Abstract
Rice SPX domain gene, OsSPX1, plays an important role in the phosphate (Pi) signaling network. Our previous work showed that constitutive overexpression of OsSPX1 in tobacco and Arabidopsis plants improved cold tolerance while also decreasing total leaf Pi. In the present study, we generated rice antisense and sense transgenic lines of OsSPX1 and found that down-regulation of OsSPX1 caused high sensitivity to cold and oxidative stresses in rice seedlings. Compared to wild-type and OsSPX1-sense transgenic lines, more hydrogen peroxide accumulated in seedling leaves of OsSPX1-antisense transgenic lines for controls, cold and methyl viologen (MV) treatments. Glutathione as a ROS scavenger could protect the antisense transgenic lines from cold and MV stress. Rice whole genome GeneChip analysis showed that some oxidative-stress marker genes (e.g. glutathione S-transferase and P450s) and Pi-signaling pathway related genes (e.g. OsPHO2) were significantly down-regulated by the antisense of OsSPX1. The microarray results were validated by real-time RT-PCR. Our study indicated that OsSPX1 may be involved in cross-talks between oxidative stress, cold stress and phosphate homeostasis in rice seedling leaves.
Introduction
The SPX domain, a domain of 180 residues in length at the N-termini of the proteins, was defined after the SYG1/Pho81/XPR1 proteins. Many proteins possessing the SPX domain have been suggested to be involved in phosphate (Pi) signaling, for example, the N-terminus of yeast SYG1 binds to the G-protein beta subunit and inhibits mating pheromone signal transduction [1]. The putative Pi-level sensors, Pho81 and NUC-2, have a SPX domain in their N-termini and may be involved in regulation of Pi transport [2–4]. The human XPR1 functions as a Pi sensor and may be involved in G-protein associated signal transduction [5,6]. The SPX domain of the yeast low-affinity Pi transporter Pho90 was reported to regulate transport activity through physical interaction with Spl2 [7]. In plants, many SPX domain proteins were also identified as involved in the Pi-related signal transduction pathway and regulation pathways. Phosphorus (P) is well-known as a major macronutrient for plant growth and development.
All SPX domain proteins in rice and Arabidopsis have been classified into four classes based on phylogenetic and domain analyses [8,9]. The four classes are differentiated by specific conserved domains: three rice and 11 Arabidopsis proteins in Class 1 (PHO1 and PHO1-like); six rice (OsSPX) and four Arabidopsis (AtSPX) proteins in Class 2; three Arabidopsis and six rice proteins (four rice genes) in Class 3; and two Arabidopsis and two rice proteins in Class 4. The PHO1 (At3g23430) and PHO1-like proteins were identified as involved in ion transport in Arabidopsis [10–13]. The Arabidopsis pho1 mutant was characterized by severe deficiency in shoot Pi but normal root Pi content. PHO1 is a gene specifically involved in the loading of Pi into the xylem in roots and is expressed in cells of the root vascular system [11]. Three members of the AtPHO1 family had possible interactions with signaling pathways involved in Pi deficiency and responses to auxin, cytokinin and abscisic acid [14]. The PHO1 gene family was also identified in Physcomitrella patens and responded to Pi deficiency [12]. Arabidopsis AtSPX family genes, encoding another class of proteins with a SPX domain, have diverse functions in plant tolerance to phosphorus starvation [8]. AtSPX1 showed 52-fold induction under Pi starvation [15]. The expression levels of AtSPX1 and AtSPX3 were induced by Pi starvation, AtSPX2 was slightly induced and AtSPX4 was suppressed. The AtSPX family may be part of the Pi-signaling pathways controlled by PHR1 and SIZ1 [8]. Three rice OsSPXs were identified as Pi-starvation response genes in rice seedlings [16]; OsSPX1 is involved in Pi homeostasis through a negative feedback loop under Pi-limited conditions in rice [17]. OsSPX1 was suggested to a regulator for the transcriptions of OsSPX2, 3 and 5; and OsSPX3 negatively regulated the PSI (Pi-starvation induced) genes [18]. OsSPX1 was reported to suppress the function of OsPHR2 in the regulation of OsPT2 expression and Pi homeostasis in rice shoots [19].
In general, plants regulate multiple metabolic processes to adapt to low Pi environments, such as altering lipid metabolism [20], increasing synthesis and activity of RNases and acid phosphatases, and changing the metabolic bypasses of glycolysis [21]. During Pi over-accumulation, some Pi-starvation related genes were reported to be involved in Pi toxicity: for example, rice plants over-expressing OsPT8 showed Pi toxicity symptoms in leaves under high Pi supply [22]; mature leaves of OsSPX1 RNAi plants showed necrotic spots under Pi-sufficient conditions [17]; OsPHR2 over-expressing lines displayed chlorosis or necrosis on leaf margins at high Pi levels [23]; and ltn1 (OsPHO2) mutant displayed leaf tip necrosis in mature leaves [24].
There is a close relationship between Pi-signaling and abiotic stresses, including cold stress, in plants. In Arabidopsis pho1-2 and pho2-1 mutants, Hurry et al. (2000) reported that low Pi played an important role in triggering cold acclimatization of leaf tissues [25]. Our previous study reported that constitutive overexpression of OsSPX1 in tobacco and Arabidopsis plants improved cold tolerance while also decreasing total leaf Pi [9]. There have been some studies on the relationship between reactive oxygen species (ROS) signaling and cold stress [26–30]. In the present study, we generated rice antisense and sense transgenic lines of OsSPX1 to study the role of OsSPX1 involved in cold response and the possible relationship with oxidative stress. Furthermore, we conducted rice whole genome GeneChip analysis to elucidate the possible molecular mechanism underlying the down-regulation of OsSPX1 causing high sensitivity to oxidative stress in rice seedling leaves.
Results
Generation of OsSPX1 transgenic rice lines
To characterize the gene function of OsSPX1 in rice plants, we applied an antisense and sense transgenic approach. The cloned full-length cDNAs for OsSPX1 (adapted from our previous paper [9]) was used to generate transgenic rice lines with Oryza sativa ssp. japonica cv. Nipponbare as the wild-type (WT) background. The down-regulation of OsSPX1 gene by antisense approach (Figure S1A) and the over-expressing OsSPX1 gene by sense approach (Figure S1B) was under the control of the ubiquitin promoter. Several independent hygromycin-resistant transgenic lines were generated for Ubi::OsSPX1-antisense and -sense. Real-time RT-PCR was applied to determine the expression levels of OsSPX1 in WT and transgenic rice lines, using different primers for Ubi::OsSPX1-antisense and -sense transgenic rice lines (primers are listed in Table S2). The T3 generation Ubi::OsSPX1-sense lines (lines S1-S5, on the right of Figure 1) accumulated a substantial amount of the transcript, whereas the level of transcript was very low in WT. For the T3 generation Ubi::OsSPX1-antisense lines (lines A1-A3, on the left of Figure 1), the expression level of OsSPX1 was significantly lower than in WT plants (about 35% - 75% lower in Ubi::OsSPX1-antisense lines; by t-test, p< 0.05).
Figure 1. Real-time RT-PCR validation of transgenic rice lines.
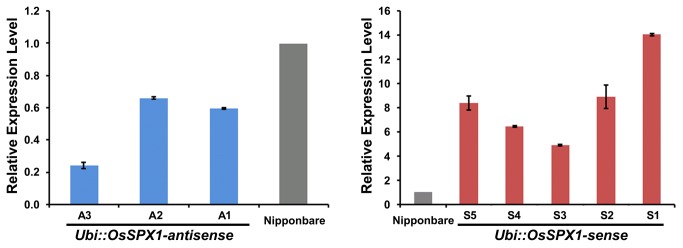
The expression levels of OsSPX1 in WT (gray bars) and transgenic rice lines (blue bars for Ubi::OsSPX1-antisense lines and red bars for Ubi::OsSPX1-sense lines). Expression levels were normalized to 18S rRNA. The results are mean values ±SD of three replicates.
