Summary
The impact of RNA viruses on the post-transcriptional regulation of cellular gene expression is unclear. Sindbis virus causes a dramatic relocalization of the cellular HuR protein from the nucleus to the cytoplasm in infected cells. This is due to the expression of large amounts of viral RNAs that contain high affinity HuR binding sites in their 3’ UTRs effectively serving as a sponge for the HuR protein. Sequestration of HuR by Sindbis virus is associated with destabilization of cellular mRNAs that normally bind HuR and rely on it to regulate their expression. Furthermore, significant changes can be observed in nuclear alternative polyadenylation and splicing events on cellular pre-mRNAs due to sequestration of HuR protein by the 3’ UTR of transcripts of this cytoplasmic RNA virus. These studies suggest a new molecular mechanism of virus-host interaction that likely has significant impact on virus replication, cytopathology and pathogenesis.
Keywords: HuR Protein, ELAVL1, alphavirus, polyadenylation, splicing, mRNA stability, RNA sponge
Introduction
Post-transcriptional processes such as splicing, polyadenylation and mRNA decay play a major role in regulating cellular gene expression and shaping the transcriptome and proteome. On average, human genes contain approximately 8 introns and ~70% of genes are alternatively spliced. This generates significant diversity in the proteome and can influence regulatory signals present in transcripts (Kornblihtt et al., 2013). Polyadenylation is also a dynamically regulated process with over 50% of pre-mRNAs containing alternative poly(A) signals that can dramatically alter the composition of regulatory elements in the 3’ untranslated region (UTR) as well as influence mRNA coding capacity (Shi, 2012). Finally in concert with transcription, regulated mRNA stability plays a major role in determining mRNA abundance and quality control of gene expression (Schoenberg and Maquat, 2012). All three of these posttranscriptional processes are regulated by RNA binding proteins (RBPs). The HuR protein, for example, is a ubiquitously expressed RNA binding protein that is predominantly nuclear but shuttles between the nucleus and cytoplasm and is differentially phosphorylated. HuR has a well-established role in the post-transcriptional regulation of gene expression and is the best-characterized mRNA stabilizing factor (Abdelmohsen and Gorospe, 2010) to date. In addition, HuR has been shown to influence translation as well as play a role in the regulation of alternative splicing and alternative polyadenylation in the nucleus (Zhu et al., 2007; Lebedeva et al, 2011). Thus the protein appears to be pivotal to achieving the coordination of changes in cellular gene expression in response to various stimuli (von Roretz et al., 2011).
A key question is how the regulators of post-transcriptional processes are themselves regulated. Important routes include changes in expression levels and post translational modifications which can lead to altered RNA binding affinity or localization. Interestingly, both miRNAs and RBPs can also be modulated through non-functional interaction with RNA molecules bearing their target sequences. These RNA decoys or sponges can have dramatic effects on gene expression. Long non-coding RNAs (Wang et al., 2013), pseudogenes (Johnsson et al., 2013), and circular RNAs (Hansen et al., 2013) have been shown to effectively sequester miRNAs in cells and significantly influence gene expression. Harnessing RNA sponge technologies has also been proposed as a way of directing the regulation of post-transcriptional gene expression (Kong et al., 2006). In addition to sequestering miRNAs, it is also possible that RNA sponges can sequester RNA binding proteins. Although there is little solid evidence for this occurring naturally, artificial decoy RNAs have been successful in interfering with RBP function in cell culture (Bevilacqua et al., 2007; Soundararajan et al., 2008; Eiring et al., 2010; Bolognani et al., 2012). We hypothesized that RNA viruses, through their ability to produce copious amounts of viral RNAs, may use this sponge approach as an integral part of their strategy to usurp cellular gene expression and interfere with targeted cellular processes. To date, only DNA herpesviruses have been shown to encode small RNAs that act as miRNA sponges (Cazalla et al., 2010; Libri et al, 2012). However it has not been investigated whether RNA viruses can act as sponges to sequester cellular RNA binding proteins and have an important, directed effect on cellular gene expression.
Alphaviruses (e.g. Sindbis virus (SinV), Chikungunya virus and Venezuelan equine encephalitis virus) are capped and polyadenylated positive-sense RNA viruses that encode a single subgenomic RNA (Schwartz and Albert, 2010). Their replication occurs exclusively in the cytoplasm. Alphavirus infection has dramatic effects on many aspects of cellular physiology. However in most cases the direct cause of these effects has not been elucidated. In particular, the influence of alphavirus infections on the cellular post-transcriptional processes of splicing, polyadenylation and mRNA stability has not been extensively investigated. We have previously demonstrated that alphavirus transcripts avidly bind to the cellular HuR protein through conserved high affinity binding sites in their 3’ UTR (Garneau et al., 2008; Sokoloski et al., 2010; Dickson et al., 2012). Furthermore, this interaction between viral RNAs and the cellular HuR protein is important for efficient viral gene expression/replication. We now wish to explore the key question of what is the impact of interactions between alphavirus RNAs and the HuR protein on cellular post-transcriptional processes.
In this study, we demonstrate that the 3’ UTR of SinV RNAs serves as an effective sponge that sequesters HuR protein in the cytoplasm during infection. Furthermore, sequestration of HuR by viral RNAs results in the dramatic destabilization of cellular mRNAs that normally rely on the protein to regulate their stability. In addition, viral RNA sequestration of HuR also significantly alters alternative polyadenylation and splicing of select cellular transcripts that normally rely on HuR binding to regulate processing site choice. Collectively these data demonstrate an important role for the concept of ‘sponging’ or sequestration of a cellular RNA binding protein by the transcripts of RNA viruses and mechanistically show how a cytoplasmic RNA virus can directly influence nuclear post-transcriptional processes.
