms the comparison methods in the MCI subgroup classification. Specifically, our method achieves accuracy of 67.83%, sensitivity of 64.88%, specificity of 70.00%, and the AUC of 0.6957, while the best performance is only 57.90% using individual modality (i.e. SSTS, when using MRI), 61.71% by MSTM, and 65.74% by MJSM, respectively. The results of MCI subgroup classification without feature selection step are also provided in Table 4.
Table 5.
MCI subgroup classification performance of all comparison methods.
| Method | MCI converter vs. MCI non-converter | ||||
|---|---|---|---|---|---|
| p-value | ACC (%) | SEN (%) | SPE (%) | AUC | |
| SSTS | <0.001 | 57.90 | 46.74 | 66.43 | 0.6158 |
| MSTM | <0.001 | 61.71 | 56.28 | 65.89 | 0.6455 |
| MJSM | 0.020 | 65.74 | 56.04 | 73.21 | 0.6828 |
| Proposed | - | 67.83 | 64.88 | 70.00 | 0.6957 |
Fig. 4.
ROC curves of different methods for classification of MCI subgroups.
3.6. The most discriminative regions
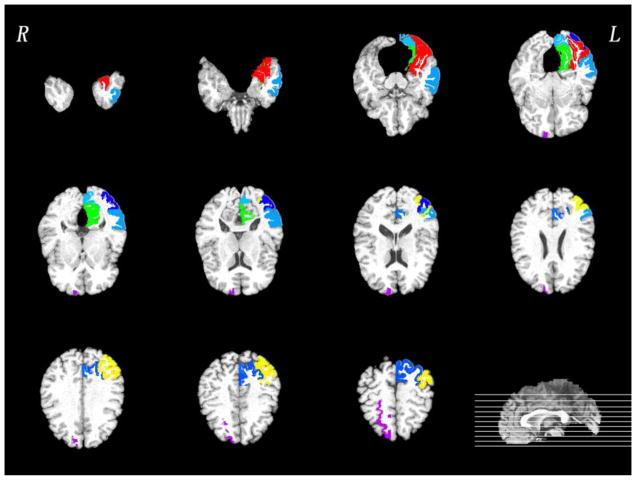
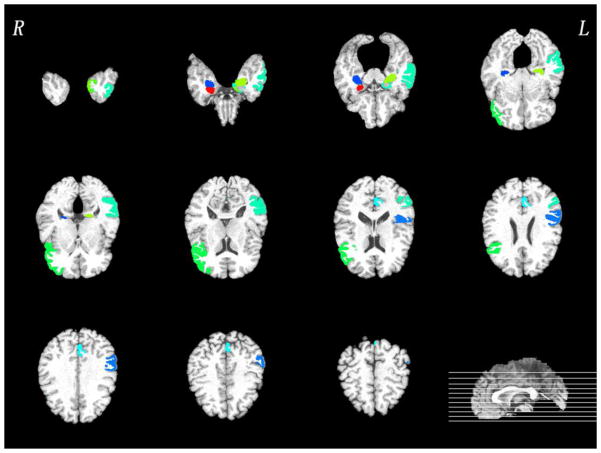
In this subsection, we investigate the most discriminative regions that were selected by the proposed feature selection method, i.e., for MCI subgroup classification. Since the feature selection in each fold is performed only based on the training set, the selected features could differ slightly across different cross-validation folds. We thus define the most discriminative regions as regions that were most frequently selected in all cross-validations. The top ten selected regions in PET and MRI were provided in Figs. 5–6, respectively. Most of selected regions, e.g., hippocampal formation, precuneus and entorhinal cortex, are highly related to AD pathology. In particular, hippocampal formation is a memory-related brain structure that was always affected in the AD.
Fig. 5.
Top ten most discriminative PET regions in the MCI subgroup classification. Of note, different colors in the figure just indicate different brain regions.
Fig. 6.