Cold tolerance analysis of OsSPX1 transgenic lines and WT rice seedlings
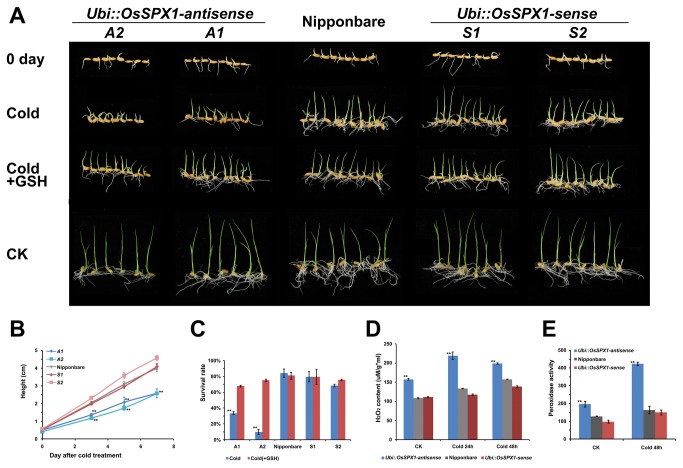
The cold stress response of the OsSPX1 transgenic rice (T4 generation plants of both antisense and sense transgenic lines) plants were tested together with WT at the seedling stage. The rice seedlings (WT and Ubi::OsSPX1-antisense and -sense lines) with 5-mm bud burst were treated at 4-5°C for 7 d and then recovered at room temperature (Figure 2A). There were no obvious differences between the WT and transgenic lines at room temperature for 6 d; however, after cold treatment the Ubi::OsSPX1-antisense plants grew slowly (or even died) and were significantly shorter than the WT and sense plants (Figure 2A and 2B). The height of seedlings was measured at 0, 3, 5 and 7 d after cold treatment. Starting from 3 d of recovery after cold treatment, the height of seedlings of two Ubi::OsSPX1-antisense transgenic lines (A1 and A2; the blue solid lines in Figure 2B) showed significant differences from WT (gray solid line in Figure 2B; t-test, p < 0.01) and sense lines (S1 and S2, the red solid lines in Figure 2B). Up to 7 d of recovery after cold treatment, the growth of several plants of the A1 and A2 lines ceased and some seedlings died (Figure 2C). The survival rates of A1 and A2 lines were significantly lower than WT plants (p < 0.01) and sense lines (S1 and S2). This phenotype indicated that the antisense plants were more cold-stress sensitive than WT and sense plants.
Figure 2. Characterization of the transgenic rice seedlings under cold treatment.
A. 0 day - germinated seeds of WT and transgenic plants; CK - seedlings of the WT and transgenic plants grown for 6 d at room temperature (28°C); Cold - seedlings with 5 mm bud burst were treated at 4-5°C for 7 d, then recovered for 4 d at room temperature (28°C); Cold+GSH - the cold-treated seedlings were recovered under 10 mg/L GSH solution for 4 d at room temperature (28°C). A1 and A2 are Ubi::OsSPX1-antisense transgenic lines (on the left), S1 and S2 are Ubi::OsSPX1-sense lines (on the right).
B. The height of the WT and transgenic plants over the recovery time after cold treatment (blue lines for Ubi::OsSPX1-antisense and red lines for Ubi::OsSPX1-sense).
C. The survival rate of the WT and transgenic plants after cold treatment, with/without GSH.
D. H2O2 content of the leaves of WT and transgenic rice plants under cold treatment (blue bars for Ubi::OsSPX1-antisense and red bars for Ubi::OsSPX1-sense).
E. Peroxidase activity in the WT and transgenic rice plants under cold treatment (blue bars for Ubi::OsSPX1-antisense and red bars for Ubi::OsSPX1-sense).
** represent significant differences (p < 0.01) between transgenic and WT rice plants according to Student’s t-test.
In addition, we measured the hydrogen peroxide (H2O2) content in leaves of WT and transgenic rice plants (Figure 2D). Before cold treatment (i.e. at room temperature), the leaves of Ubi::OsSPX1-antisense plants accumulated much more H2O2 than did WT and sense plants. After 24 h of cold treatment, the leaf H2O2 content of the antisense plants increased about 40%, but that of WT plants increased about 20% and that of sense plants only increased about 5%. The leaf H2O2 content difference between the antisense transgenic lines and WT plants was significant (p < 0.01), as was the difference between sense line and WT plants. There was a similar trend after 48 h of cold treatment (Figure 2D).
We also measured the peroxidase activity in the WT and transgenic plants (Figure 2E), and this showed a similar trend to leaf H2O2 content. Peroxidase activity was much higher in antisense plants than in WT and sense plants, both before and after cold treatment.
When adding the antioxidant, glutathione (GSH), into the cold treated rice plants, the cold-sensitive phenotype of the Ubi::OsSPX1-antisense transgenic plants was recovered (Figure 2A and C), including the growth rate and survival rate.
Differential response of OsSPX1 transgenic and WT rice seedlings under methyl viologen (MV, or Paraquat) treatment
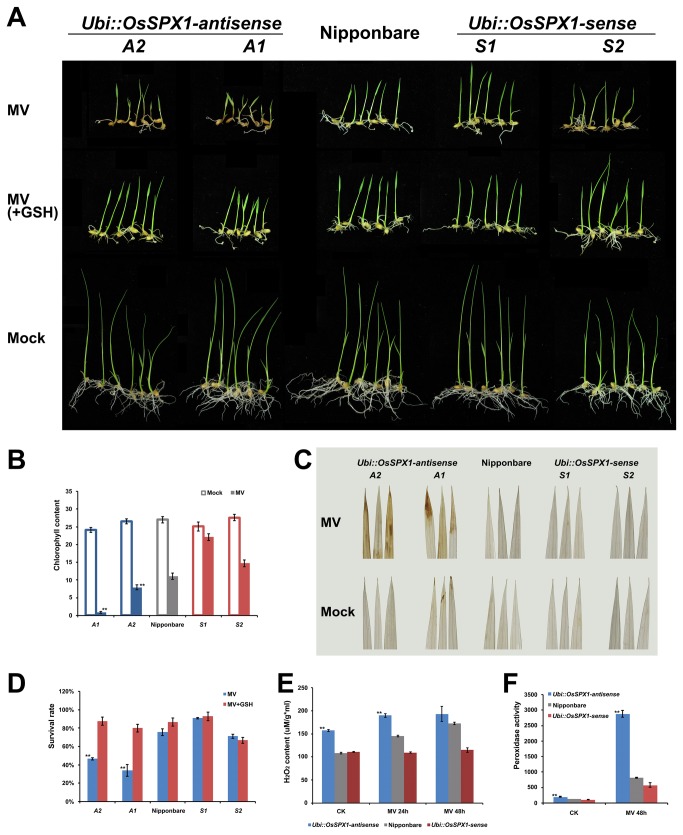
The significant difference in H2O2 content and peroxidase activity between WT and transgenic rice plants under normal condition and cold treatment indicated that the differential cold-stress responses of OsSPX1 transgenic lines may be related to oxidant response activity. We performed a MV treatment experiment to test our hypothesis. Four-day-old seedlings of WT, Ubi::OsSPX1-antisense and -sense transgenic lines were transferred to 10 μM MV solution, with water as mock. After 3 d of growth, the differences in leaf phenotype became significant between antisense lines (A1 and A2) and the WT and sense lines (S1 and S2) (Figure 3A). In the mock condition, WT and transgenic seedlings grew normally, and all leaves were green. Under MV treatment, both WT and transgenic seedlings were dwarfed. However, there was significant phenotype divergence: the leaves of A1 and A2 seedlings became scorched and yellow with little green, their seedling leaves presented lesions and then died; in contrast, S1 and S2 plants grew better, and the seedling leaves were still green after 3 d of MV treatment (Figure 3A). The detected leaf chlorophyll contents also confirmed the leaf phenotype (Figure 3B). Leaf chlorophyll contents were similar in WT and transgenic plants during mock condition; however, under MV solution, the leaf chlorophyll contents of A1 and A2 lines were significantly lower than that of WT (p < 0.01) and lines S1 and S2. The survival rate of A1 and A2 lines under MV treatment were also significantly lower than that of WT (p < 0.01) and sense lines (Figure 3D).