Results
The 3’ UTR of SinV RNA Acts as a Sponge to Sequester the Cellular HuR Protein in the Cytoplasm of Infected Cells
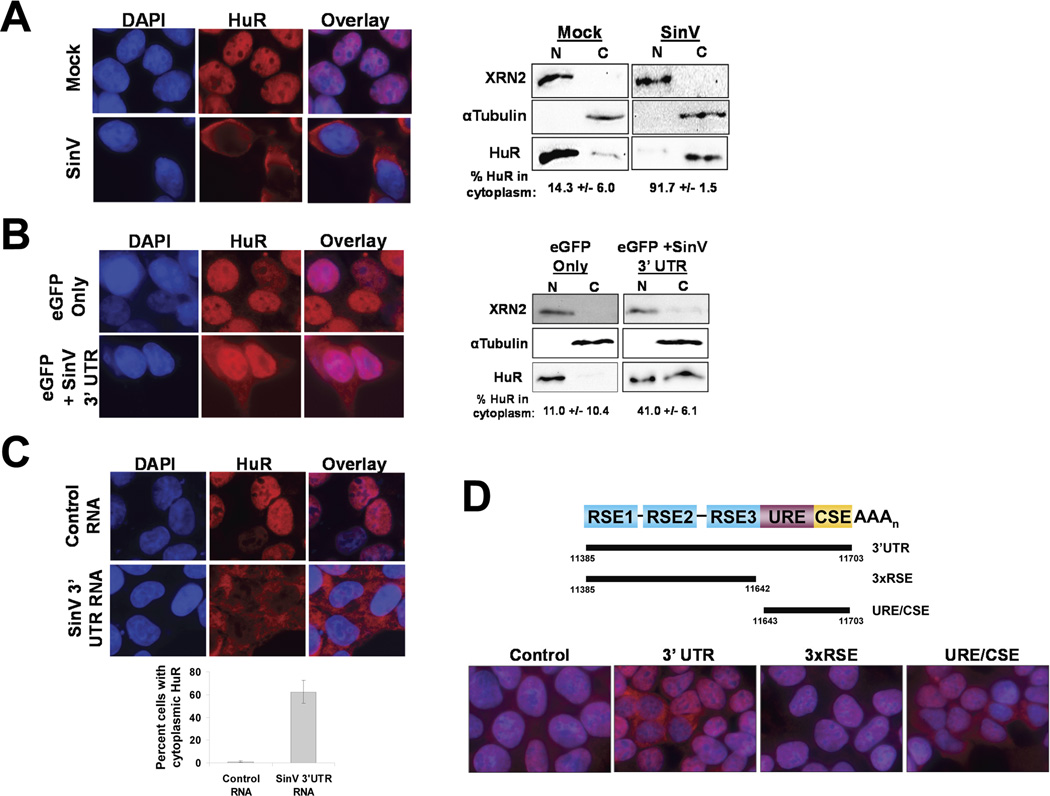
We have previously shown that infection by SinV, as well as several other alphaviruses, results in a redistribution of the cellular HuR protein to the cytoplasm (Sokoloski et al., 2010; Dickson et al., 2012). The underlying mechanism for this phenomenon, however, is not known. As seen in Figure 1A, HuR protein goes from being a predominantly nuclear protein in uninfected cells (14.3 +/− 6.0% in the cytoplasm) to a mainly cytoplasmic protein (91.7 +/− 1.5%) 24 hours post infection with SinV in 293T cells. The redistribution of HuR protein to the cytoplasm required SinV replication/gene expression and appears to be caused by a mechanism distinct from the shuttling of HuR that is observed during cellular stress responses (Dickson et al., 2012). Furthermore, the cytoplasmic redistribution of HuR was not observed during infections with other cytoplasmic RNA viruses, suggesting that it was due to an alphavirus-specific function.
Figure 1. The SinV 3’ UTR is sufficient to induce the relocalization of HuR protein from the nucleus to the cytoplasm.
A. 293T cells were mock-treated or infected with SinV. At 24 hrs post treatment, cells were analyzed for HuR protein localization by immunofluorescence (left panel) or by subcellular fractionation and western blotting (right panel) using antibodies specific for the indicated proteins. DAPI was used to identify the nuclear compartment. Panel B. Cells were transfected with plasmids encoding an eGFP reporter bearing either the default 3’ UTR (eGFP only) or the SinV 3’ UTR (eGFP + SinV 3’ UTR) and analyzed for HuR protein subcellular localization by immunofluorescence (left panel) or by subcellular fractionation and western blotting (right panel). Panel C: 293T cells were transfected with a control RNA or with a SinV 3’ UTR RNA. At 6 hrs post transfection, cells were analyzed for HuR protein subcellular localization by immunofluorescence. Quantification is shown with standard deviations. Panel D: 293T cells were transfected with a control RNA or with the indicated fragment of the SinV 3’ UTR (diagrammed at the top of the panel). At 6 hrs post transfection, cells were analyzed for HuR protein subcellular localization by immunofluorescence.
A survey of viral protein mutants failed to identify a viral ORF required for HuR redistribution to the cytoplasm (data not shown). However based on studies with SinV mutants, we noticed a correlation between the overall abundance of viral RNA and the relocalization of HuR to the cytoplasm of infected cells. Thus we hypothesized that HuR relocalization may be an RNA-dependent phenomenon. SinV genomic and subgenomic RNAs both contain two high affinity HuR binding sites (the U-rich Element (URE) and Conserved Sequence Element (CSE)) in their 3’ UTRs (Sokoloski et al., 2010). In order to assess if the SinV 3’ UTR could mediate HuR relocalization to the cytoplasm, plasmids encoding GFP reporter mRNAs with or without the SinV 3’ UTR were transfected into 293T cells. Based on fluorescence microscopy, the two plasmid constructs produced similar overall amounts of GFP in the transfections (data not shown). As seen in Fig. 1B, transfection of a plasmid that expressed only the GFP reporter mRNA had no effect on HuR localization. Interestingly, transfection of a plasmid expressing a GFP reporter mRNA containing the SinV 3’ UTR caused a ~4-fold increase in the proportion of HuR protein in the cytoplasm. Thus we conclude that the SinV 3’ UTR on its own appears to have a propensity to affect HuR localization in the cell.