Top ten most discriminative MRI regions in the MCI subgroup classification. Of note, different colors in the figure just indicate different brain regions.
3.7. Evaluation using a longitudinal dataset
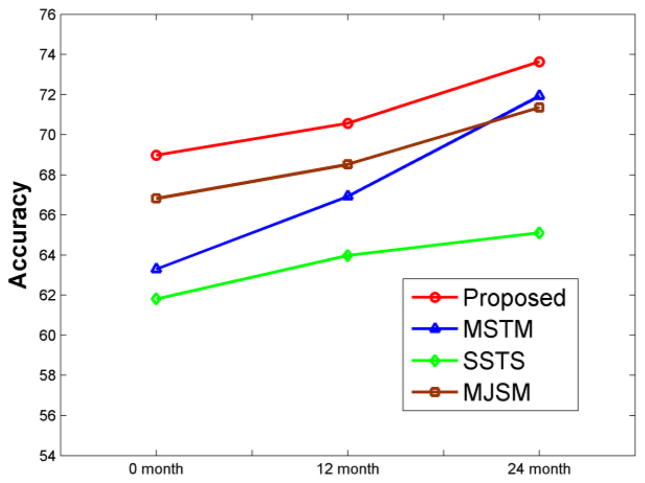
We use the same longitudinal dataset as used in our previous study (Zhang and Shen, 2012) to further evaluate the performance of our proposed method. The same procedure, including feature selection, parameter determination, and cross-validation, is used for comparing the performances of four methods in differentiating MCI converters from MCI non-converters. The classification accuracies of the four methods are shown in Fig. 7, with the x-axis indicating the scan time for the MCI subjects. It can be observed that the proposed method consistently performs better than other 3 methods, and also the performances of all methods increase for the late scanned images, since the separation between MCI converters and MCI non-converters becomes larger and larger when subjects become older and older.
Fig. 7.
Performances of four different methods on a longitudinal dataset. Our proposed method achieves the best results, and the performances of all four methods are improved for the later scans since the difference between MCI converters and MCI non-converters becomes larger and larger with aging.
4. Discussion
This paper has presented a novel multi-modality multi-task feature selection approach for brain disease diagnosis. Different from the conventional single-task feature selection, which selects features independently from each modality, and also the joint feature selection using the L2,1 norm, the proposed method considers the relationship of features from each modality and then preserves the inter-modality relationship during the feature selection. Experimental results show that our method can improve the performance of AD/MCI diagnosis.
Several recent studies used multimodal imaging features for diagnosis of AD, and demonstrated that the complementary information from different modalities could significantly improve the classification results (Dai et al., 2012b; Fan et al., 2007; Walhovd et al., 2010). However, feature selection in these studies was performed independently in each modality. Thus, it may overlook the complementary information conveyed in different modalities. Multi-task learning approach is a paradigm that learns a number of supervised learning tasks simultaneously by exploiting the commonalities between them. Because it allows the learner to use the relationship among different tasks, this often leads to a better model than the case of learning these tasks independently (Evgeniou and Pontil, 2007). The key of multi-task learning is to capture the intrinsic relationship among different tasks. Actually, tasks can be related in various ways. The relationship of tasks was modeled by different assumptions, e.g., (1) all tasks are close to each other in some norm (Bakker and Heskes, 2003), (2) all tasks share a common underlying characteristic (Ben-David and Schuller, 2003), or (3) all task have a common set of features (Argyriou et al., 2008).