Figure 3. Differential response of WT and transgenic rice lines under MV treatment.
A. Four-day-old seedlings treated with 10 μM MV (MV) and 10 μM MV plus 10 mg/L GSH (MV+GSH) for 3 d, water as mock. A1 and A2 are Ubi::OsSPX1-antisense transgenic lines (on the left), S1 and S2 are Ubi::OsSPX1-sense lines (on the right).
B. The chlorophyll content of seedlings under MV treatment (blue bars for Ubi::OsSPX1-antisense and red bars for Ubi::OsSPX1-sense).
C. DAB staining of the seedling leaves treated with 10 μM MV, water as mock. A1 and A2 are Ubi::OsSPX1-antisense transgenic lines (on the left), S1 and S2 are Ubi::OsSPX1-sense lines (on the right).
D. The survival rate of WT and transgenic plants treated with 10 μM MV, with/without GSH.
E. H2O2 content of the leaves of WT and transgenic plants under MV treatment (blue bars for Ubi::OsSPX1-antisense and red bars for Ubi::OsSPX1-sense).
F. Peroxidase activity in WT and transgenic rice plants under MV treatment (blue bars for Ubi::OsSPX1-antisense and red bars for Ubi::OsSPX1-sense).
** represent significant differences (p < 0.01) between transgenic and WT rice plants according to Student’s t-test.
In addition, we conducted 3,3’-diaminobenzidine (DAB) staining to detect the accumulation of H2O2 in rice leaves (Figure 3C). Under MV treatment, the leaves of Ubi::OsSPX1-antisense transgenic plants were significantly stained dark brown while WT and sense leaves were lighter or not stained. Moreover, the direct measurement of H2O2 also indicated that the leaves of antisense plants accumulated much more H2O2 than did WT and sense plants with and without MV treatment (p < 0.01, Figure 3E). The peroxidase activity of rice seedlings showed a similar trend to the H2O2 content (Figure 3F). Overall, the WT and sense seedlings showed strong oxidative stress resistance compared to antisense seedlings in the MV experiment. As for cold treatment, when GSH solution was added to the MV, the sensitive phenotype of Ubi::OsSPX1-antisense transgenic plants under MV treatment was recovered (Figure 3A and D).
In general, H2O2 is one of the ROS that leads to programmed cell death (PCD), but it also acts as a signal molecule in various biological processes including cold response. The high level of H2O2 in Ubi::OsSPX1-antisense transgenic plants led us to hypothesize that down-regulation of OsSPX1 resulted in a reduced ability to eliminate of ROS. Thus we designed gene expression experiments to determine molecular relationships between cold stress and ROS controlled by OsSPX1.
Transcriptome analysis of Ubi::OsSPX1-antisense transgenic and WT rice seedlings
GeneChip® microarray transcriptome experiments were performed with seedlings collected from the WT rice and two Ubi::OsSPX1-antisense transgenic lines (A1 and A2) under cold treatment and normal conditions. The change in expression level for each probe set between antisense lines and WT was conducted by MAS5 algorithm using the GCOS baseline tool. Then one-sample t-test were applied to the log2ratios to identify differentially expressed probe sets. Using the cut-off (p ≤ 0.05 and log2ratio ≥ 0.6), we identified a total of 1266 probe sets, including 748 up-regulated and 518 down-regulated in Ubi::OsSPX1-antisense transgenic lines (detailed information on these probe sets is listed in Table S1).
We further applied gene ontology (GO) enrichment analysis (by EasyGO tool [31]) on these differentially expressed probe sets and determined the significant GO terms (FDR p ≤ 0.05 as cut-off; Table 1). For the 518 probe sets down-regulated in Ubi::OsSPX1-antisense transgenic lines, several GO terms including response to toxin, response to chemical stimulus, potassium ion symporter activity, and glutathione transferase activity were significantly enriched. These enriched GO categories indicated that the differentially expressed probe sets may be related to oxidative process. For the 748 probe sets up-regulated in antisense lines, the enriched GO terms included sugar mediated signaling, photosynthesis, NADH dehydrogenase activity, and hormone-related GO terms (GA and ABA).
Table 1. GO analysis of differentially expressed probe sets between Ubi::OsSPX1-antisense transgenic lines and WT (Nipponbare).