Plasmid-generated RNAs are made in the nucleus while SinV transcripts are produced in the cytoplasm. Thus a limitation of the plasmid-based transfection study described in Fig. 1B is that the reporter RNAs have a nuclear experience and thus do not necessarily recapitulate the scenario of expression of viral 3’ UTR sequences during the cytoplasmic-only replication of SinV. Therefore we transfected RNAs into cells and assayed HuR localization to directly assess the ability of the SinV 3’ UTR to induce the redistribution of HuR protein to the cytoplasm. Transfection of a control RNA had no effect on HuR localization in cells (Fig. 1C; Control RNA panels). However, transfection of a SinV 3’ UTR RNA caused a dramatic redistribution of HuR protein from the nucleus to the cytoplasm within 6 hours post-transfection (Fig. 1C, SinV 3’ UTR RNA panels). As seen in Fig. 1D, transfection of RNA fragments of the SinV 3’ UTR mapped the region of the 3’ UTR required for HuR relocalization to a 60 base region that contains a U-Rich Element (URE) and the Conserved Sequence Element (CSE). This 60 base region was demonstrated previously to contain high affinity HuR binding sites (Sokoloski et al., 2010). In order to determine if the amount of SinV 3’UTR-derived RNA that was transfected into cells was biologically relevant, we used qRT-PCR and converted values to numbers of RNA molecules using a standard curve generated by titration of known amounts of linearized SinV 3’ UTR-containing plasmid DNA. Not surprisingly, the functional concentration of transfected RNA – the amount of RNA that actually gets into 293T cells and is stable at the time of analysis (6 hours post transfection) - was very much less than the overall amount of RNA that is used in the transfection experiment (1 ug or ~3×1012 molecule). While there was variation in the amount of RNA that was present inside of cells due to natural variability in transfection efficiencies, 1.54 ×109 copies/ug of total RNA (or ~75,000 copies of transfected RNA per cell) was sufficient to see the effects shown in Fig. 1D. This amount of RNA was similar to the amount of SinV 3’ UTR that is produced during infection (~1.0×109 copies per ug of total RNA or ~50,000 copies of SinV 3’ UTR per cell) in Fig. 1A. Similar values were seen in two independent experiments. Furthermore these transcript numbers per cell are similar to values reported previously (160,000 genomes per infected cell) for SinV at late times post infection (Wang et al., 1991). Therefore we conclude that cytoplasmic expression of the SinV 3’ UTR RNA, in the absence of any viral proteins, is sufficient to induce the relocalization of the cellular HuR protein. These observations suggest that the large amount of viral transcripts produced during an infection act like a sponge to sequester HuR protein in the cytoplasm.
The Sequestration of HuR by SinV has Dramatic Effects on the Relative Stability of a Subset of Cellular mRNAs
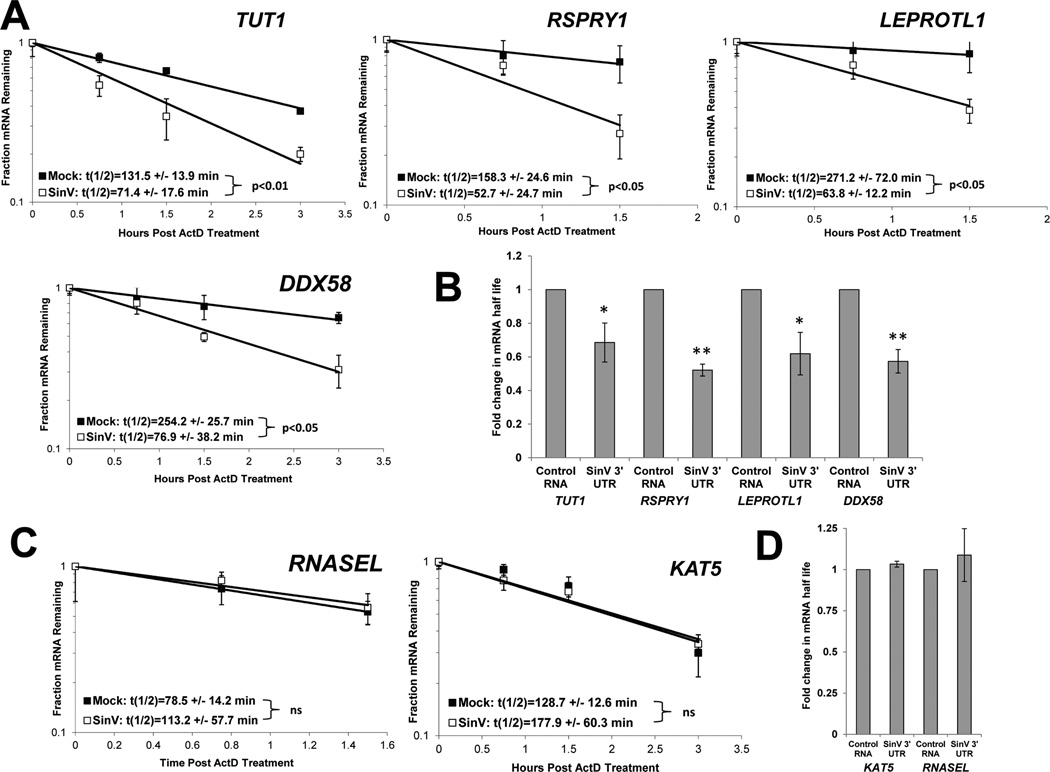
We hypothesized that the dramatic relocalization and sequestration of HuR protein by SinV RNAs would have major effects on the post-transcriptional regulation of cellular gene expression in infected cells. One of the best described roles of the cellular HuR protein is to regulate and mediate the stability of cellular mRNAs by direct binding through U- rich elements (von Roretz et al., 2011). In order to assess whether SinV infection influenced the stability of cellular mRNAs, the relative half-lives of a set of transcripts was compared between infected and mock-infected cells. 293T cells were either mock-treated or infected at a multiplicity of infection (MOI) of 10 with SinV. At 24 hrs post infection, cells were treated with actinomycin D to halt further cellular transcription. Total RNA samples were obtained at designated times post shut-off of cellular transcription and the relative abundances of selected mRNAs assessed. As seen in Fig. 2A, numerous cellular mRNAs such as DDX58 (RIG I), TUT1, LEPROTL1 and RSPRY1 were significantly destabilized during SinV infection, with mRNA half-lives reduced between 2 to 4 fold. Furthermore, the transfection of RNA containing only the 3’ UTR of SinV was sufficient to cause a significant decrease in half-lives of these mRNAs (Fig. 2B). However, as seen in Figs. 2C and 2D, not all cellular mRNAs were destabilized during viral infection or upon transfection of the SinV 3’ UTR RNA. KAT5 and RNASEL mRNAs showed no significant change in half-life. Thus we conclude that there was a selective destabilization of cellular mRNAs during SinV infection or RNA transfection of the 3’ UTR of SinV.
