Summary
Chromosomal integrity has been known for many years to affect the ability of mouse embryonic stem cells (mESCs) to contribute to the germline of chimeric mice. Abnormal chromosomes are generally detected by standard cytogenetic karyotyping. However, this method is expensive, time consuming, and often omitted prior to blastocyst injection, consequently reducing the frequency of mESC-derived offspring. Here, we show a fast, accurate, and inexpensive screen for identifying the two most common aneuploidies (Trisomy 8 and loss of chromosome Y) in genetically manipulated mESCs using quantitative real-time PCR (qPCR). Screening against these two aneuploidies significantly increases the fraction of normal mESC clones. Our method is extremely sensitive and can detect as low as 10% aneuploidy among a large population of mESCs. It greatly expedites the generation of mutant mice and provides a quick tool for assessing the aneuploidy percentages of any mESC line.
Highlights
-
•
Fast aneuploidy detection of mESCs using quantitative real-time PCR
-
•
Simultaneous processing of multiple cell lines
-
•
Highly sensitive: identifies low percentage of aneuploidy within an ESC clone
-
•
Method can detect loss or gain of any chromosomal region of interest
D’Hulst et al. show a fast, sensitive, and inexpensive screen for identifying the two most common aneuploidies (Trisomy 8 and loss of chromosome Y) in genetically manipulated mouse embryonic stem cells (mESCs) using qPCR, replacing expensive standard karyotyping procedures. Presence of aneuploidy as low as 10% can be detected. Screening against these two aneuploidies significantly increases germline transmission rates for mESCs. This method greatly expedites the generation of mutant mice.
Introduction
Techniques for manipulating genes in the germline of experimental mammals provide a powerful means to study the pathophysiology of human disease (Misra and Duncan, 2002). Genes can be altered or introduced by the genetic manipulation of embryonic stem cells (ESCs). ESCs, derived from the inner cell mass of blastocysts, can contribute to all embryonic tissues, including the germ cells of developing mice. “Gene targeting” of a specific genetic locus in ESCs has been a staple in the field of genetic manipulation for the last 20 years. Mutations are introduced through homologous recombination by endogenous mechanisms, by TALENs, or by CRISPR/Cas9 (Capecchi, 2005; Mussolino and Cathomen, 2012; Wang et al., 2013; Zhang et al., 2011). Genetically manipulated ESCs are subsequently injected into wild-type blastocysts, which are then surgically transferred to a pseudopregnant surrogate mother to generate chimeric mice that are able to transmit the mutant genetic locus to their progeny. Recent advances allow genetically modified ESCs to be injected immediately into eight-cell embryos, and, when F0 mice are produced, 100% germline transmission efficiency ensues. Either procedure requires that ESCs follow the correct developmental program and contribute to the germline (Liu et al., 1997). However, ESCs often have a low efficiency of germline transmission. It has been shown that mouse ESC (mESC) lines carrying three copies of chromosome 8 (chr8), i.e., Trisomy 8, have a higher growth rate and a diminished efficiency of contribution to the germline as observed in 19/29 karyotyped ESCs (Liu et al., 1997). A comprehensive analysis of mESC clones in Japan revealed only 66% of 540 mESC clones contained normal chromosome counts. When a subset of 88 mESC clones was further analyzed, 40% (35 mESCs) of them contained chromosomal aneuploidy with 34/35 having aneuploidy containing either chr8 and/or chrY (Sugawara et al., 2006). A recent study, in which 97 mESC lines were analyzed using conventional cytogenetic analysis and fluorescent in situ hybridization (FISH), showed that chromosomal aneuploidy occurs in 32% (31) of the specimens with 30/31 containing either chr8 and/or chrY (Kim et al., 2013). Aneuploidy for all other chromosomes was almost always associated with chr8 and/or chrY (83/85 mESC clones). Only in two cases out of 214 (29+88+97) total analyzed mESC clones, a trisomy was identified (chr11) that did not contain either chr8 and/or chrY. In sum, removal of 39% (83/214) of potentially detrimental mESC clones by karyotype analysis is crucial before injecting modified mESCs into blastocysts.
Unfortunately, existing cytogenetic karyotyping methods are labor intensive, requiring at least 15 fixed metaphase preparations, subsequent DAPI staining, and analysis with an epi-fluorescence microscope equipped with specialized digital imaging software. Conventional karyotyping is expensive (∼$400/clone) and often needs to be outsourced to specialized core facilities. Therefore, we designed a simple, fast, and cost-effective (∼$1/clone) SYBR-Green-I-based qPCR procedure to identify common chromosomal aneuploidies found in genetically manipulated mESCs. In contrast to regular PCR, qPCR combines PCR amplification and detection into a single step using detection chemistries such as target-specific hydrolysis probes and SYBR Green I dye, which binds double-stranded DNA and emits fluorescence only when bound. To date, qPCR is the benchmark for gene expression analysis, but recent developments also encourage the use of qPCR for copy number determination (D’haene et al., 2010b). In this regard, qPCR is currently used for diagnostic copy number profiling of the SHOX gene region in idiopathic short stature (ISS) and allied disorders (D’haene et al., 2010a). In addition, it has been shown to be an elegant tool to accurately screen for correctly targeted mESC clones (using the so-called loss-of-allele assay or LOA) (Frendewey et al., 2010) and for genotyping of gene-targeted and transgenic mice (Haurogné et al., 2007; Sakurai et al., 2008). These studies show that qPCR can be applied to discriminate 2-fold differences in the copy number of a specific transgene, allowing discrimination between homozygous and hemizygous transgenic animals. We have applied and optimized this method to discriminate among chromosomal aneuploidies, ranging from loss of a chromosome to extra copies of a particular chromosome of interest.