| GO term | GO name | Qnum | B/Rnum | FDR p-value |
|---|---|---|---|---|
| 518 probe sets down-regulated in Ubi::OsSPX1-antisense transgenic lines | ||||
| Biological Process | 265 | 21427 | ||
| GO:0009407 | toxin catabolic process | 8 | 89 | 5.08E-06 |
| GO:0009404 | toxin metabolic process | 8 | 113 | 4.01E-04 |
| GO:0009626 | plant-type hypersensitive response | 11 | 218 | 1.32E-03 |
| GO:0009636 | response to toxin | 8 | 130 | 1.83E-03 |
| GO:0009812 | flavonoid metabolic process | 6 | 105 | 3.26E-02 |
| GO:0009813 | flavonoid biosynthetic process | 6 | 105 | 3.26E-02 |
| GO:0042221 | response to chemical stimulus | 34 | 1509 | 4.02E-02 |
| Molecular Function | 320 | 27148 | ||
| GO:0008393 | fatty acid (omega-1)-hydroxylase activity | 3 | 5 | 2.82E-12 |
| GO:0009674 | potassium:sodium symporter activity | 3 | 10 | 8.22E-07 |
| GO:0022820 | potassium ion symporter activity | 3 | 10 | 8.22E-07 |
| GO:0004364 | glutathione transferase activity | 9 | 179 | 7.28E-03 |
| GO:0008493 | tetracycline transporter activity | 3 | 21 | 8.84E-03 |
| Cellular Component | 338 | 29111 | ||
| GO:0012505 | endomembrane system | 90 | 5277 | 8.84E-03 |
| 748 probe sets up-regulated in Ubi::OsSPX1-antisense transgenic lines | ||||
| Biological Process | 370 | 21427 | ||
| GO:0006270 | DNA replication initiation | 8 | 19 | 1.49E-22 |
| GO:0009687 | abscisic acid metabolic process | 6 | 21 | 3.16E-11 |
| GO:0009688 | abscisic acid biosynthetic process | 6 | 21 | 3.16E-11 |
| GO:0015979 | photosynthesis | 24 | 387 | 9.08E-08 |
| GO:0009405 | pathogenesis | 13 | 157 | 4.20E-06 |
| GO:0006306 | DNA methylation | 4 | 20 | 2.83E-04 |
| GO:0009686 | gibberellin biosynthetic process | 5 | 33 | 3.03E-04 |
| GO:0007018 | microtubule-based movement | 8 | 85 | 4.26E-04 |
| GO:0009685 | gibberellin metabolic process | 5 | 44 | 9.08E-03 |
| GO:0010182 | sugar mediated signaling | 4 | 32 | 2.92E-02 |
| GO:0009814 | defense response, incompatible interaction | 11 | 202 | 3.55E-02 |
| GO:0015712 | hexose phosphate transport | 4 | 33 | 3.55E-02 |
| Molecular Function | 439 | 27148 | ||
| GO:0001882 | nucleoside binding | 3 | 5 | 4.95E-08 |
| GO:0003954 | NADH dehydrogenase activity | 33 | 656 | 5.14E-08 |
| GO:0003886 | DNA (cytosine-5-)-methyltransferase activity | 3 | 7 | 4.03E-06 |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | 7 | 55 | 1.73E-05 |
| GO:0016651 | oxidoreductase activity, acting on NADH or NADPH | 36 | 898 | 2.82E-05 |
| GO:0051539 | 4 iron, 4 sulfur cluster binding | 3 | 11 | 6.24E-04 |
| GO:0003777 | microtubule motor activity | 9 | 127 | 6.61E-03 |
| GO:0003989 | acetyl-CoA carboxylase activity | 14 | 275 | 1.09E-02 |
| GO:0008094 | DNA-dependent ATPase activity | 8 | 109 | 1.13E-02 |
| GO:0008254 | 3'-nucleotidase activity | 3 | 18 | 4.30E-02 |
| GO:0019133 | choline monooxygenase activity | 3 | 18 | 4.30E-02 |
| Cellular Component | 491 | 29111 | ||
| GO:0009512 | cytochrome b6f complex | 13 | 101 | 2.74E-12 |
| GO:0043601 | nuclear replisome | 3 | 5 | 9.53E-09 |
| GO:0009579 | thylakoid | 29 | 549 | 7.03E-08 |
| GO:0009523 | photosystem II | 21 | 367 | 2.22E-06 |
| GO:0009522 | photosystem I | 12 | 167 | 5.86E-05 |
| GO:0044436 | thylakoid part | 16 | 306 | 7.08E-04 |
| GO:0009539 | photosystem II reaction center | 6 | 66 | 4.93E-03 |
| GO:0005875 | microtubule associated complex | 8 | 131 | 3.57E-02 |
| GO:0009941 | chloroplast envelope | 13 | 295 | 4.91E-02 |
| GO:0043245 | extraorganismal space | 10 | 197 | 4.91E-02 |
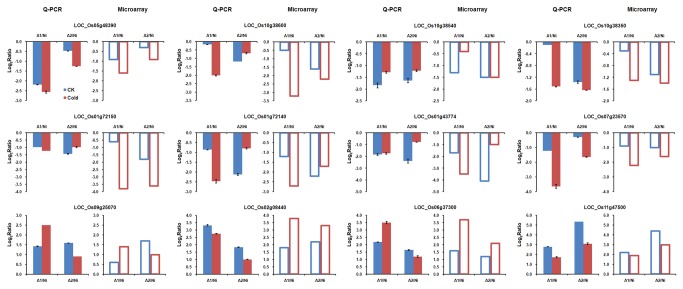
Furthermore, we selected several genes for real-time RT-PCR confirmation, such as OsPHO2, GSTs, P450s and WRKYs. Additional biological samples were collected for real-time RT-PCR validation. For each selected gene, the log2ratio between two Ubi::OsSPX1-antisense transgenic lines (A1 and A2) and the WT under control and cold conditions are shown in Figure 4 - the corresponding log2ratio of microarray results is also shown. One-by-one comparison validated the majority of microarray results with real-time RT-PCR results in these selected genes, most of them showed similar patterns.
Figure 4. Real-time RT-PCR validation for selected probe sets.
Twelve genes were selected for real-time RT-PCR to validate the expression patterns of Ubi::OsSPX1-antisense transgenic lines (A1 and A2) and WT rice (Ni) seedlings under control condition and cold treatment. These genes are: LOC_Os05g48390 - OsPHO2, ubiquitin conjugating enzyme; LOC_Os10g38600 - glutathione S-transferase GSTU6; LOC_Os10g38540 - glutathione S-transferase GSTU6; LOC_Os10g38350 - glutathione S-transferase GSTU6; LOC_Os01g72150 - glutathione S-transferase; LOC_Os01g72140 - glutathione S-transferase; LOC_Os01g43774 - cytochrome P450 CYP72A23; LOC_Os07g23570 - cytochrome P450 CYP709C9; LOC_Os09g25070 - OsWRKY62; LOC_Os02g08440 - OsWRKY71; LOC_Os06g37300 - ent-kaurene oxidase; LOC_Os11g47500 - xylanase inhibitor protein 1 precursor. The error bars of real-time RT-PCR results represent the standard error from three independent biological replicates.
From these GO terms and some important gene families, some key genes were highlighted. There were nine glutathione S-transferase (GST) genes in the differentially expressed probe sets, but only one was up-regulated in Ubi::OsSPX1-antisense transgenic lines (Table S1). Among the other eight down-regulated GSTs, six were up-regulated by MV treatment (Table 2); moreover, four of the six GSTs were located on the rice chromosome 10, including the GST gene cluster: LOC_Os10g38340, LOC_Os10g38350 and LOC_Os10g38360. Of cytochrome P450 genes, several family members were up-regulated in Ubi::OsSPX1-antisense transgenic lines, such as CYP71 and CYP94 families; while several family members were down-regulated in antisense lines, such as CYP72A, CYP709 and CYP86A subfamilies (Table S1). Notably two P450 genes, CYP72A23 and CYP709C9, were down-regulated in antisense lines and up-regulated by MV treatment (Table 2). There were some other family genes shown preference in the differentially expressed probe sets, for examples, many peroxidases (including OsAPx3) and UDP-glucoronosyl transferases were down-regulated in antisense lines, and several WRKY family members were up-regulated in antisense lines.
Table 2. Differentially expressed probe sets between Ubi::OsSPX1-antisense transgenic lines and WT (Nipponbare) related to MV treatment.
|
|
Ubi::OsSPX1-antisense vs Ni
|
MV vs CK (Ni)(a)
|
||||||
|---|---|---|---|---|---|---|---|---|
|
Probe Set ID
|
Log2Ratio | p-value | Log2Ratio | p-value | Locus ID | Annotation | ||
| Os.40000.1.S1_at | -1.63 | 4.07E-02 | 1.58 | 1.28E-05 | LOC_Os10g38600 | glutathione S-transferase GSTU6 | ||
| Os.40000.1.S1_x_at | -1.88 | 4.51E-02 | 1.70 | 2.58E-05 | LOC_Os10g38600 | glutathione S-transferase GSTU6 | ||
| Os.13015.1.S1_at | -1.25 | 4.80E-02 | 2.92 | 1.31E-03 | LOC_Os10g38360 | glutathione S-transferase GSTU6 | ||
| Os.46635.1.S1_x_at | -1.03 | 2.61E-02 | 2.41 | 2.66E-04 | LOC_Os10g38350 | glutathione S-transferase GSTU6 | ||
| Os.9013.1.S1_at | -2.00 | 3.77E-02 | 4.22 | 7.38E-04 | LOC_Os10g38340 | glutathione S-transferase GSTU6 | ||
| OsAffx.24011.1.S1_at | -2.45 | 4.90E-02 | 4.31 | 4.60E-05 | LOC_Os01g72150 | glutathione S-transferase | ||
| Os.22957.1.S1_at | -1.95 | 9.09E-03 | 3.36 | 3.09E-06 | LOC_Os01g72140 | glutathione S-transferase | ||
| Os.9017.1.S1_x_at | -1.43 | 1.79E-02 | 3.79 | 4.10E-05 | LOC_Os07g23570 | cytochrome P450 CYP709C9 | ||
| Os.41199.1.A1_at | -2.58 | 3.90E-02 | 6.35 | 5.21E-04 | LOC_Os01g43774 | cytochrome P450 CYP72A23 | ||
| Os.56112.1.A1_at | -2.08 | 3.12E-02 | 1.93 | 3.94E-02 | LOC_Os01g43774 | cytochrome P450 CYP72A23 | ||
(a) The methyl viologen (MV) transcriptome data from [32]
We further used real-time RT-PCR to investigate the expression levels of GST and P450 genes affected by OsSPX1 under oxidative stress. We detected the expression pattern of these genes in antisense line (A1), sense line (S1), and WT plants under MV treatment (Table 3). All these genes - including GST genes in chromosome 10 GST cluster, CYP72A23 and CYP709C9 - were significantly down-regulated in Ubi::OsSPX1-antisense transgenic line compared to WT and Ubi::OsSPX1-sense transgenic line (t-test, p < 0.05).