Figure 2. SinV infection influences the stability of some but not all cellular mRNAs.
At 24 hpi with SinV, 293T cells were treated with actinomycin D and the relative levels of the indicated mRNAs were assessed at the designated time points following shut off of transcription using qRT-PCR to determine mRNA half-lives. Panel A depicts mRNAs that were destabilized during SinV infection while panel C contains mRNAs whose stability was not affected. Representative decay curves are shown with standard deviation of experimental measurements and the average half-lives are reported with standard deviations from three independent experiments. In Panels B and D, 293T cells were transfected with equimolar amounts of either a control RNA (GemA60) or the SinV 3’UTR RNA. At 4.5 hrs post transfection, cells were treated with actinomycin D and the relative levels of the indicated mRNAs were assessed by qRT-PCR. Average fold change in half-lives are reported with standard deviations from two independent experiments. * p<0.05; ** p<0.01.
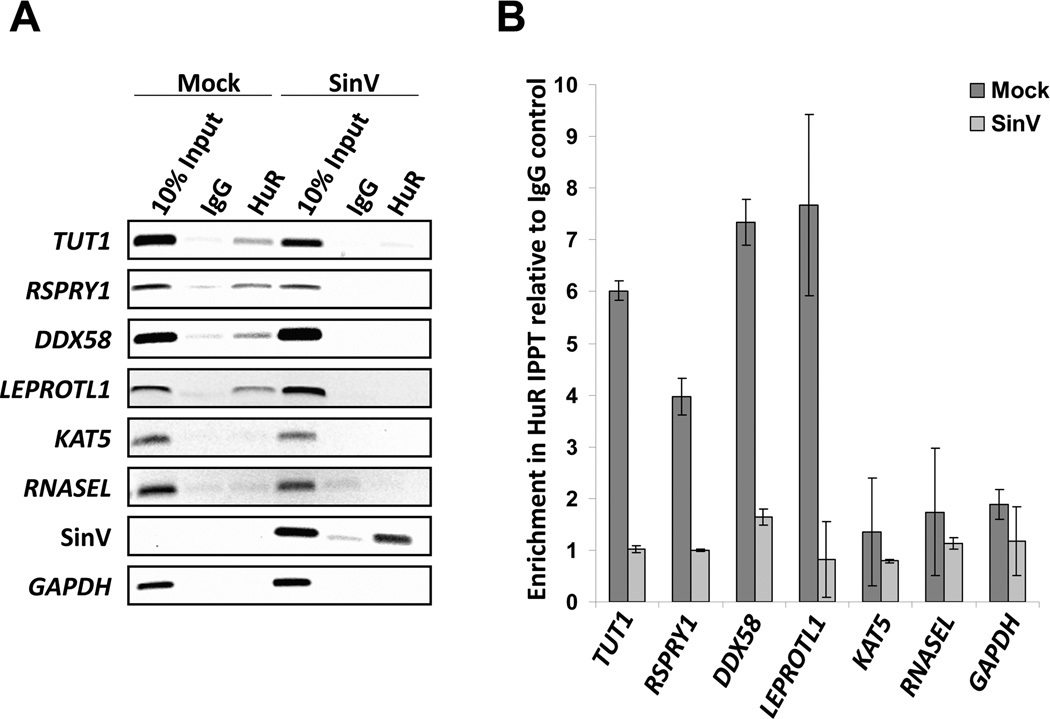
In order to elucidate the underlying mechanism for the selective destabilization of cellular mRNAs, we hypothesized that differential binding of the cellular HuR protein may be involved due to sequestration of the protein by viral RNAs. In order to assess this hypothesis, formaldehyde-stabilized HuR-containing ribonucleoprotein complexes were immunoprecipitated from mock-infected and SinV infected cells and co-precipitating mRNAs were assessed by RT-PCR and qRT-PCR. As seen in Figure 3A, HuR protein normally associated with four of the mRNAs tested (TUT1, RSPRY1, DDX58 and LEPROTL1), but HuR interactions with these transcripts were dramatically reduced in SinV infected cells. Interestingly, all four of these mRNAs were also destabilized in SinV infection (Fig. 2). Both the KAT5 and RNASEL mRNAs failed to substantially interact with HuR protein under any condition tested – and notably neither of these mRNAs experienced a change in stability during SinV infection. Fig. 3B demonstrates an independent replicate of the experiment in Fig. 3A but analyzed by qRT-PCR with samples normalized to the IgG control immunoprecipitation to allow a quantitative analysis. In addition, SinV RNAs were effectively co-precipitated with the HuR-specific antisera from infected samples (Fig. 3A), consistent with the notion that they are largely bound by HuR protein. Finally, we note that RNASEL has been previously shown to be regulated by HuR protein and that endogenous RNASEL mRNA was shown to bind HuR in murine C2C12 cells (Li et al., 2007). We assume that differences in cell type used may account for the observations reported here.