The accumulation of fluorescent signal during the exponential phase of a PCR results in a fast, precise quantification of PCR products and allows for objective data analysis. The quantification cycle or Cq value represents the fractional PCR cycle that is characteristic for the amplification curve (e.g., where increase in fluorescence is maximum) or at which the fluorescence crosses a certain threshold (D’haene et al., 2010b). Cq value and initial amount of input DNA are inversely related: a sample that contains more copies of template will cross the threshold at an earlier cycle compared to one containing fewer copies of template (Schmittgen et al., 2000). Consequently, the theoretical difference in Cq (dCq) between two and three copies of an autosome is about a half cycle, whereas the dCq between one and two copies is one cycle. With 53 mESC lines (all male), we were able to validate the sensitivity and accuracy of our approach in great detail, which resulted in a quick and user-friendly paradigm to test the quality of any genetically manipulated mESC clone in less than 1 day. We confirmed our approach with the two most common chromosome aneuploidies found in mESCs: chr8 and chrY, and we further validated the approach with rare chr11 and partial chr1 aneuploidies among our collection of cytogenetically karyotyped clones.
Results
Extensive Karyotyping of 141 ESC Clones
A large karyotyping project in our lab revealed that out of 141 mESC lines analyzed with conventional DAPI-band karyotyping, 48.2% of the clones (68 total) were “normal” and do not contain chromosomal breakage or low mitotic index; 51.8% of the clones (73 in total) had chromosomal abnormalities including Trisomy 8 (24.8%), loss of chrY (9.2%), both Trisomy 8 and 11 (2.1%), both Trisomy 8 and an extra copy of chrY (0.7%), and other structural abnormalities (14.2%). Overall, our data revealed that 38% (53/141) of mESC clones contained either chr8 and/or chrY aneuploidy (Table S1 available online) consistent with data reported in the literature (Kim et al., 2013; Liu et al., 1997; Sugawara et al., 2006). Only 1/141 mESC clones (ESC 8) contained a trisomy that did not also contain chr8 and/or chrY (31% of ESC 8 cells contained an extra chr1). The overall rate derived from the three published studies and our study suggests that non-chr8 and/or non-chrY aneuploidies appear at an equal or lower rate than 3/355 mESC clones (0.84%). Two anomalies that cannot be identified by any method other than cytogenetic karyotyping (chromosomal breakage and low mitotic index) occurred in 4/141 ESC clones (2.8%). Thus, chr8 and chrY aneuploidies are by far the most common cytogenetic anomalies identified.
Quantitative Real-Time PCR Using SYBR Green I Dye
We set up a qPCR approach to identify the most common aneuploidies (chr8 and chrY) and the rare aneuploidies (chr1 and chr11) using two chromosomes without aneuploidy (chr7 and chrX), which serve as our references. DNA isolated from 53 mESC lines is utilized in the qPCR validation experiments (Table 1). We use the SYBR Green I dye as the fluorogenic marker to keep the method simple and easily applicable for any lab. Hydrolysis probes, at considerable costs, may also function; however, our approach requires only one well-designed and characterized primer set per target chromosome and no sequence-specific probe, significantly reducing assay setup. For the qPCR, we designed primers that specifically generate exonic amplicons located on mouse chr1 (olfr16), chr7 (omp), chr8 (olfr370), chr11 (olfr1), chrX (obp1a), and chrY (sry) and extensively validated them in silico and empirically according to the latest Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009) (Table 2). Intronic region amplicons, with reduced genomic complexity, gave inconsistent results (data not shown) and were avoided. To increase PCR efficiency and accessibility of the genomic DNA, we digested all mESC DNA with restriction endonucleases before qPCR screening, reducing the genomic DNA (gDNA) viscosity. All samples were run in triplicate, and the average Cq value per sample was used in the calculations. The absolute average of the difference between the triplicate Cq values of all our samples was 0.0732 ± 0.0737. If any absolute Cq difference (|dCq|) was greater than 0.2206 (i.e., 0.0732 + 2 SDs) between triplicates, then this PCR was repeated.
Table 1.
Cytogenetic Karyotypes of Mouse ESC Lines Used for qPCR
| Cell Line | ISCN | Cell Line | ISCN |
|---|---|---|---|
| Normal | Trisomy 8 | ||
| ESC 3 | 40,XY [15] | ESC 1 | 41,XY,+8 [15] |
| ESC 5 | 40,XY [14] | ESC 2 | 41,XY,+8 [15] |
| ESC 19a | 40,XY [15] | ESC 4 | 41,XY,+8 [15] |
| ESC 24 | 40,XY [14] | ESC 7 | 41,XY,+8 [15] |
| ESC 25 | 40,XY [14] | ESC 9 | 41,XY,+8 [15] |
| ESC 31a | 40,XY [12] | ESC 20 | 41,XY,+8 [14] |
| ESC 32 | 40,XY [13] | ESC 27 | 41,XY,+8 [13] |
| ESC 33 | 40,XY [15] | ESC 28 | 41,XY,+8 [14] |
| ESC 52a | 40,XY [14] | ESC 29 | 41,XY,+8 [15] |
| ESC 53a | 40,XY [15] | ESC 30 | 41,XY,+8 [15] |
| ESC 54a | 40,XY [15] | ESC 51 | 41,XY,+8 [13] |
| 31% Trisomy 1 | Trisomy 8 and Extra chrY | ||
| ESC8b | See Table S1 | ESC12 | 42,XYY,+8 [15] |
| Loss Of chrY | Trisomy 8 and 11 | ||
| ESC 10 | 39,X,-Y [15] | ESC 13 | 42,XY,+8,+11 [14] |
| ESC 14 | 39,X,-Y [15] | ESC 16 | 42,XY,+8,+11 [13] |
| ESC 15 | 39,X,-Y [14] | ESC 18 | 42,XY,+8,+11 [15] |
| ESC 17 | 38∼39,X,-Y [16] | ||
| ESC 21 | 39,X,-Y [15] | ||
| ESC 22 | 39,X,-Y [15] | ||
| ESC 23 | 39,X,-Y [14] | ||
The following lines had unknown cytogenetic karyotypes and were used to validate our qPCR approach: ESC 6, ESC 11, and ESC 34–ESC 50. Karyotypes of lines ESC 40, ESC 45, and ESC 48 were confirmed by DAPI-banded karyotyping after obtaining the qPCR results. Karyotypes were the following: ESC 40, 37∼40,XY[20]; ESC 45, 38∼39,X,-Y[15]/40,XY[1]; ESC48, 39∼40,XY[15]. See also Table S1.