We propose to preserve the relative distance between feature vectors of different modalities of the same subject, before and after their projection to the low-dimensional feature space. Promising classification performance we obtained also indicates a good diagnostic power and generalizability of our proposed framework. As we can see from Table 2, the SSTS method does not use the complementary information from other modalities and thus leads to the lowest classification accuracy when using only single-modality features for classification. Our finding, along with other multi-modality classification studies (Dai et al., 2012b; Fan et al., 2007; Walhovd et al., 2010), demonstrates that the complementary information in different modalities can help diagnose AD and MCI. On the other hand, both MSTM and MJSM methods have better classification results than SSTS, but still lower than our results. MSTM method performs feature selection in each modality independently without considering the complementary information between modalities. On the other hand, MJSM method forces too strongly for joint feature selection from different modalities, i.e., selecting same regional features from different modalities. Actually, the abnormal regions in PET and MRI data could be different, e.g., some regions could have decreased GM volume while still the normal metabolism, and other regions could have metabolic abnormality while the normal GM volume (Ishii et al., 2005). Therefore, the complementary information conveyed by different modalities might be eliminated after this too-strong joint feature selection, thus finally affecting the classification performance.
Furthermore, as shown in Table 3, our method is better than three other recent studies, even though they used more modalities, which further shows the efficacy of our proposed method in both AD and MCI classifications. Although direct comparison with the aforementioned studies is not appropriate due to the possible use of different subjects (although from the same ADNI dataset), the obtained results validate the promising performance of our method for classification to some extent. For fair comparison, we also compared the results with our previous study which used the same dataset. Better performance (compared to our previous work) further validates the efficacy of our proposed method.
Classifying MCI converters from MCI non-converters has recently received a significant amount of attention due to its importance for early AD diagnosis. In MCI patients, there are typically two types of clinical changes: 1) MCI patients who will convert to AD in the future time point, i.e., MCI-converters, and 2) MCI patients who will not convert after a certain period of time, i.e., MCI non-converters. Early diagnosis of MCI conversion is of tremendously important for possible delay of the disease, and prediction of progression of the disease. For classifying MCI converters from MCI non-converters, as shown in Table 5, our method achieves an accuracy of 67.83%, sensitivity of 64.88%, and specificity of 70.00%. Our accuracy and AUC are both higher than the comparison methods. Besides, better classification results on a longitudinal dataset further demonstrate the robustness and superiority of our proposed method.
Identification of objective biomarkers is of great interest as it could, ultimately, inform clinical decisions of individual patients. With this consideration, our proposed feature selection method seeks to identify those features that are the most discriminative in classifying the MCI converters from MCI non-converters. These features include the hippocampal formation, frontal, parietal, and occipital region. Specifically, hippocampal formation plays an important role in the consolidation of information from short-term memory to long-term memory (Eichenbaum et al., 1994; Jaffard and Meunier, 1993). This region is one of the first brain regions to suffer damage, with memory loss and disorientation, which are included among the early AD symptoms (Van Hoesen and Hyman, 1990). In addition, voxel-based analysis studies demonstrate significant changes in perirhinal cortex (Leube et al., 2008), middle temporal gyrus (Busatto et al., 2003), amygdala (Matsuda, 2013), entorhinal cortex (Derflinger et al., 2011), uncus (Yang et al., 2012), and inferior frontal gyrus (Kim et al., 2011). Besides, PET studies showed significant abnormalities in the precuneus (Del Sole et al., 2008), middle occipital gyrus (Smith et al., 2009), superior parietal lobule (Potkin et al., 2002), inferior occipital gyrus (Melrose et al., 2009), lingual gyrus (Eustache et al., 2004), superior frontal gyrus (Desgranges et al., 2002), medial occipitotemporal gyrus (Matsuda, 2001), and angular gyrus (Hunt et al., 2006). The fact that our results are consistent with the previous findings suggests the effectiveness of our method in identifying biomarkers for MCI subgroup classification.