Figure 3. SinV infection significantly reduced the association of HuR with cellular mRNAs.
293T cells were either mock-infected or infected with SinV. At 24 hrs post treatment, HuR protein-RNA complexes were isolated by immunoprecipitation using HuR-specific antibodies or a control normal mouse IgG. Co-precipitating mRNAs were analyzed by RT-PCR (panel A) or qRT-PCR (panel B). The two panels depict results from independent infections.
Therefore we conclude that the destabilization of cellular mRNAs during SinV infection is associated with a reduction of HuR binding due to sequestration of the cellular stability factor by viral RNAs. Given the importance of several of these proteins in innate immunity (e.g. RIG I), cellular gene expression (e.g. TUT1) and overall cellular biology, it is likely that SinV RNA-induced alterations in cellular mRNA stability will have important biological consequences on cytopathology, host-response to the virus and overall viral replication.
Alternative pre-mRNA Polyadenylation and Splicing in the Nucleus is also Influenced by SinV Infection
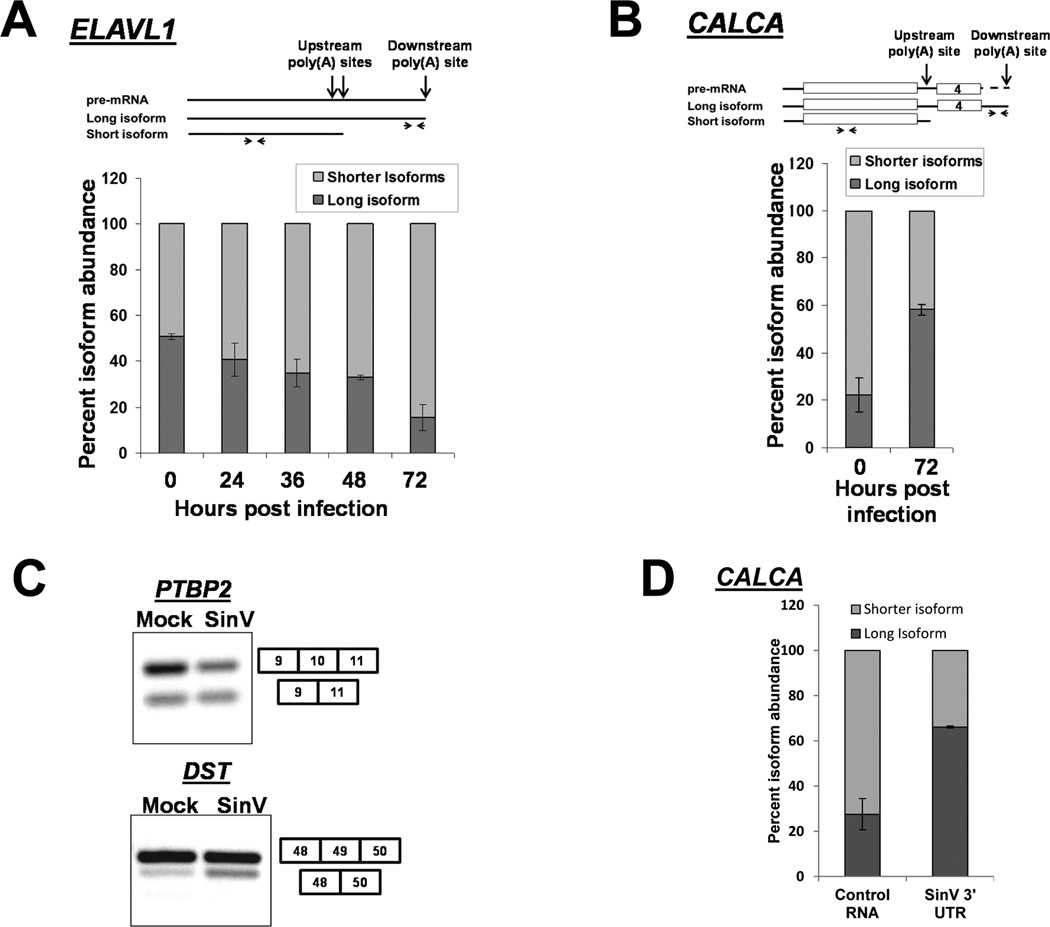
In addition to its well-studied role in mRNA stability, the cellular HuR protein also influences mRNA processing events in the nucleus. Through competition for binding sites on pre-mRNAs, HuR has been described to affect both alternative polyadenylation and alternative splicing (Zhu et al., 2007; Lebedeva et al, 2011). Interestingly, HuR also autoregulates the polyadenylation site choice of its own mRNA in certain cell/tissue types. The differential use of polyadenylation sites changes the length of the 3’ UTR of the HuR mRNA which influences the stability/translatability of the mRNA isoform that is produced (Dai et al., 2012). Thus we hypothesized that SinV may influence nuclear mRNA processing events via the sequestration of HuR in the cytoplasm. In order to test this hypothesis, we assessed alternative polyadenylation or splicing for a number of mRNAs known to be influenced by HuR protein. As seen in Fig. 4A, the HuR mRNA is normally present in a ~50/50 ratio of a long (6 Kb) isoform that uses the downstream poly(A) site versus a set of shorter (1.5–2.7 Kb) isoforms that use an upstream poly(A) site. There are HuR binding sites in the vicinity of the upstream poly(A) site that regulate its usage (Dai et al., 2012). Interestingly during a SinV infection, the relative usage of the upstream poly(A) sites on the HuR pre-mRNA increases dramatically. By 72 hpi, over 80% of the HuR transcripts that are made represent the more translatable forms that utilize the upstream poly(A) site. A similar alteration in the regulation of alternative polyadenylation in the calcitonin mRNA during SinV infection was also observed (Fig. 4B). In this case, calcitonin premRNA polyadenylation site usage is normally influenced by HuR regulation of alternative splicing of exon 4 which determines the inclusion of the upstream poly(A) site in the transcript (Lebedeva et al, 2011). As seen in Fig. 4B, SinV infection promotes the accumulation of the longer isoform that is processed at the downstream poly(A) site – likely by sequestering HuR protein and preventing its function in the combinatorial regulation that promotes usage of the upstream polyadenylation site. Finally, HuR is also known to be a regulator of alternative splicing in the mammalian cell nucleus. As seen in Fig. 4C, SinV infection caused significant increases in the splicing of exon 10 of PCBP2 (1.28 +/− 0.09 fold; p<0.05) and exon 49 of DST (2.1 +/− 0.46 fold; p<0.05), both of which are known to be regulated by HuR interactions (Lebedeva et al, 2011). Notably, the effect of SinV infection on nuclear RNA processing could also be observed in transfections of RNA containing the SinV 3’ UTR. As seen in Fig. 4D, transfection of RNAs containing the SinV 3’ UTR caused a ~3-fold difference in the relative abundance of isoforms of the calcitonin mRNA compared to cells transfected with a control RNA. Therefore we conclude that SinV infection can influence regulated pre-mRNA processing events in the nucleus, presumably through the sequestration of HuR protein in the cytoplasm. Given the importance of post-transcriptional RNA processing in determining the breadth of the cellular proteome, these SinV-induced alterations can have a dramatic effect on cellular biology. Furthermore, to our knowledge this is the first time that a cytoplasmic positive-sense RNA virus has been shown to selectively influence alternative mRNA processing events in the nucleus.