These cell lines produced germline animals.
ESC8 is of mixed genetic background and was derived in the lab.
Table 2.
In Silico and Empirical Evaluation of the qPCR Primer Setsa
| Chr | Location | Gene | Primers (5′-3′) | Amplicon Length (bp) | RTprimerDB ID | Standard Curve |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Slope | E (%) | r2 | Linear Dynamic Range (ng) | LOD (ng) | ||||||
| 1 | 172956826–172956929 | olfr16 | GAGTTCGTCTTCCTGGGATTC | 103 | 8651 | −2.904 | 120 | 0.997 | 0.84–13.5 | 0.09b |
| TAATGATGTTGCCAGCCAGA | ||||||||||
| 7 | 98145342–98145482 | omp | GCCCACTTGATTCCCTGA | 140 | 8652 | −2.907 | 120 | 0.997 | 0.84–13.5 | 0.07 |
| GCATCTGCTGGGTCAGGTCC | ||||||||||
| 8 | 83541895–83542057 | olfr370 | CACTATGGATGTGCCTCTTTTATCT | 162 | 8653 | −3.137 | 108 | 0.999 | 0.84–13.5 | 0.08 |
| TACCCTATGAAGAGCTGATTTGAAG | ||||||||||
| 11 | 73395777–73395941 | olfr1 | TCTATGCCCTGTTCCTGGTC | 164 | 8654 | −3.439 | 95 | 0.996 | 0.84–13.5 | 0.02 |
| CAACTTGGGCATTGTGACAG | ||||||||||
| X | 78085543–78085693 | obp1a | GGATCAGAATTATGGATCATGTG | 150 | 8655 | −2.994 | 115 | 0.994 | 0.84–13.5 | 0.15 |
| GATCATGAGAAGGGGAAGGA | ||||||||||
| Y | 2663266–2663432 | sry | CTCATCGGAGGGCTAAAGTG | 166 | 8656 | −3.235 | 103 | 0.985 | 0.84–13.50 | 0.28c |
| AAGCTTTGCTGGTTTTTGGA | ||||||||||
See also Figure S2.
MIQE guidelines compliant.
A LOD of 0.09 ng refers to about 150 cells when screening for autosomes.
A LOD of 0.28 ng refers to about 1,000 cells when screening for sex chromosomes (XO or XY).
Data Analysis Using the 2−ddCq Method
Our assay uses relative quantification to calculate the target chromosome copy number by the so-called comparative Cq method or 2−ddCq method (Pfaffl, 2001; Schmittgen et al., 2000). This method compares the Cq value of one target gene to another internal control or reference gene from a single sample. We chose genes on chr7 (omp) and chrX (obp1a) as reference genes because no aneuploidy was detected for either in any of our 141 karyotyped male mESCs. For the comparative Cq method to be valid, the amplification efficiency of the target and the reference genes must be approximately equal. The amplification efficiencies of each of the primer sets was determined based upon the generation of standard curves using gDNA dilution series and for all primer sets analyzed the PCR efficiencies ranged between 95% and 120% (Figure S2). For the initial calibration of our data, we used three calibrator mESC lines with previously determined, normal cytogenetic karyotypes, which produced germline animals (ESC 31, 52, and 53). We had numerous germline animals produced from mESCs and chose three to calculate an average normalized relative quantity (NRQ). Theoretically, only one calibrator is required for accurate analysis of the data. In the case that no such lines are readily available in the lab, mESCs with a normal cytogenetic karyotype are commercially available (http://www.atcc.org/). After initial screening of mESCs using this qPCR method, additional calibrators can be identified and added to future analyses.
The final NRQ value is presented as the fold change in gene copy number normalized to an endogenous reference gene (omp on chr7 or obp1A on chrX) and relative to one or more samples of known karyotype, the calibrators (Sakurai et al., 2008). For example, a cell line with a normal karyotype has a final NRQ that is approximately 1 (including our calibrator), whereas loss of chrY-cells results in a NRQ of approximately 0 (for the sry gene) and Trisomy 8 or 11 cells show a NRQ of 1.5 (for olfr370 or olfr1, respectively). We tested our method by calculating the NRQ value for chr8 (olfr370), chr11 (olfr1), and chrY (sry) in 34 ESC lines with known cytogenetic karyotypes and normalized the results against the NRQ value for our reference genes chr7 (omp) and chrX (obp1a), see Experimental Procedures. To obtain the absolute copy number for each of the chromosomes of interest, the following formula (Frendewey et al., 2010) was applied (Figures 1, S1A, and S1B):
Figure 1.
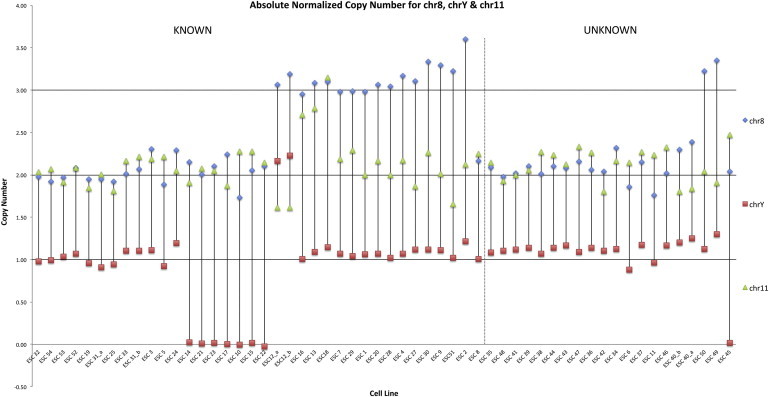
Normalized Chromosome Copy Numbers
The absolute copy number for the autosomes is calculated by multiplying the NRQ values by 2 according to this formula: Copy number = calibrator copy number X NRQ or (2−ddCq). A copy number of 2 reflects normal autosome ploidy, and a copy number of 3 reflects trisomy. The absolute copy number of chromosome Y equals the NRQ value. A copy number of 1 reflects normal ploidy, a copy number of 0 reflects loss of chrY, and a copy number of 2 reflects duplication of chrY. A dotted line marks the border between mESCs with previously determined (known) and unknown karyotypes at the time of qPCR analysis. See also Figure S1.