Table 3. Real-time RT-PCR for selected genes in transgenic lines and Nipponbare under MV treatment.
|
A1 vs Ni
(a)
|
A1 vs S1
(b)
|
||||
|---|---|---|---|---|---|
| Locus ID | Log2Ratio | p-value | Log2Ratio | p-value | Annotation |
| LOC_Os06g40120 | -2.40 | 1.08E-02 | -6.83 | 1.30E-06 | OsSPX1 |
| LOC_Os10g38600 | -0.75 | 2.60E-04 | -0.74 | 9.93E-03 | glutathione S-transferase GSTU6 |
| LOC_Os10g38540 | -1.72 | 3.43E-03 | -2.39 | 9.26E-04 | glutathione S-transferase GSTU6 |
| LOC_Os10g38360 | -0.86 | 4.30E-03 | -0.40 | 1.77E-02 | glutathione S-transferase GSTU6 |
| LOC_Os10g38350 | -1.15 | 2.09E-03 | -0.78 | 4.02E-02 | glutathione S-transferase GSTU6 |
| LOC_Os10g38340 | -1.99 | 1.45E-04 | -1.76 | 9.54E-03 | glutathione S-transferase GSTU6 |
| LOC_Os01g72150 | -1.22 | 5.83E-03 | -1.77 | 1.11E-03 | glutathione S-transferase |
| LOC_Os01g72140 | -1.01 | 3.18E-02 | -1.47 | 1.40E-02 | glutathione S-transferase |
| LOC_Os07g23570 | -1.63 | 8.26E-05 | -0.84 | 5.08E-02 | cytochrome P450 CYP709C9 |
| LOC_Os01g43774 | -0.87 | 8.52E-04 | -0.38 | 1.46E-02 | cytochrome P450 CYP72A23 |
(a) A1 is Ubi::OsSPX1-antisense transgenic line and Ni is WT rice
(b) S1 is Ubi::OsSPX1-sense transgenic line
Discussion
OsSPX1 plays an important role in Pi homeostasis by controlling several biological processes [8,9,17,19]. It was reported that excessive Pi accumulation in seedling leaves of OsSPX1-RNAi rice plants caused toxicity [17]. Our previous work reported that increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana [9]. In the present study, we further elucidated the role of OsSPX1 in cold response and the possible relationship with oxidative stress in rice seedlings. We applied an antisense and sense transgenic approach, and generated several individual Ubi::OsSPX1-antisense and -sense transgenic lines (Figure 1). The cold tolerance experiment on rice seedlings showed that Ubi::OsSPX1-antisense lines were significantly sensitive to cold stress (Figure 2A, 2B and 2C). We also detected more H2O2 accumulation in the seedling leaves of Ubi::OsSPX1-antisense lines, both in control and cold conditions (Figure 2D). H2O2, as a key ROS molecule, is induced by various biotic or abiotic stresses (including cold stress) and damages cellular macromolecules such as lipids, enzymes and DNA. The oxidative resistant ability of plants may be correlated with their resistance to cold/chilling stress. We further used MV, a widely used oxidant forming the toxic superoxide radical [32–34], to test the oxidative response of Ubi::OsSPX1-antisense and -sense transgenic and WT rice plants (Figure 3). There was a positive correlation between the cold and oxidation response in these rice plants, the antisense lines were highly sensitive to cold and oxidative stresses compared with sense lines and WT plants. H2O2 accumulation was significantly higher in seedling leaves of antisense than in sense lines and WT plants, both in control and MV treatment (Figure 3E).
We conducted the transcriptome analysis with rice whole genome GeneChip to elucidate the possible molecular mechanism underlying the down-regulation of OsSPX1 that caused high sensitivity to cold and oxidative stress in rice seedlings. We identified 1266 differentially expressed probe sets (including 748 up-regulated and 518 down-regulated in Ubi::OsSPX1-antisense lines). Transcriptome data analysis and real-time RT-PCR confirmed that several Pi-signaling pathway related genes were affected by down-regulation of OsSPX1, for example, OsPHO2 (LOC_Os05g48390) was down-regulated in Ubi::OsSPX1-antisense transgenic lines (Figure 4 and Table S1). The mutant of OsPHO2, also named ltn1, displayed leaf tip necrosis predominantly and increased the uptake and translocation of Pi [24]. Arabidopsis pho2 mutants, with increased shoot Pi, were shown to be more sensitive to freezing than WT after cold acclimatization [25]. Besides OsPHO2, there were several phospholipid biosynthesis-related genes also down-regulated in Ubi::OsSPX1-antisense transgenic lines (shown in Table S1), such as glycerol-3-phosphate acyltransferases, phospholipase D and patatin T5 precursors. Two rice RNase T2 [35] genes (LOC_Os09g36680 and LOC_Os09g36700) were up-regulated in Ubi::OsSPX1-antisense transgenic lines. Their orthologs, RNS2 in Arabidopsis and AhSL28 in Antirrhinum, were all induced during leaf senescence and Pi starvation [36,37]. The RNase activities were also activated in ltn1 (ospho2) mutant under Pi-sufficient conditions [24].
GO analysis showed that several GO terms related to oxidative process were enriched in the differentially expressed probe sets. For examples, GSTs (EC 2.5.1.18) were included in GO terms ‘response to toxin’ and ‘glutathione transferase activity’, and enriched in the 518 probe sets down-regulated in the Ubi::OsSPX1-antisense transgenic lines. The substrate of GSTs, GSH is an antioxidant [38] and helps to clear the harmful components (including H2O2) in the cell [39]. We highlight the GST genes, especially for the hotspot GST gene cluster region in rice chromosome 10 (Table 2) - most of these genes were down-regulated in the Ubi::OsSPX1-antisense transgenic lines and induced by MV treatment in seedlings [32]. Moreover, these genes were also down-regulated in the antisense line under MV treatment (Table 3). Interestingly, GSH recovered the sensitive phenotype of antisense plants under cold and MV treatment (Figures 2A and 3A) and increased their survival rate (Figures 2C and 3D). This indicated that the lower expression level of GST genes in the Ubi::OsSPX1-antisense transgenic plants affected the GSH level and ROS homeostasis, and caused the antisense lines to be highly sensitive to cold and oxidative stresses.