Figure 4. SinV infection influences alternative polyadenylation and splicing of cellular pre-mRNAs.
293T cells were infected with SinV for the indicated times. Short or long isoforms of the ELAVL1 (HuR) mRNA formed by alternative polyadenylation (Panel A) or the indicated isoforms of CALCA (calcitonin) were quantified by qRT-PCR using the primers illustrated at the top of the panels. Note that the level of ‘shorter isoforms’ was determined by subtraction of the amount of the longer isoform from the level of total RNA detected by the upstream primer. Quantification is shown with standard deviations calculated from three independent experiments. Panel C. Total RNA was isolated 24 hpi, probed for the presence of the splicing isoforms of the indicated genes by RT-PCR and analyzed on a 2% agarose gel. Panel D: 293T cells were transfected with equimolar amounts of either a control RNA (GemA60) or the SinV 3’UTR RNA. Total RNA was isolated at 6 hours post transfection and analyzed for CALCA isoforms as described in Panel B above. Quantification is shown with standard deviations calculated from two independent experiments.
Discussion
This study demonstrates that the 3’ UTR of SinV on its own is capable of causing the relocalization of the normally predominately nuclear HuR protein into the cytoplasm. This sequestration of HuR protein by viral RNAs has several dramatic effects of post-transcriptional regulation of cellular mRNAs. Cellular mRNAs that are normally stabilized by HuR protein have significantly reduced half lives in virus infected cells. In addition, changes can be observed in alternative polyadenylation and splicing patterns of pre-mRNAs normally regulated by nuclear HuR protein. These findings highlight novel aspects of virus-host interactions that are likely to be highly significant to viral pathogenesis and cytopathology.
Given the potentially large number of RNA binding proteins in a cell, an RNA must be expressed in large amounts to act as an effective sponge for a specific protein with biological consequences. Alphaviral genomic/subgenomic RNAs are obviously very highly expressed in infected cells. Furthermore, RNA sponges may interact with more than one molecule of an RNA binding protein (RBP) at a time. Along these lines, the SinV 3’ UTR contains multiple binding sites for HuR (Sokoloski et al., 2010) and the protein itself can oligomerize on RNA substrates (Benoit et al., 2010), thus increasing the potential effectiveness of such a sponge. The sponging phenomenon can also be observed when the SinV 3’ UTR is expressed as part of a GFP reporter mRNA construct from transfected plasmid DNA (Fig. 1B). Thus it is possible that the sponging of RBPs by overexpressed mRNAs in standard transfection experiments may need to be routinely considered when interpreting results (Westmark et al., 2006). Finally, this RNA sponge phenomenon may also need to be taken into account when using alphavirus vectors as a platform strategy for vaccine delivery (White et al., 2013).
In addition to HuR, alphavirus RNAs might also effectively sequester other RNA binding proteins in the cell during infection. The viral subgenomic RNA, for instance, is known to interact with hnRNP K (Burnham et al., 2007) and unknown cellular factors interact with the translational enhancer in the 5’ UTR (Patel et al., 2013). In addition to viral RNAs, viral non-structural proteins could also serve as de facto sponges for cellular factors. Pull-downs with tagged non-structural proteins have isolated a number of host factors (Atasheva et al 2007; Frolova et al 2006, Cristea et al 2006). The viral nsp3 protein interacts with G3BP, 14-3-3 proteins, YBX1, HSC70 and amphiphysin-1 and -2 (Cristea et al., 2006; Cristea et al., 2010; Neuvonen et al., 2011; Gorchakov et al., 2008). Finally, it may be important to consider that transcripts from other RNA viruses may also act as sponges of cellular RBPs. Thus it is tempting to speculate that the sequestration of cellular RNA binding proteins may be a very important aspect of virus-host interactions/viral pathogenesis that needs additional investigation.