The Interchromosomal SD
In order for this method to be readily applicable in any lab, we defined a standardized confidence level to assess aneuploidy of any given mESC clone. This cutoff level is mathematically imposed by the relative differences between Cq values, not likely to vary between labs or instruments. A sample is considered to be “normal” (normal chromosomal count) when the NRQ values for chr8, chr11, and chrY are all approximately 1. If any value becomes significantly greater or smaller than 1, then aneuploidy is present. We further defined this divergence away from 1 by determining the likelihood that an mESC clone had significant NRQ values greater or less than 1. An interchromosomal SD (IC SD) was derived by determining the SD between the NRQs of each amplicon for the different chromosomes within each ESC clone (with the NRQ for our reference chromosomes 7 and X being “1”). To identify an acceptable IC SD, we used 11 “normal” mESC lines to calculate a 95% confidence level (or cutoff line) for any mESC clone to be defined as normal. This cutoff line was derived by averaging the IC SD for the 11 normal ESC lines and adding twice its SD. This [AVG IC SD + 2 × SD (IC SD)] = 0.0455 + 2 × 0.0226, which equals 0.0906, can be used to assess all mESCs (Figure 2). We confirmed all previously identified chr8, chr11, and chrY aneuploidies using our screening method (Figure 1, left of dotted line) and validated the anomalous IC SD (Figure 2, above dotted line) of their respective ESC clones. A more stringent cutoff would be the [AVG IC SD + 1 × SD (IC SD)], which equals 0.0681. All mESC clones that generated germline animals (ESC 19, 31, 52, 53, and 54) show IC SD below this stringent cutoff (Figure 2, green bars).
Figure 2.
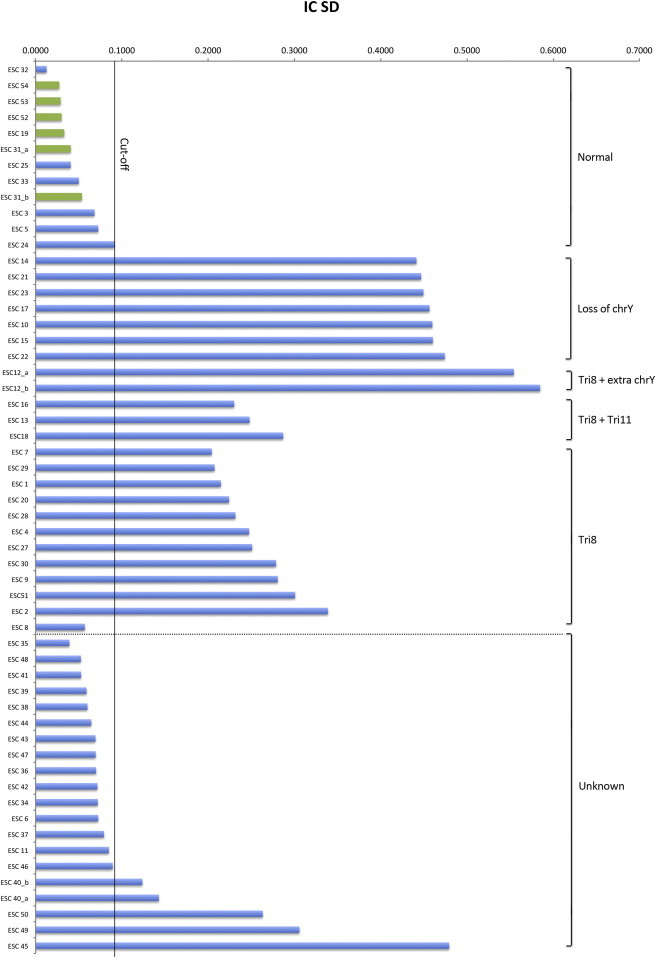
Interchromosomal SDs
The IC SD represents the interchromosomal SD of NRQ values between all chromosomes within an mESC line. The confidence level (CL) of 0.0906 was calculated using 11 known, normal mESC lines and can be used as a standardized cutoff line (shown in black) to assess karyotypes of any cell lines. mESC lines that produced germline animals are highlighted in green, and all show IC SD values even below our stringent cutoff of 0.0681. The IC STDEV refers to the interchromosomal SD or IC SD.
We further validated this method using 19 additional mESC lines with unknown cytogenetic karyotypes. We identified 15 normal mESC lines, one loss of chrY (ESC 45) and two Trisomy 8 (ESC 49, 50) (Figure 1, right of dotted line, and Figure 2, below dotted line). We confirmed the cytogenetic karyotypes of ESC 48 (normal) and ESC 45 (loss of chrY) using conventional DAPI banding (Table 1). ESC 40 consistently yielded IC SDs >0.0906 (Figure 2) and subsequent cytogenetic karyotyping revealed chromosomal aberrations in 20% of the cells (Table S1).
Detection of Low-Percentage Chromosomal Imbalances
To determine the limits of the aneuploidy detection by our method, we created mixtures between the normal line ESC 31 and the robust Trisomy 8 line ESC 9 or the loss of chrY line ESC 21. The NRQ values were determined for mixtures with 0%, 5%, 10%, 15%, 20%, 40%, 60%, 80%, and 100% loss of chrY or Trisomy 8 (Figures 3A and 3B). The IC SD values show detectable chromosomal imbalances for 10% mixtures and above for both chr8 and chrY (Figure 3C). Thus, our data suggest that 10% Trisomy 8 or loss of chrY is the limit of detection in our qPCR assay. However, using our most stringent IC SD cutoff, mESC clones with 5% Trisomy 8 would be discarded.