Although our proposed method demonstrates a good performance from cross-validations, several limitations should be considered in the present study. First, the proposed feature selection method requires the same number of features computed from different modalities. Indeed, in addition to MRI and PET data, there are other modalities in ADNI database, such as CSF and genetic data, which have different number of features. These modalities carry important pathological information that can help further improve classification performance. In the future work, we plan to extend our proposed method for including more modalities. Second, we use 10-fold cross-validation strategy on ADNI dataset to evaluate performance of our proposed method. Although we do not use separate dataset to test our method, 10-fold cross validation, as used popularly in the machine learning field for classification performance evaluation, somehow tests the performance of our method on separate datasets. Third, there is no consensus of a time boundary for MCI converters and non-converters. Here, we use 18 months for an exploratory analysis. Further classification analysis should be performed by using different time boundary in MCI subgroup classification. Finally, although we use a cross-validation to evaluate the generalizability of our method, it is important to test on a completely independent dataset in the future.
5. Conclusion
We have proposed a novel multi-task learning based feature selection method to effectively preserve the complementary information from multi-modal neuroimaging data for AD/MCI identification. Specifically, we treat the selection of features from each modality as a task and then propose a new constraint to preserve the inter-modality relationship during the feature selection. Experimental results on ADNI database demonstrate that our multi-task feature selection method, after integrated with multi-kernel SVM, outperforms the state-of-the-art methods. In the future, we will extend our work to include more modalities (such as CSF and genetic features) for further improving AD/MCI classification performance.
Highlight.
A novel multi-task feature selection method is proposed for AD/MCI diagnosis.
Proposed feature selection preserves potential strong inter-modality relationship.
Integration of multimodality data using multi-kernel SVM.
High accuracy of 94.37% for AD classification and 78.80% for MCI classification.
Perform significantly better in distinguishing MCI-converters from non-converters.
Acknowledgments
This work was supported in part by NIH grants EB006733, EB008374, EB009634, AG041721, MH100217 and AG042599. This work was also partially supported by the National Research Foundation grant (No. 2012-005741) funded by the Korean government. In addition, H. Chen was supported by the 973 project (No. 2012CB517901), the Natural Science Foundation of China (Nos. 61125304 and 61035006), and the Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20120185110028). F. Liu was supported by China Scholarship Council (No. 2011607033) and the Scholarship Award for Excellent Doctoral Student granted by Ministry of Education (No. A03003023901010). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argyriou A, Evgeniou T, Pontil M. Convex multi-task feature learning. Machine Learning. 2008;73:243–272. [Google Scholar]
- Bakker B, Heskes T. Task clustering and gating for bayesian multitask learning. The Journal of Machine Learning Research. 2003;4:83–99. [Google Scholar]
- Ben-David S, Schuller R. Exploiting task relatedness for multiple task learning. Learning Theory and Kernel Machines. 2003:567–580. [Google Scholar]
- Bischkopf J, Busse A, Angermeyer MC. Mild cognitive impairment--a review of prevalence, incidence and outcome according to current approaches. Acta Psychiatr Scand. 2002;106:403–414. doi: 10.1034/j.1600-0447.2002.01417.x. [DOI] [PubMed] [Google Scholar]
- Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Busatto GF, Garrido GE, Almeida OP, Castro CC, Camargo CH, Cid CG, Buchpiguel CA, Furuie S, Bottino CM. A voxel-based morphometry study of temporal lobe gray matter reductions in Alzheimer’s disease. Neurobiol Aging. 2003;24:221–231. doi: 10.1016/s0197-4580(02)00084-2. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2:27. [Google Scholar]