The alteration of cellular RNA stability/post-transcriptional control through the sequestration of HuR protein by SinV may be a very significant factor in viral pathogenesis. HuR protein has been implicated in the regulation of a variety of cellular responses including stress, differentiation, apoptosis, cell cycle progression, immune responses and coordination of inflammatory reactions (Yiakouvaki et al., 2012; Srikantan and Gorospe, 2012; Khabar, 2010). HuR affects these processes through regulating mRNA stability, translation and through its interplay with miRNAs (Srikantan et al., 2012). Previous work has also shown that every alphavirus we have tested to date interacts with HuR protein through high affinity sites in its 3’ UTR, indicating that this is a highly conserved host interaction of this family of viruses (Dickson et al., 2012). We are currently performing global analyses to assess the full impact of SinV infection and HuR sequestration on cellular mRNA abundance, stability and translation. The studies reported here also highlight the potential value of HuR protein-viral RNA interactions as a therapeutic target to reduce alphavirus replication as well as perhaps pathological sequelae of infection.
To our knowledge, the data reported here are the first to document changes in alternative polyadenylation and splicing of cellular pre-mRNAs in a cytoplasmic RNA virus infection. Some changes in pre-mRNA processing may simply be an effect of HuR sequestration and not have a direct positive impact on viral infection. However, the SinV-induced changes in HuR pre-mRNA polyadenylation that we have observed favor the production of the shorter HuR isoforms that are inherently more translatable than the longer isoforms (Dai et al., 2012). Thus SinV is not only commandeering the available HuR protein in the cell, but also possibly dysregulating its synthesis to perhaps make more HuR protein for use in its own replication (Sokoloski et al., 2010).
In closing, this study highlights a novel set of effects of a cytoplasmic RNA virus on cellular post-transcriptional regulation of gene expression by viral RNAs acting to sequester a specific host RNA binding protein. Thus further studies of mechanisms and implications of the interference of RNA virus transcripts with the combinatorial regulation of host cell gene expression afforded by cellular RNA binding proteins should yield interesting insights into viral replication and cellular pathogenesis.
Experimental Procedures
Cells and Viruses
293T and BHK-21 cells were grown in 5% CO2 at 37°C and maintained in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum. Sindbis viral stocks were created by the electroporation of infectious viral RNA (transcribed from the clone pMRE16) into BHK-21 cells. All infections were performed at an MOI of 10.
DNA and RNA Transfections
The 3’ UTR of SinV was inserted between the BsrG1 and Not1 sites of pEGFP-n1 (Clontech). Capped and polyadenylated RNAs for the RNA transfections were generated by in vitro SP6 transcription of linearized pGEMA60 (Control) or pMREA60 (SinV 3’ UTR) or plasmids containing the 3xRSE and URE/CSE fragments of the SinV 3’ UTR (Garneau et al, 2008) as previously described (Wilusz and Shenk, 1988). Transfections of plasmids or equimolar amounts of purified RNAs were done using Opti-MEM® I Reduced Serum Medium and Lipofectamine® 2000 or TransIT mRNA transfection reagent (Mirus). Cells were incubated for 6 hours for RNA transfections or 72 hours for plasmid transfections prior to analysis.
Immunofluorescence Assays
293T cells grown on glass coverslips were either mock-infected or infected with SinV. At 24 hpi, the coverslips were rinsed with PBS and fixed with 4% paraformaldehyde in PBS. Coverslips were then sequentially treated with methanol and ethanol prior to blocking with a 6% solution of bovine serum albumin in PBS. HuR primary antibody (Santa Cruz 3A2) was added followed by anti-mouse Alexa Fluor® 594 as secondary antibody. Prolong® Gold with DAPI was then added and coverslips were cured overnight in the dark. Samples were analyzed via fluorescence microscopy using an Olympus IX71 inverted microscope equipped with a Q imaging retiga 2000R digital camera.
Immunoprecipitation and Western Blotting
At 24 hpi, mock-infected or SinV-infected 293T cells were washed with PBS and protein- RNA interactions were stabilized by adding 1% formaldehyde in PBS. The cross-linking reaction was quenched by the addition of glycine and cells were lysed by sonication after suspension in RIPA buffer (50mM Tris-Cl pH 7.5 / 1% v/v NP40 / 0.5% w/v sodium deoxycholate / 0.05% w/v sodium dodecyl sulfate (SDS) / 1mM EDTA / 150mM NaCl) with 1U of RNase inhibitor. Clarified lysates were incubated with anti-HuR (Santa Cruz 3A2) or normal mouse IgG (Santa Cruz sc-2025) and 1U of RNase inhibitor at 4°C. Complexes were bound to Protein G Sepharose beads and washed extensively with RIPA buffer containing 1M urea. Precipitated complexes were resuspended in TEDS Buffer (50mM Tris-Cl pH 7.0 / 5mM EDTA / 1% v/v SDS) and cross-links were reversed by incubation at 70°C. Co-precipitated RNAs were extracted using TRIzol and analyzed by PCR using the primers indicated in Table S1.
Western blots were performed using a Trans-Blot Semi-Dry Transfer Cell and nitrocellulose membranes. Primary antibodies used were Tubulin (Sigma T9026), XRN2 (NB 100-57541) and HuR (Santa Cruz Biotechnologies 3A2). Secondary antibodies were conjugated to horseradish peroxidase. Proteins were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and imaged with VersaDoc / Image Lab software (Bio-Rad).
Biochemical Subcellular Fractionation
293T cells were rinsed with 1× PBS, swollen in EBKL Buffer (25mM HEPES pH7.6 / 5mM MgCl2 / 1.5mM KCl / 2mM DTT / 0.1% v/v NP-40) for 15 min. and the cytoplasmic membrane was selectively lysed using a dounce homogenizer. Nuclei and cytoplasm were separated via centrifugation. The nuclear pellet was washed with EMBK Buffer (25mM HEPES pH 7.6 / 5mM MgCl2 / 1.5mM KCl / 75mM NaCl / 175mM sucrose / 2mM DTT), resuspended in 0.5% NP-40, and genomic DNA was sheared via brief sonication.
mRNA Half-life Analysis
293T cells were either mock-infected or infected with SinV. 24 hours post infection, actinomycin D (5 µg/µL) was added. For mRNA half-life analysis in RNA transfected cells, actinomycin D was added at 4.5 hours post transfection. Following a 30 minute incubation to ensure transcriptional shut off, samples were collected at the indicated times using TRIzol. Total RNA from these samples was analyzed via qRT-PCR using the standardized primers indicated in Table S1. Relative transcript abundances were determined using the ΔΔCt method and GAPDH as a reference. The average half-life of each transcript from three independent experiments is reported +/− the standard deviation.