Figure 3.
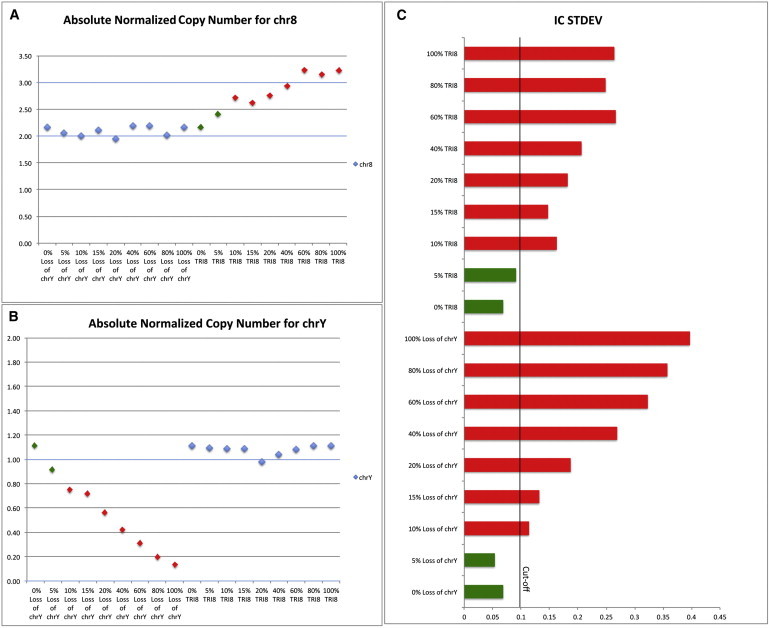
Detection of Low-Percentage Chromosomal Imbalances
Normalized chromosome copy numbers for percentage changes in chromosome 8 (A), chromosome Y (B), and corresponding IC SD values (C). To obtain a gradual decrease in loss of chrY or a gradual increase in chr8, we mixed appropriate volumes of a normal control and calibrator (ESC 31) with a 100% loss of chrY cell line (ESC 21) or a robust 100% Tri8 line (ESC 9), respectively. The IC SD values show detectable chromosomal imbalances for 10% mixtures and above for both chr8 and chrY, reflecting the sensitivity of our method. Blue squares indicate the internal controls, green squares (A and B) or bars (C) indicate dilutions below the limit of detection (and are considered “normal” according to our cutoff level), and red squares (A and B) or bars (C) indicate partial Trisomy 8 (A) or partial Loss of chrY (B) are considered “aneuploidy” according to our cutoff level. The IC STDEV refers to the interchromosomal SD or IC SD.
Discussion
We are able to rapidly identify the common Trisomy 8 and chrY aneuploidies as well as the rare Trisomy 11 and partial Trisomy 1 aneuploidies in mESCs. Conventional cytogenetic karyotyping requires direct inspection of chromosomal images through human visual interpretation. The SYBR Green qPCR-based assay is numerical with fixed confidence levels and not dependent on such subjectivity (Frendewey et al., 2010). Our method provides a standardized cutoff IC SD of 0.0906 by which all mESC lines can be determined as “normal.” This number might vary slightly, but will not exceed 0.1.
Using a set of 141 karyotyped gene-targeted mESC clones, we found that ∼50% (73 clones) contain abnormal cytogenetic karyotypes that might preclude them from usage in blastocyst injections and prevent the production of germline mutations. From this set of 73 clones, 73% (53) contained aneuploidies of which Trisomy 8 was the most frequent (39/53). The chromosome Y (chrY) aneuploidies (14/53, either loss or gain) were the only other consistent chromosomal aneuploidy identified in our study. ChrY aneuploidies generate (male) mESCs unable to contribute to the germline of male chimeric animals and thus progeny cannot be obtained. The germ cells of male chimeras are typically used to autoexcise a selectable marker cassette. Removal of the selectable marker in a mutation is mandatory because it interferes with gene expression of the surrounding genomic locus.
A cumulative analysis of cytogenetic karyotyping from four groups, including ours, reveals that out of 355 mESC clones only 2 (0.5%) contained Trisomy 11 without accompanying aneuploidies (Liu et al., 1997; Sugawara et al., 2006). In our study, Trisomy 11 only occurred in the presence of Trisomy 8 (ESC 13, 16, 18). Therefore, screening for Trisomy 11 aneuploidy is redundant. In addition, these findings are consistent with our single ESC clone containing 31% Trisomy 1 cells without accompanying aneuploidies (ESC 8). In order to identify this 31% Trisomy 1 frequency in ESC 8, we had to cytogenetically karyotype 54 cells (Table S1). To confirm these data, we screened for partial Trisomy 1 with primers to olfr16 (located on chr1, Table 2) and show a NRQ value of 1.4, which represents an absolute chr1 copy number of 2.8 (data not shown), similar to where the NRQ value would lay for a 30% Trisomy 8 (Figure 3A). Our method is straightforward, and the Trisomy 1 data show the ease of adding extra primer sets to the analysis if necessary. In addition, the IC SD for ESC 8 including the chr1 analysis was 0.1699, which is greater than our cutoff of 0.0906, and clearly identifies aneuploidy. Importantly, adding the NRQ values for chr1 to the IC SD calculation for the calibrators (ESC 31, ESC 52, and ESC 53) maintained their IC SD values below 0.05 (data not shown).