In additional, several P450 genes (e.g. LOC_Os07g23570 - CYP709C9) were also down-regulated in Ubi::OsSPX1-antisense transgenic lines in control and cold and MV treatments, and induced by MV treatment (Tables 2 and 3). Rice CYP709C9 was considered a possible ortholog of wheat P450 genes CYP709C1 and CYP709C3v2 [32], both of which were related to defence response [40,41]. These GSTs and P450s were reported as potential marker genes for rice oxidative stress tolerance [32]. In Ubi::OsSPX1-antisense transgenic lines, all these genes were down-regulated compared to WT either in normal condition or under cold treatment. This suggested that the high sensitivity to cold and oxidative stresses from down-regulation of OsSPX1 may be caused by the influence on the expression level of these GSTs and P450s. Furthermore, these genes were also down-regulated in Ubi::OsSPX1-antisense transgenic lines compared to sense lines and WT under MV treatment (Table 3), suggesting that OsSPX1 may be involved in broad biological processes related to oxidative stress.
For two other detoxification enzymes, peroxidases and UDP-glucoronosyl transferase, the majority of differentially expressed genes were down-regulated in Ubi::OsSPX1-antisense transgenic lines (Table S1). Peroxidases are one group of H2O2-scavenging enzymes [42]; and both gene expression and activity of cytosolic ascorbate peroxidase (APX) strongly decreased during the activation of PCD [43]. We also noted that OsAPx3 (LOC_Os04g14680, peroxisomal ascorbate peroxidase) and L-ascorbate oxidase (LOC_Os11g42220) were down-regulated in Ubi::OsSPX1-antisense lines, which may be correlated with the higher H2O2 level in these transgenic lines. Two NADP-dependent oxidoreductases (LOC_Os11g14910 and LOC_Os12g12470) were also down-regulated in Ubi::OsSPX1-antisense transgenic lines, indicating a weak antioxidant capacity. There are some reports concerning the relationship between UGT (UDP-glucoronosyl transferase) genes and oxidative stresses [33,44].
Of the up-regulated genes in Ubi::OsSPX1-antisense transgenic lines, several GO terms, such as ‘sugar mediated signalling’ and ‘hexose phosphate transport’, were enriched - this may be related to the crosstalk among the cold acclimatization, Pi starvation and sugar signalling pathway [45]. Some GO terms related to abscisic acid, gibberellin and pathogenesis were also enriched. Several CYP94 members (subfamily of P450 genes; Table S1) were also up-regulated in antisense lines, which are related to the JA signalling pathway [46,47]. JA antagonizes the ABA pathway in response to multiple abiotic and biotic stresses [48].
We identified about 100 cold-stress related genes through gene annotation, GO annotation and from the literature. We studied the expression pattern of these genes between Ubi::OsSPX1-antisense transgenic lines and WT plants under control and cold treatment (Table S3). Only one gene (OsICE1, LOC_Os11g32100) belonged to the 1266 differentially expressed probe sets, and other 17 genes showed differential expression under cold treatment, including several OsDREBs which were up-regulated in antisense lines. During the experiment, we also studied the effects of OsSPX1 in other abiotic stresses, such as salt and osmotic. However, our preliminary results showed no significantly different phenotypes among Ubi::OsSPX1-antisense and -sense lines and WT seedlings (data not shown). It seems that OsSPX1 may play a specific role in cross-talks between oxidative stress and cold stress.
In summary, we generated rice antisense and sense transgenic lines for OsSPX1 and discovered that down-regulation of OsSPX1 caused more H2O2 accumulation in leaves and high sensitivity to cold and oxidative stresses in rice seedlings. Rice whole genome GeneChip and GO enrichment analyses indicated that the GO terms ‘response to toxin’ and oxidative process genes such as GST genes were significantly enriched in the probe sets down-regulated by the antisense of OsSPX1. This may provide a clue concerning the possible molecular mechanism explaining why OsSPX1 down-regulation caused high sensitivity to cold and oxidative stress in rice seedling leaves. GSH could protect the Ubi::OsSPX1-antisense transgenic lines from cold and MV treatments. There may be some signaling links between cold hypersensitivity and toxicity induced by Pi accumulation in plant leaves, both of which may be related to H2O2 abundance causing oxidative stress. Our study may greatly benefit in plant protection from Pi toxicity and abiotic stresses, and improving growth and crop yield.
Materials and Methods
Plant materials
Seeds of rice (Nipponbare as WT, and Ubi::OsSPX1-antisense and Ubi::OsSPX1-sense transgenic lines) were surface-sterilized in 5% (w/v) sodium hypochlorite for 20 min and then washed in distilled water three or four times, then germinated in water for 2 d at room temperature and 1 d at 37°C. The seedlings were transferred to water-saturated Whatman filter paper and grown in a greenhouse (28°C / 25°C and 12 /12 h of day/night, and 83% relative humidity).
For RNA isolation
fresh seedlings were harvested after 24h of cold (4°C) treatment, or 10μM MV treatment; control plants under normal condition were also harvested at the same time.
For phenotype evaluation
to identify the phenotype of rice (Nipponbare as WT, and Ubi::OsSPX1-antisense and -sense transgenic lines) under cold treatment, the germinated seeds with 5-mm bud burst were transferred to 4-5°C for 7 d, then the seedlings were recovered at room temperature in a greenhouse. For identification of the rice phenotype under MV treatment, we transferred the four-day-old seedlings of rice lines to mock (water) and 10 μM MV solution, and the phenotype of each line was investigated and recorded. To protect from cold and MV treatment, 10mg/L GSH was added.
Construction of transgenic rice lines
We mainly followed a previous method [9] to clone the sense and antisense of the full-length cDNA of OsSPX1 into the binary vector pCOU controlled by the ubiquitin promoter. The recombinant plasmids were then introduced into Agrobacterium tumefaciens EHA105 strain following the freeze-thaw method. Transgenic rice lines were obtained by using calli derived from mature embryos of Nipponbare [49]. The concentration of the selected antibiotic, hygromycin B, was 50 mg/L.
RNA isolation and real-time RT-PCR
All seedling samples from Ubi::OsSPX1-antisense transgenic lines and WT under cold treatment and normal condition were homogenized in liquid nitrogen before isolation of the RNA. Total RNA was isolated using TRIZOL® reagent (Invitrogen, CA, USA) and purified using Qiagen RNeasy columns (Qiagen, Hilden, Germany). Reverse transcription was performed using Moloney murine leukemia virus (M-MLV; Invitrogen). We heated 10-µl samples containing 2 µg of total RNA, and 20 pmol of random hexamers (Invitrogen) at 70°C for 2 min to denature the RNA, and then chilled the samples on ice for 2 min. We added reaction buffer and M-MLV to a total volume of 20 µL containing 500 µM dNTPs, 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5mM dithiothreitol, 200 units of M-MLV and 20 pmol random hexamers. The samples were then heated at 42°C for 1.5 h. The cDNA samples were diluted to 8 ng/µL for real-time RT-PCR analysis.
For real-time RT-PCR, triplicate quantitative assays were performed on 1µL of each cDNA dilution using the SYBR Green Master Mix (Applied Biosystems, PN 4309155) with an ABI 7900 sequence detection system according to the manufacturer’s protocol (Applied Biosystems). The gene-specific primers were designed by using PRIMER3 (http://frodo.wi.mit.edu/primer3/input.htm). The amplification of 18S rRNA was used as an internal control to normalize all data (forward primer, 5’-CGGCTACCACATCCAAGGAA-3’; reverse primer, 5’- TGTCACTACCTCCCCGTGTCA-3’). Gene-specific primers are listed in Table S2. The relative quantification method (ΔΔCT) was used to evaluate quantitative variation between replicates examined.