- Dai D, Wang J, Hua J, He H. Classification of ADHD children through multimodal magnetic resonance imaging. Frontiers in systems neuroscience. 2012a:6. doi: 10.3389/fnsys.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Yan C, Wang Z, Wang J, Xia M, Li K, He Y. Discriminative analysis of early Alzheimer’s disease using multi-modal imaging and multi-level characterization with multi-classifier (M3) Neuro Image. 2012b;59:2187–2195. doi: 10.1016/j.neuroimage.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Del Sole A, Clerici F, Chiti A, Lecchi M, Mariani C, Maggiore L, Mosconi L, Lucignani G. Individual cerebral metabolic deficits in Alzheimer’s disease and amnestic mild cognitive impairment: an FDG PET study. Eur J Nucl Med Mol Imaging. 2008;35:1357–1366. doi: 10.1007/s00259-008-0773-6. [DOI] [PubMed] [Google Scholar]
- Derflinger S, Sorg C, Gaser C, Myers N, Arsic M, Kurz A, Zimmer C, Wohlschlager A, Muhlau M. Grey-matter atrophy in Alzheimer’s disease is asymmetric but not lateralized. J Alzheimers Dis. 2011;25:347–357. doi: 10.3233/JAD-2011-110041. [DOI] [PubMed] [Google Scholar]
- Desgranges B, Baron JC, Giffard B, Chetelat G, Lalevee C, Viader F, de la Sayette V, Eustache F. The neural basis of intrusions in free recall and cued recall: a PET study in Alzheimer’s disease. Neuro Image. 2002;17:1658–1664. doi: 10.1006/nimg.2002.1289. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H, Otto T, Cohen NJ. Two functional components of the hippocampal memory system. Behavioral and Brain Sciences. 1994;17:449–472. [Google Scholar]
- Eustache F, Piolino P, Giffard B, Viader F, De La Sayette V, Baron JC, Desgranges B. ‘In the course of time’: a PET study of the cerebral substrates of autobiographical amnesia in Alzheimer’s disease. Brain. 2004;127:1549–1560. doi: 10.1093/brain/awh166. [DOI] [PubMed] [Google Scholar]
- Evgeniou A, Pontil M. Multi-task feature learning. Advances in neural information processing systems: Proceedings of the 2006 conference; The MIT Press; 2007. p. 41. [Google Scholar]
- Fan Y, Rao H, Hurt H, Giannetta J, Korczykowski M, Shera D, Avants BB, Gee JC, Wang J, Shen D. Multivariate examination of brain abnormality using both structural and functional MRI. Neuro Image. 2007;36:1189–1199. doi: 10.1016/j.neuroimage.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D. Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. Neuro Image. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA, Foster NL, Jack CR, Jr, Galasko DR, Doody R. Mild cognitive impairment can be distinguished from Alzheimer disease and normal aging for clinical trials. Archives of neurology. 2004;61:59. doi: 10.1001/archneur.61.1.59. [DOI] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuro Image. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A, Schonknecht P, Henze M, Toro P, Haberkorn U, Schroder J. CSF tau protein and FDG PET in patients with aging-associated cognitive decline and Alzheimer’s disease. Neuropsychiatr Dis Treat. 2006;2:207–212. doi: 10.2147/nedt.2006.2.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Sasaki H, Kono AK, Miyamoto N, Fukuda T, Mori E. Comparison of gray matter and metabolic reduction in mild Alzheimer’s disease using FDG-PET and voxel-based morphometric MR studies. Eur J Nucl Med Mol Imaging. 2005;32:959–963. doi: 10.1007/s00259-004-1740-5. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, JLW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffard R, Meunier M. Role of the hippocampal formation in learning and memory. Hippocampus. 1993;3(Spec No):203–217. [PubMed] [Google Scholar]
- Kabani NJ. 3D anatomical atlas of the human brain. Neuro Image. 1998:7. [Google Scholar]
- Karas GB, Burton EJ, Rombouts SA, van Schijndel RA, O’Brien JT, Scheltens P, McKeith IG, Williams D, Ballard C, Barkhof F. A comprehensive study of gray matter loss in patients with Alzheimer’s disease using optimized voxel-based morphometry. Neuro Image. 2003;18:895–907. doi: 10.1016/s1053-8119(03)00041-7. [DOI] [PubMed] [Google Scholar]