Using our screening, we were able to confirm the previously determined cytogenetic karyotypes of 34 mESC lines and to further validate our method using 19 additional mESC lines with unknown cytogenetic karyotypes. Our study includes the screening of five mESC lines that produced germline transmission (ESC 19, 31, 52, 53, and 54), all with an IC SD ranging from 0.0268 to 0.0401, well below our determined cutoff value. Consequently, we believe that the smaller an IC SD value, the greater the likelihood of the mESC line being passaged through the germline. Using mixtures of a normal mESC line with mESC lines containing Trisomy 8 or loss of chrY, we show that our method is able to detect chromosomal imbalances as low as 10% aneuploidy. By contrast, standard cytogenetic karyotyping requires analysis of 29–31 cells to detect 10% aneuploidy or 59–73 cells to detect 5% aneuploidy with a 95% confidence level (Hook, 1977); far fewer (15–20 cells) are usually analyzed by a standard cytogenetic core facility. In this regard, ESC 24 shows a borderline IC SD of 0.0912, yet the line gave a normal cytogenetic karyotype when 14 cells were analyzed (Table 1). Based on our method, this clone would be excluded from blastocyst injection because the elevated IC SD may reflect low-percentage aneuploidies.
Many laboratories are exploring reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) used for drug and disease modeling and directed differentiation experiments. It was previously shown that ∼75% of iPSC lines contain aberrant cytogenetic karyotypes (Boland et al., 2009; Chen et al., 2011). We have tested our method on one mouse iPSC line with results consistent to cytogenetic karyotyping (data not shown). It has not escaped our attention that this qPCR-based method will significantly reduce the number of iPSC lines requiring cytogenetic karyotyping, greatly reducing costs. In addition, it lays the groundwork for assessing the aneuploidy while passaging mouse iPSC lines, not usually done because of the costs involved in karyotyping numerous iPSC cell lines.
A similar qPCR-based method for comprehensive chromosomal aneuploidy screening of human blastocysts has recently been shown to improve the success of in vitro fertilization (Treff and Scott, 2013). Although these data demonstrate that the qPCR technology is capable of fast and accurate screening of all 24 human chromosomes, the technique is not readily applicable to mouse ESCs or iPSCs. In contrast, our approach is (1) simple: only one carefully designed primer set per chromosome is required, and all primers are available to the scientific community (RTPrimerDB IDs are shown in Table 2); (2) cost effective: SYBR Green I is used as the fluorescent dye instead of gene-specific hydrolysis probes, which would increase the cost of the screen tremendously; (3) standardized: we provide a cutoff level by which any clone can be assessed; and (4) sensitive: our technique is able to detect slight percentage differences in aneuploidies. Finally, our method has proved to be robust, as the three controls that were included were shown to be true: (1) interrun Cq differences for the same sample are smaller than the average |dCq| between triplicates, not requiring interrun calibration and again simplifying data analysis; (2) independent restriction endonuclease digests for the same DNA sample yield similar IC SD values (ESC 12a and b and ESC 31a and b, Figure 2); and (3) subsequent passages for a given ESC line yield similar NRQ and IC SD values (ESC 49 and 50 and ESC 7 and 9).
In conclusion, our method is fast and extremely inexpensive and will greatly expedite many researchers’ work in generating mESC germline transmission. Furthermore, it is of broad interest to every scientist using genetically modified mice because mESC clones that will produce germline animals can be readily identified.
Experimental Procedures
This study is written according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines (MIQE) to ensure its technical qualities and to allow for correct interpretation and repeatability (Bustin et al., 2009; D’haene et al., 2010a). A MIQE compliance checklist is available in the Supplemental Information (Table S2).
Cytogenetic Karyotyping of Mouse Embryonic Stem Cell Lines
Subconfluent cultures were treated with 0.05 μg/ml Colcemid (Karyomax, Invitrogen) for 30–40 min before harvesting according to standard cytogenetics procedures. Briefly, cells were trypsinized (0.025% Trypsin: EDTA, EMD Millipore) to a single-cell suspension, pelleted at 180 × g for 8 min, and resuspended in warm 0.075M KCl. After 8 min incubation at 37°C, approximately 1/4 volume of 3:1 methanol/glacial acetic acid fixative was added and gently mixed, and the cells were pelleted as before. The cells were then fixed in three changes of fixative. Fixed cell suspensions were stored at −20°C. Fixed metaphase preparations were dropped onto dry slides, and the quality of spreading was assessed by phase microscopy. Slides were then air-dried and aged (at 37°C for several days, or at 60°C for several hours). Aged slides were immersed in 0.08 μg/ml DAPI in 2 × saline sodium citrate for 3 min, rinsed, air-dried, and then mounted in antifade solution (Vectashield, Vector Laboratories) and stored at 4°C. DAPI-stained slides were scanned using a Nikon E800 epi-fluorescence microscope equipped with a digital imaging system (Applied Spectral Imaging). Metaphase images were inverted to resemble conventional G-banding and karyotyped using BandView software (ASI). Where possible, a minimum of 15 metaphases was examined for each sample. All metaphases were fully karyotyped. To avoid confusion, abnormal karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN), rather than the Standardized Genetic Nomenclature for Mice, because the experimental systems are intended to model human disease.