Affymetrix GeneChip Analysis
For each sample, 8 μg of total RNA was used for making biotin-labeled cRNA targets. All processes concerning cDNA and cRNA synthesis, cRNA fragmentation, hybridization, washing and staining, and scanning, followed the GeneChip Standard Protocol (Eukaryotic Target Preparation). In this experiment, Poly-A RNA Control Kit and the One-Cycle cDNA Synthesis kit were applied. Affymetrix GCOS software was used for data normalization and comparative analysis. The change of expression level for each probe set between each Ubi::OsSPX1-antisense transgenic line and WT rice sample was calculated by MAS5 algorithm through GCOS baseline tool. One-sample t-test was applied onto the log2ratio values to identify the differentially expressed probe sets.
In order to map the probe set ID to the locus ID in the rice genome, the consensus sequence of each probe set was compared by BLAST (Basic Local Alignment and Search Tool) against the TIGR Rice Genome version 5. The cut-off e-value was set as 1e-20. EasyGO [31] tool was used for gene functional categorization.
Chlorophyll content and H2O2/peroxidase measurements
Chlorophyll relative content was measured using a SPAD-502 Chlorophyll Meter. Each rice leaf was measured 3-4 times, and about 20 leaves were detected and calculated.
Five days after 10 μM MV treatment, staining with DAB (a H2O2 staining agent) was performed as described in the references 50,51. Seedlings were stained with DAB for 12 h in the darkness, then destained with acetic acid/glycerol/ethanol (1:1:3) at 100°C for 15 min. Representative leaves were viewed under a stereo fluorescence microscope.
Using the Amplex Red hydrogen peroxide/peroxidase assay kit (Molecular Probes), we followed the manufacturer’s instructions to measure the quantity of H2O2 production in rice leave samples and the peroxidase activity in rice samples.
Supporting Information
Schematic diagram of the draft structure of binary vector structures used for rice transformation. Construction scheme of plasmids with OsSPX1 in sense and antisense orientation.
A: The binary vector pCOU OsSPX1-antisense was used for transgenic rice transformation.
B: The binary vector pCOU OsSPX1-sense was used for transgenic rice transformation, which was adapted from our previous work [9].
LB and RB correspond to the T-DNA left and right borders.
(JPG)
The 1266 probe sets showing differential expression between Ubi::OsSPX1-antisense transgenic lines and WT seedlings. Including the raw intensity, log2ratio, p-value, and additional annotation of each probe set.
(XLS)
Primer list of probe sets for real-time RT-PCR.
(DOC)
Cold-stress related probe sets showing differential expression between Ubi::OsSPX1-antisense transgenic lines and WT seedlings under cold treatment. Including the raw intensity, log2ratio, p-value, and additional annotation of each probe set.
(XLS)
Acknowledgments
We thank Qunlian Zhang, Qi You, Liwei Zhang, and Xin Yi for their technical support.
Funding Statement
This work was supported by grants from the Ministry of Science and Technology of China (31071125) and the Ministry of Education of China (NCET-09-0735). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Spain BH, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J (1995) Truncated forms of a novel yeast protein suppress the lethality of a G protein alpha subunit deficiency by interacting with the beta subunit. J Biol Chem 270: 25435-25444. doi: 10.1074/jbc.270.43.25435. PubMed: 7592711. [DOI] [PubMed] [Google Scholar]
- 2. Lenburg ME, O'Shea EK (1996) Signaling phosphate starvation. Trends Biochem Sci 21: 383-387. doi: 10.1016/S0968-0004(96)10048-7. PubMed: 8918192. [DOI] [PubMed] [Google Scholar]
- 3. Lee M, O'Regan S, Moreau JL, Johnson AL, Johnston LH et al. (2000) Regulation of the Pcl7-Pho85 cyclin-cdk complex by Pho81. Mol Microbiol 38: 411-422. doi: 10.1046/j.1365-2958.2000.02140.x. PubMed: 11069666. [DOI] [PubMed] [Google Scholar]
- 4. Voicu PM, Petrescu-Danila E, Poitelea M, Watson AT, Rusu M (2007) In Schizosaccharomyces pombe the 14-3-3 protein Rad24p is involved in negative control of pho1 gene expression. Yeast 24: 121-127. doi: 10.1002/yea.1433. PubMed: 17173334. [DOI] [PubMed] [Google Scholar]
- 5. Tailor CS, Nouri A, Lee CG, Kozak C, Kabat D (1999) Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci U S A 96: 927-932. doi: 10.1073/pnas.96.3.927. PubMed: 9927670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Battini JL, Rasko JE, Miller AD (1999) A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci U S A 96: 1385-1390. doi: 10.1073/pnas.96.4.1385. PubMed: 9990033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hürlimann HC, Pinson B, Stadler-Waibel M, Zeeman SC, Freimoser FM (2009) The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep 10: 1003-1008. doi: 10.1038/embor.2009.105. PubMed: 19590579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan K, Yi K, Dang L, Huang H, Wu W et al. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965-975. doi: 10.1111/j.1365-313X.2008.03460.x. PubMed: 18315545. [DOI] [PubMed] [Google Scholar]
- 9. Zhao L, Liu F, Xu W, Di C, Zhou S et al. (2009) Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol J 7: 550-561. doi: 10.1111/j.1467-7652.2009.00423.x. PubMed: 19508276. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Ribot C, Rezzonico E, Poirier Y (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135: 400-411. doi: 10.1104/pp.103.037945. PubMed: 15122012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889-902. doi: 10.1105/tpc.000745. PubMed: 11971143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Secco D, Poirier Y (2008) Characterization of the PHO1 gene family and the responses to phosphate deficiency of Physcomitrella patens. Plant Physiol 146: 646-656. PubMed: 18055586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J et al. (2007) Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J 50: 982-994. doi: 10.1111/j.1365-313X.2007.03108.x. PubMed: 17461783. [DOI] [PubMed] [Google Scholar]
- 14. Ribot C, Wang Y, Poirier Y (2008) Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 227: 1025-1036. doi: 10.1007/s00425-007-0677-x. PubMed: 18094993. [DOI] [PubMed] [Google Scholar]
- 15. Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988-999. doi: 10.1104/pp.106.079707. PubMed: 16679424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Yi K, Tao Y, Wang F, Wu Z et al. (2006) Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant Cell Environ 29: 1924-1935. doi: 10.1111/j.1365-3040.2006.01568.x. PubMed: 16930318. [DOI] [PubMed] [Google Scholar]
- 17. Wang C, Ying S, Huang H, Li K, Wu P et al. (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895-904. doi: 10.1111/j.1365-313X.2008.03734.x. PubMed: 19000161. [DOI] [PubMed] [Google Scholar]
- 18. Wang Z, Hu H, Huang H, Duan K, Wu Z et al. (2009) Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol 51: 663-674. doi: 10.1111/j.1744-7909.2009.00834.x. PubMed: 19566645. [DOI] [PubMed] [Google Scholar]
- 19. Liu F, Wang Z, Ren H, Shen C, Li Y et al. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508-517. doi: 10.1111/j.1365-313X.2010.04170.x. PubMed: 20149131. [DOI] [PubMed] [Google Scholar]
- 20. Misson J, Raghothama KG, Jain A, Jouhet J, Block MA et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci U S A 102: 11934-11939. doi: 10.1073/pnas.0505266102. PubMed: 16085708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nilsson L, Müller R, Nielsen TH (2010) Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol Plant 139: 129-143. doi: 10.1111/j.1399-3054.2010.01356.x. PubMed: 20113436. [DOI] [PubMed] [Google Scholar]