Analysis of Alternative Splicing/Polyadenylation
Alternative splicing of PTBP2, ZNF207 and DST was assessed as described in Lebedeva et al., (2011) and RT-PCR products were separated on 2% agarose gels. Products were visualized using ethidium bromide staining. Isoform abundances of HuR and calcitonin were assessed using primers upstream and downstream of the predicted polyadenylation sites and qRT-PCR.
Supplementary Material
Highlights.
The 3’ UTR of Sindbis virus RNAs act as a sponge for the cellular HuR protein
HuR-mediated cellular mRNA stability is dysregulated upon Sindbis virus infection
HuR regulated alternative splicing and polyadenylation is altered by Sindbis virus
Sequestration of proteins by viral RNAs can disrupt post-transcriptional control
Acknowledgements
We thank Aaron Phillips and Ken Olson (Colorado State University) for supplying us with the MRE16 clone of SinV and members of the Wilusz laboratories for helpful discussions and suggestions. These studies were supported by NIH grant AI063434 and an NIAID award through the Rocky Mountain Regional Center of Excellence (U54 AI-065357) to J.W. MDB received support through the United States Air Force AFIT/CIP Program, SLM received support from a USDA NIFA NNF training grant (2010-38420-20367), and AWE received support from an ASM Undergraduate Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 2007;81:5046–5057. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit RM, Meisner NC, Kallen J, Graff P, Hemmig R, Cèbe R, Ostermeier C, Widmer H, Auer M. The x-ray crystal structure of the first RNA recognition motif and site-directed mutagenesis suggest a possible HuR redox sensing mechanism. J. Mol. Biol. 2010;397:1231–1244. doi: 10.1016/j.jmb.2010.02.043. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A, Ghisolfi L, Franzi S, Maresca G, Gherzi R, Capaccioli S, Nicolin A, Canti G. Stabilization of cellular mRNAs and up-regulation of proteins by oligoribonucleotides homologous to the Bcl2 adenine-uridine rich element motif. Mol. Pharmacol. 2007;71:531–538. doi: 10.1124/mol.106.029041. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Gallani AI, Sokol L, Baskin DS, Meisner-Kober N. mRNA stability alterations mediated by HuR are necessary to sustain the fast growth of glioma cells. J. Neurooncol. 2012;106:531–542. doi: 10.1007/s11060-011-0707-1. [DOI] [PubMed] [Google Scholar]
- Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 2007. 2007;367:212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. Tracking and elucidating alphavirus-host protein interactions. J. Biol, Chem. 2006;281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, Rout MP, Chait BT, MacDonald MR. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 2010;84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Zhang G, Makeyev EV. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2012;40:787–800. doi: 10.1093/nar/gkr783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson AM, Anderson JR, Barnhart MD, Sokoloski KJ, Oko L, Opyrchal M, Galanis E, Wilusz CJ, Morrison TE, Wilusz J. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J. Biol. Chem. 2012;287:36229–36238. doi: 10.1074/jbc.M112.371203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R. [Google Scholar]
- Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. The 3' untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and Mammalian cells. J. Virol. 2008;82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R, Garmashova N, Frolova E, Frolov I. Different types of nsP3- containing protein complexes in Sindbis virus-infected cells. J. Virol. 2008;82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell. Mol. Life Sci. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell. Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Kong J, Sumaroka M, Eastmond DL, Liebhaber SA. Shared stabilization functions of pyrimidine-rich determinants in the erythroid 15-lipoxygenase and alpha-globin mRNAs. Mol. Cell. Biol. 2006;26:5603–5614. doi: 10.1128/MCB.01845-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein. HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3'-untranslated region of its mRNA. J. Biol. Chem. 2007;282:7950–6790. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U.S.A. 2012;109:279–284. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog. 2011;7:e1002383. doi: 10.1371/journal.ppat.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Burnham AJ, Gebhart NN, Sokoloski KJ, Hardy RW. Role for subgenomic mRNA in host translation inhibition during Sindbis virus infection of mammalian cells. Virology. 2013;441:171–181. doi: 10.1016/j.virol.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe. 2010;8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- Srikantan S, Gorospe M. HuR function in disease. Front. Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip. Rev. RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- Wang Y-F, Sawicki SG, Sawicki DL. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 1991;65:985–988. doi: 10.1128/jvi.65.2.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Westmark PR, Shin HC, Westmark CJ, Soltaninassab SR, Reinke EK, Malter JS. Decoy mRNAs reduce beta-amyloid precursor protein mRNA in neuronal cells. Neurobiol. Aging. 2006;27:787–796. doi: 10.1016/j.neurobiolaging.2006.03.003. [DOI] [PubMed] [Google Scholar]
- White LJ, Sariol CA, Mattocks MD, Wahala MPBW, Yingsiwaphat V, Collier ML, Whitley J, Mikkelsen R, Rodriguez IV, Martinez MI, de Silva A, Johnston RE. An alphavirus vector-based tetravalent dengue vaccine induces a rapid and protective immune response in macaques that differs qualitatively from immunity induced by live virus infection. J. Virol. 2013;87:3409–3424. doi: 10.1128/JVI.02298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J, Shenk T. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, Kontoyiannis DL. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. J. Clin, Invest. 2012;122:48–61. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou HL, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.