Tissue Culture and Genomic DNA Preparation
Gene-targeted mESC lines from 13 different electroporations (into E14 parental ESC line, 129ola) grown on a 4 cm2 growth area (Gene Targeting Facility, The Rockefeller University) were thawed and plated on 9.6 cm2 wells (6-well plate) onto mitotically inactivated mouse embryonic fibroblasts (MEFs) (Millipore) in a standard ESC medium (EmbryoMax Dulbecco’s Modified Eagle Medium [DMEM], High Glucose, Low Bicarbonate without sodium pyruvate [EMD Millipore], supplemented with 15% ESC-approved fetal bovine serum [FBS, Gibco], EmbryoMax Nucleosides [100×] 1%, EmbryoMax Penicillin-Streptomycin [100×] 1%, EmbryoMax Non-Essential Amino Acids [100×] 1%, EmbryoMax L-Glutamine Solution [100×] 1%, EmbryoMax 2-Mercaptoethanol (100×) 1%, and ESGRO mLIF Medium Supplement 1000, units/ml [all from EMD Millipore]). Cells were maintained at 37°C and 5% CO2 in a humidified incubator. At 90%–95% confluency, the cells were passaged into a 25 cm2 flask (T-25) on MEFs. At 90%–95% confluency, the cells were trypsinized and transferred into a new T-25 flask without MEFs. After 30 min, the medium containing the cells was gently removed and plated into a new flask without MEFs. The MEFs adhere to the flask in about 30 min, whereas the ESCs require longer time to attach and remain in the suspension. Thus, a significant number of MEFs can be removed by this differential adhesion step. The MEFs removal protocol was repeated on the following day to ensure a complete lack of MEFs, which is crucial to the accuracy of the qPCR karyotyping. 30 min posttrypsinization, the medium containing the ESCs was collected into a sterile 15 ml centrifuge tube and spun at 1000 rpm for 5 min. The supernatant was removed, and the pellet was resuspended in 6 ml of lysis buffer (100 ml protocol: 10 mM Tris-HCl [pH 7.5] = 0.5 ml of 2 M, 10 mM EDTA = 2 ml of 0.5 M, 10 mM NaCl = 0.2 ml of 5 M, 0.5% [w/v] Sarkosyl = 0.5 g N-lauroylsarcosine [Sigma #L-9150]) and 40 μl of proteinase K (200 mg/ml, Fisher Scientific) and incubated overnight at 55°C. The DNA was precipitated by adding 6 ml of isopropanol and gently inverting the tube four to five times. The DNA pellet was resuspended in 400 μl of Tris-EDTA buffer (Fisher Scientific), and the tube was air-dried at 42°C for 1–2 hr. A second 42°C closed-cap incubation was performed overnight. The gDNA concentrations of all samples ranged between 55.44 ng/μl (ESC 37) and 1,897.12 ng/μl (ESC 50) with a median of 316.79 ng/μl and an average 260/280 ratio of 1.90 ± 0.0474. All concentrations are listed in the Table S6. Genomic ESC DNA was digested with EcoRI (New England Biolabs) to remove the viscosity of the sample, which is critical for both the accessibility of the primers and the reproducibility of the Cq values. None of the amplicons contain EcoRI restriction sites. Restriction conditions were the following for a total volume of 100 μl:
-
1.
gDNA: 3 μl (∼1 μg)
-
2.
EcoRI buffer: 10 μl
-
3.
EcoRI: 2 μl
-
4.
PCR grade water: 85 μl
-
5.
Cut for 3 hr at 37°C (no heat inactivation)
SYBR Green I Real-time ESC genomic DNA PCR
Six PCR primer pairs were designed with Primer3 Software (http://bioinfo.ut.ee/primer3-0.4.0/), in silico validated by OligoAnalyzer 3.1 (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/), and synthesized by Integrated DNA Technologies with standard desalting purification. The BLAST program from the NCBI browser (http://blast.ncbi.nlm.nih.gov/) was used for in silico specificity analysis and primer sequences were submitted to the RTPrimerDB (http://medgen.ugent.be/rtprimerdb/). Primer IDs are shown in Table 2). The absence of secondary structures in the region in which the primers anneal was verified using MFOLD (http://mfold.rna.albany.edu/?q=mfold). The in silico validation was followed by an extensive empirical validation. The specificity of each amplicon was tested based upon melting curve analysis (single peaks) and gel electrophoresis (Figure S2B). Amplification efficiencies were calculated based upon the generation of standard curves using a 2-fold gDNA dilution series (Figure S2A). The linear dynamic range was five dilutions and standard curve slopes, PCR efficiencies, r2 values, linear dynamic range and limit of detection (LOD) for all amplicons analyzed is summarized in Table 2. The qPCR mixture (10 μl) contained 4.25 μl Lightcycler 480 SYBR Green I Master (Roche Diagnostics), 1.7 μl of each primer (at 10 pM), 0.85 μl of deionized H2O, and 1.5 μl of digested genomic ESC DNA (i.e., ±15 ng). qPCRs were set up manually using a Matrix Electronic Multichannel Pipette with 12.5 μl Impact 384 Tips (Thermo Scientific, cat#7421), and the reactions were carried out in white 384-well plates (E&K Scientific, cat# 486384) covered with ThermaSeal optical covers (Excel Scientific, cat# TSS-RTQ-100). All plates were spun down prior to covering to eliminate air bubbles. This step is also critical for the reproducibility of the Cq values. Reaction were run on a Roche Lightcycler 480 using the following cycling conditions: an initial denaturation step at 95°C for 5 min, followed by a 35 cycle amplification step of 95°C for 20 s, 60°C for 15 s, and 72°C for 15 s and a final cooling step at 40°C for 30 s. PCR products were detected in the presence of SYBR Green, and the Cq value for the no template controls was >30 for all primer sets.
Data analysis
Quantification cycle values were extracted using the Lightcycler 480 software (version 1.5.0 SP4) using the second derivative maximum algorithm. Further data analysis was carried out in Excel (see Tables S3, S4 and S5) on the exported data as follows:
To calculate the normalized relative quantities (NRQs or 2-ddCq values) for each sample within each gene of interest:
-
1.
Calculate the intertriplicate difference in Cq values (dCqtriplicates). Samples with |dCq|triplicates >0.2206 should be removed from the analysis and repeated.
-
2.
Calculate the average Cq value per sample per gene using the triplicate Cq values.
-
3.
Calculate the difference in Cq between the gene of interest (GOI) and the reference gene on chr7, i.e., dCq(GOI-chr7).
-
4.
Calculate for each gene the difference between the dCq (GOI-chr7) of each sample and the dCq (GOI-chr7) of the chosen calibrator (i.e., ddCq). In case of multiple calibrators, repeat the calculation for each calibrator separately.
-
5.
Calculate within each chromosome the 2−ddCq for each sample using the previously obtained ddCq values. This is the NRQ value.
-
6.
In case of using multiple calibrators, calculate the mean 2−ddCq value for each sample by averaging all NRQ values obtained using the different calibrators.