- 22. Jia H, Ren H, Gu M, Zhao J, Sun S et al. (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164-1175. doi: 10.1104/pp.111.175240. PubMed: 21502185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Jiao F, Wu Z, Li Y, Wang X et al. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673-1686. doi: 10.1104/pp.107.111443. PubMed: 18263782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu B, Zhu C, Li F, Tang J, Wang Y et al. (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101-1115. doi: 10.1104/pp.110.170209. PubMed: 21317339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hurry V, Strand A, Furbank R, Stitt M (2000) The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. Plant J 24: 383-396. doi: 10.1046/j.1365-313x.2000.00888.x. PubMed: 11069711. [DOI] [PubMed] [Google Scholar]
- 26. Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847-856. doi: 10.1104/pp.105.068254. PubMed: 16183833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller G, Shulaev V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481-489. doi: 10.1111/j.1399-3054.2008.01090.x. PubMed: 18346071. [DOI] [PubMed] [Google Scholar]
- 28. Mittler R, Kim Y, Song L, Coutu J, Coutu A et al. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580: 6537-6542. doi: 10.1016/j.febslet.2006.11.002. PubMed: 17112521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M et al. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734-2746. doi: 10.1104/pp.104.046599. PubMed: 15333755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K et al. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141-152. doi: 10.1016/j.molcel.2004.06.023. PubMed: 15225555. [DOI] [PubMed] [Google Scholar]
- 31. Zhou X, Su Z (2007) EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics 8: 246. doi: 10.1186/1471-2164-8-246. PubMed: 17645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu F, Xu W, Wei Q, Zhang Z, Xing Z et al. (2010) Gene expression profiles deciphering rice phenotypic variation between Nipponbare (Japonica) and 93-11 (Indica) during oxidative stress. PLOS ONE 5: e8632. doi: 10.1371/journal.pone.0008632. PubMed: 20072620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim CE, Choi JN, Kim IA, Lee SA, Hwang YS et al. (2008) Improved resistance to oxidative stress by a loss-of-function mutation in the Arabidopsis UGT71C1 gene. Mol Cells 25: 368-375. PubMed: 18443422. [PubMed] [Google Scholar]
- 34. Shin JH, Yoshimoto K, Ohsumi Y, Jeon JS, An G (2009) OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol Cells 27: 67-74. doi: 10.1007/s10059-009-0006-2. PubMed: 19214435. [DOI] [PubMed] [Google Scholar]
- 35. MacIntosh GC, Hillwig MS, Meyer A, Flagel L (2010) RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol Genet Genomics 283: 381-396. doi: 10.1007/s00438-010-0524-9. PubMed: 20182746. [DOI] [PubMed] [Google Scholar]
- 36. Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ (1993) RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci U S A 90: 5118-5122. doi: 10.1073/pnas.90.11.5118. PubMed: 8506358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang L, Lai Z, Ma W, Zhang Y, Xue Y (2002) AhSL28, a senescence- and phosphate starvation-induced S-like RNase gene in Antirrhinum. Biochim Biophys Acta 1579: 64-71. doi: 10.1016/S0167-4781(02)00507-9. PubMed: 12401221. [DOI] [PubMed] [Google Scholar]
- 38. Wingsle G, Hallgren J-E (1993) Influence of SO2 and NO2 Exposure on Glutathione, Superoxide Dismutase and Glutathione Reductase Activities in Scots Pine Needles. Journal of Experimental Botany: 463-470. [Google Scholar]
- 39. Marrs KA (1996) The Functions and Regulation of Glutathione S-Transferases in Plants. Annu Rev Plant Physiol Plant Mol Biol 47: 127-158. doi: 10.1146/annurev.arplant.47.1.127. PubMed: 15012285. [DOI] [PubMed] [Google Scholar]
- 40. Kandel S, Morant M, Benveniste I, Blée E, Werck-Reichhart D et al. (2005) Cloning, functional expression, and characterization of CYP709C1, the first sub-terminal hydroxylase of long chain fatty acid in plants. Induction by chemicals and methyl jasmonate. J Biol Chem 280: 35881-35889. doi: 10.1074/jbc.M500918200. PubMed: 16120613. [DOI] [PubMed] [Google Scholar]
- 41. Kong L, Anderson JM, Ohm HW (2005) Induction of wheat defense and stress-related genes in response to Fusarium graminearum. Genome 48: 29-40. doi: 10.1139/g04-097. PubMed: 15729394. [DOI] [PubMed] [Google Scholar]
- 42. Asada K (1999) THE WATER-WATER CYCLE IN CHLOROPLASTS: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601-639. doi: 10.1146/annurev.arplant.50.1.601. PubMed: 15012221. [DOI] [PubMed] [Google Scholar]
- 43. de Pinto MC, Paradiso A, Leonetti P, De Gara L (2006) Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. Plant J 48: 784-795. doi: 10.1111/j.1365-313X.2006.02919.x. PubMed: 17092315. [DOI] [PubMed] [Google Scholar]
- 44. Wang Y, Peng X, Xu W, Luo Y, Zhao W et al. (2012) Transcript and protein profiling analysis of OTA-induced cell death reveals the regulation of the toxicity response process in Arabidopsis thaliana. J Exp Bot 63: 2171-2187. PubMed: 22207617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Secco D, Wang C, Arpat BA, Wang Z, Poirier Y et al. (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193: 842-851. doi: 10.1111/j.1469-8137.2011.04002.x. PubMed: 22403821. [DOI] [PubMed] [Google Scholar]
- 46. Koo AJ, Cooke TF, Howe GA (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci U S A 108: 9298-9303. doi: 10.1073/pnas.1103542108. PubMed: 21576464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P et al. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287: 6296-6306. doi: 10.1074/jbc.M111.316364. PubMed: 22215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523-3543. doi: 10.1093/jxb/ers100. PubMed: 22467407. [DOI] [PubMed] [Google Scholar]
- 49. Toki S, Hara N, Ono K, Onodera H, Tagiri A et al. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969-976. doi: 10.1111/j.1365-313X.2006.02836.x. PubMed: 16961734. [DOI] [PubMed] [Google Scholar]
- 50. Jambunathan N (2010) Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol Biol 639: 292-298. PubMed: 20387054. [DOI] [PubMed] [Google Scholar]
- 51. Jiang J, Su M, Wang L, Jiao C, Sun Z et al. (2012) Exogenous hydrogen peroxide reversibly inhibits root gravitropism and induces horizontal curvature of primary root during grass pea germination. Plant Physiol Biochem 53: 84-93. doi: 10.1016/j.plaphy.2012.01.017. PubMed: 22342943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic diagram of the draft structure of binary vector structures used for rice transformation. Construction scheme of plasmids with OsSPX1 in sense and antisense orientation.
A: The binary vector pCOU OsSPX1-antisense was used for transgenic rice transformation.
B: The binary vector pCOU OsSPX1-sense was used for transgenic rice transformation, which was adapted from our previous work [9].
LB and RB correspond to the T-DNA left and right borders.
(JPG)
The 1266 probe sets showing differential expression between Ubi::OsSPX1-antisense transgenic lines and WT seedlings. Including the raw intensity, log2ratio, p-value, and additional annotation of each probe set.
(XLS)
Primer list of probe sets for real-time RT-PCR.
(DOC)
Cold-stress related probe sets showing differential expression between Ubi::OsSPX1-antisense transgenic lines and WT seedlings under cold treatment. Including the raw intensity, log2ratio, p-value, and additional annotation of each probe set.
(XLS)