- Kim S, Youn YC, Hsiung GY, Ha SY, Park KY, Shin HW, Kim DK, Kim SS, Kee BS. Voxel-based morphometric study of brain volume changes in patients with Alzheimer’s disease assessed according to the Clinical Dementia Rating score. J Clin Neurosci. 2011;18:916–921. doi: 10.1016/j.jocn.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Leube DT, Weis S, Freymann K, Erb M, Jessen F, Heun R, Grodd W, Kircher TT. Neural correlates of verbal episodic memory in patients with MCI and Alzheimer’s disease--a VBM study. Int J Geriatr Psychiatry. 2008;23:1114–1118. doi: 10.1002/gps.2036. [DOI] [PubMed] [Google Scholar]
- Liu J, Ji S, Ye J. Proceedings of the Twenty-Fifth Conference on Uncertainty in Artificial Intelligence. AUAI Press; 2009. Multi-task feature learning via efficient 1, 2, 1-norm minimization; pp. 339–348. [Google Scholar]
- Matsuda H. Cerebral blood flow and metabolic abnormalities in Alzheimer’s disease. Ann Nucl Med. 2001;15:85–92. doi: 10.1007/BF02988596. [DOI] [PubMed] [Google Scholar]
- Matsuda H. Voxel-based Morphometry of Brain MRI in Normal Aging and Alzheimer’s Disease. Aging Dis. 2013;4:29–37. [PMC free article] [PubMed] [Google Scholar]
- Melrose RJ, Campa OM, Harwood DG, Osato S, Mandelkern MA, Sultzer DL. The neural correlates of naming and fluency deficits in Alzheimer’s disease: an FDG-PET study. Int J Geriatr Psychiatry. 2009;24:885–893. doi: 10.1002/gps.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterov Y. Introductory lectures on convex optimization: A basic course. Springer; 2003. [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Alva G, Keator D, Carreon D, Fleming K, Fallon JH. Brain metabolic effects of Neotrofin in patients with Alzheimer’s disease. Brain Res. 2002;951:87–95. doi: 10.1016/s0006-8993(02)03140-2. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith GS, Kramer E, Ma Y, Hermann CR, Dhawan V, Chaly T, Eidelberg D. Cholinergic modulation of the cerebral metabolic response to citalopram in Alzheimer’s disease. Brain. 2009;132:392–401. doi: 10.1093/brain/awn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996:267–288. [Google Scholar]
- Van Hoesen GW, Hyman BT. Hippocampal formation: anatomy and the patterns of pathology in Alzheimer’s disease. Prog Brain Res. 1990;83:445–457. doi: 10.1016/s0079-6123(08)61268-6. [DOI] [PubMed] [Google Scholar]
- Walhovd K, Fjell A, Brewer J, McEvoy L, Fennema-Notestine C, Hagler D, Jennings R, Karow D, Dale A. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. American Journal of Neuroradiology. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nie J, Yap PT, Shi F, Guo L, Shen D. Robust deformable-surface-based skull-stripping for large-scale studies. Med Image Comput Comput Assist Interv. 2011;14:635–642. doi: 10.1007/978-3-642-23626-6_78. [DOI] [PubMed] [Google Scholar]
- Wee CY, Yap PT, Zhang D, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Identification of MCI individuals using structural and functional connectivity networks. Neuroimage. 2012;59:2045–2056. doi: 10.1016/j.neuroimage.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee CY, Yap PT, Zhang D, Wang L, Shen D. Group-constrained sparse fMRI connectivity modeling for mild cognitive impairment identification. Brain Struct Funct. 2013 doi: 10.1007/s00429-013-0524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Pan P, Song W, Huang R, Li J, Chen K, Gong Q, Zhong J, Shi H, Shang H. Voxelwise meta-analysis of gray matter anomalies in Alzheimer’s disease and mild cognitive impairment using anatomic likelihood estimation. J Neurol Sci. 2012;316:21–29. doi: 10.1016/j.jns.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Zhang D, Shen D. Predicting future clinical changes of mci patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuro Image. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. Medical Imaging, IEEE Transactions on. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yuan L, Liu J, Ye J. A multi-task learning formulation for predicting disease progression. Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining; ACM. 2011. pp. 814–822. [Google Scholar]