-
7.
This average NRQ value indicates the fold change in dosage of the GOI as compared to the “normal” calibrator(s).
-
8.
Calculate the normalization factor using the NRQ values from chrX (NFX). The NFX is obtained by subtracting the NRQ (X) from 1 for each sample within chrX. The data are normalized by adding the NFX to the NRQ of each sample within each gene of interest. This sum will yield a NRQ of 1 for all samples within chrX.
-
9.
Repeat steps 3–8 for the other endogenous control chrX and normalize the data using the chr7.
-
10.
Calculate the average NRQ for each sample within each GOI using both NRQ values.
-
11.
This average NRQ value indicates the fold change in dosage of the GOI as compared to the “normal” calibrator(s) and normalized against two stable chromosomes, i.e., chr7 and chrX (i.e., the normalized relative quantity or NRQ).
-
12.
This value may be multiplied by 2 to obtain the absolute copy number for the autosomes or X in case of female ESCs.
To calculate the interchromosomal SD (IC SD):
-
1.
Calculate the IC SD per sample by determining the SD between the NRQs for each amplicon of the five different chromosomes within each ESC clone. The NRQ values for chr7 and chrX are 1 and are included in the IC SD calculation.
-
2.
When the IC SD is greater than 0.0681, our 67% stringent confidence level (CL), samples may be considered abnormal. If the IC SD is greater than our 95% CL of 0.0906, then samples should be considered abnormal. The corresponding NRQ can subsequently be used to evaluate which of the chromosomes is aneuploid.
Acknowledgments
We thank Thomas Bozza and Jaclyn Bubnell for helping this project reach fruition and for comments on the science or manuscript. We would like to thank Pieter Dom for the graphic work. We would also like to thank Margaret Leversha at the Memorial Sloan-Kettering Cancer Center Cyotogenetics Core Facility for the DAPI-banded karyotyping, which is supported by NIH Cancer Center Support Grant P30 CA008748. P.F. is funded by the NIH SC1 Grant SC1 GM088114-01A1 and by the Facilities Service Award and infrastructure support: RCMI 41398-01-27.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information
References
- Boland M.J., Hazen J.L., Nazor K.L., Rodriguez A.R., Gifford W., Martin G., Kupriyanov S., Baldwin K.K. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Capecchi M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Chen Q., Shi X., Rudolph C., Yu Y., Zhang D., Zhao X., Mai S., Wang G., Schlegelberger B., Shi Q. Recurrent trisomy and Robertsonian translocation of chromosome 14 in murine iPS cell lines. Chromosome Res. 2011;19:857–868. doi: 10.1007/s10577-011-9239-y. [DOI] [PubMed] [Google Scholar]
- D’haene B., Hellemans J., Craen M., De Schepper J., Devriendt K., Fryns J.P., Keymolen K., Debals E., de Klein A., de Jong E.M. Improved molecular diagnostics of idiopathic short stature and allied disorders: quantitative polymerase chain reaction-based copy number profiling of SHOX and pseudoautosomal region 1. J. Clin. Endocrinol. Metab. 2010;95:3010–3018. doi: 10.1210/jc.2009-2218. [DOI] [PubMed] [Google Scholar]
- D’haene B., Vandesompele J., Hellemans J. Accurate and objective copy number profiling using real-time quantitative PCR. Methods. 2010;50:262–270. doi: 10.1016/j.ymeth.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Chernomorsky R., Esau L., Om J., Xue Y., Murphy A.J., Yancopoulos G.D., Valenzuela D.M. The loss-of-allele assay for ES cell screening and mouse genotyping. Methods Enzymol. 2010;476:295–307. doi: 10.1016/S0076-6879(10)76017-1. [DOI] [PubMed] [Google Scholar]
- Haurogné K., Bach J.M., Lieubeau B. Easy and rapid method of zygosity determination in transgenic mice by SYBR Green real-time quantitative PCR with a simple data analysis. Transgenic Res. 2007;16:127–131. doi: 10.1007/s11248-006-9024-4. [DOI] [PubMed] [Google Scholar]
- Hook E.B. Exclusion of chromosomal mosaicism: tables of 90%, 95% and 99% confidence limits and comments on use. Am. J. Hum. Genet. 1977;29:94–97. [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Lee J.Y., Xia L., Mulvihill J.J., Li S. Trisomy 8: a common finding in mouse embryonic stem (ES) cell lines. Mol. Cytogenet. 2013;6:3. doi: 10.1186/1755-8166-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu H., Loring J., Hormuzdi S., Disteche C.M., Bornstein P., Jaenisch R. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev. Dyn. 1997;209:85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Misra R.P., Duncan S.A. Gene targeting in the mouse: advances in introduction of transgenes into the genome by homologous recombination. Endocrine. 2002;19:229–238. doi: 10.1385/ENDO:19:3:229. [DOI] [PubMed] [Google Scholar]
- Mussolino C., Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr. Opin. Biotechnol. 2012;23:644–650. doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T., Kamiyoshi A., Watanabe S., Sato M., Shindo T. Rapid zygosity determination in mice by SYBR Green real-time genomic PCR of a crude DNA solution. Transgenic Res. 2008;17:149–155. doi: 10.1007/s11248-007-9134-7. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Zakrajsek B.A., Mills A.G., Gorn V., Singer M.J., Reed M.W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- Sugawara A., Goto K., Sotomaru Y., Sofuni T., Ito T. Current status of chromosomal abnormalities in mouse embryonic stem cell lines used in Japan. Comp. Med. 2006;56:31–34. [PubMed] [Google Scholar]
- Treff N.R., Scott R.T., Jr. Four-hour quantitative real-time polymerase chain reaction-based comprehensive chromosome screening and accumulating evidence of accuracy, safety, predictive value, and clinical efficacy. Fertil. Steril. 2013;99:1049–1053. doi: 10.1016/j.fertnstert.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Cong L., Lodato S., Kosuri S., Church G.M., Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


