Abstract
Beckwith-Wiedemann syndrome (BWS) is a congenital cancer-predisposition syndrome associated with embryonal cancers, macroglossia, macrosomia, ear pits or ear creases, and midline abdominal-wall defects. The most common constitutional abnormalities in BWS are epigenetic, involving abnormal methylation of either H19 or LIT1, which encode untranslated RNAs on 11p15. We hypothesized that different epigenetic alterations would be associated with specific phenotypes in BWS. To test this hypothesis, we performed a case-cohort study, using the BWS Registry. The cohort consisted of 92 patients with BWS and molecular analysis of both H19 and LIT1, and these patients showed the same frequency of clinical phenotypes as those patients in the Registry from whom biological samples were not available. The frequency of altered DNA methylation of H19 in patients with cancer was significantly higher, 56% (9/16), than the frequency in patients without cancer, 17% (13/76; P=.002), and cancer was not associated with LIT1 alterations. Furthermore, the frequency of altered DNA methylation of LIT1 in patients with midline abdominal-wall defects and macrosomia was significantly higher, 65% (41/63) and 60% (46/77), respectively, than in patients without such defects, 34% (10/29) and 18% (2/11), respectively (P=.012 and P=.02, respectively). Additionally, paternal uniparental disomy (UPD) of 11p15 was associated with hemihypertrophy (P=.003), cancer (P=.03), and hypoglycemia (P=.05). These results define an epigenotype-phenotype relationship in BWS, in which aberrant methylation of H19 and LIT1 and UPD are strongly associated with cancer risk and specific birth defects.
Introduction
Beckwith-Wiedemann syndrome (BWS [MIM 130650]) is a model for the understanding of the role of epigenetics in cancer, since it involves frequent alterations in the imprinting and methylation of several genes on chromosomal band 11p15, including insulin-like growth factor-II (IGF2 [MIM 147470]), H19 (MIM 103280), p57KIP2 (or CDKN1C [MIM 600856]), KvLQT1 (MIM 192500), and LIT1 (MIM 604115). Genomic imprinting is a gene modification that typically involves DNA methylation but may include other factors as well and that leads to the relative silencing of a specific parental allele in somatic cells of the offspring. The phenotypic features of BWS are macrosomia, neonatal hypoglycemia, midline abdominal defects (e.g., omphalocele, diastasis recti, and umbilical hernia), macroglossia, and ear pits or ear creases, as well as an increased incidence of embryonal cancers in infants and young children. The most frequent cancer is Wilms tumor, occurring in 11% of children with BWS before 4 years of age, followed in frequency by other embryonal tumors, including hepatoblastoma and neuroblastoma (DeBaun and Tucker 1998). Other cancers occurring with lower frequency in this population include rhabdomyosarcoma and adrenocortical carcinoma.
The epigenetics of BWS is complex. Six genetic or epigenetic alterations in patients with BWS have been described:
-
1.
Loss of imprinting (LOI) of IGF2 in embryonal tumors and in the normal cells of patients with BWS, which, in BWS, involves activation of the normally silent maternally inherited allele of IGF2 (Ogawa et al. 1993; Rainier et al. 1993; Weksberg et al. 1993). Most investigators report a frequency of ∼10%–15% (Elias et al. 1998; Lee et al. 1999), but one study suggests a higher frequency (Joyce et al. 1997).
-
2.
Abnormal methylation of the normally unmethylated allele of a differentially methylated region (DMR) upstream from H19 (Steenman et al. 1994; Reik et al. 1995). The H19 DMR serves as a site for the binding of the insulator protein CTCF, and it inhibits access of the maternal allele of IGF2 to a shared enhancer downstream from H19 (reviewed by Feinberg [2001]). In the case of Wilms tumor, LOI of IGF2 is coupled to abnormal H19 methylation (Moulton et al. 1994; Steenman et al. 1994). In the case of BWS, there may also be an independent mechanism for regulating IGF2 in these patients (Joyce et al. 1997). The frequency of this alteration is 15% (reviewed by Elias et al. [1998]).
-
3.
Germline chromosomal rearrangements. We showed that balanced rearrangements interrupt the KvLQT1 gene (Lee et al. 1997b), and there are also rare unbalanced duplications (reviewed by Elias et al. [1998]).
-
4.
Mutations of the p57KIP2 gene, which are rare (Lee et al. 1997a; O’Keefe et al. 1997).
-
5.
LOI of LIT1, shown directly by us (Lee et al. 1999) and indirectly by others (Smilinich et al. 1999). LIT1 is an antisense transcript that is normally expressed from the paternal allele and lies within the KvLQT1 gene (Lee et al. 1999; Smilinich et al. 1999), and LOI of LIT1 is completely linked to abnormal methylation of a DMR upstream from the gene (Lee et al. 1999). This is the most frequent abnormality in BWS, accounting for 40%–50% of patients.
-
6.
Paternal uniparental disomy (UPD) of 11p15, occurring in ∼10% of patients (reviewed by Elias et al. [1998]).
We hypothesized that there may be a relationship between cancer risk and LOI of IGF2 and/or aberrant methylation of the H19 DMR, because we and others have described both defects in the general population, in the same type of embryonal cancers found in children with BWS (Ogawa et al 1993; Rainier et al. 1993; Steenman et al. 1994). In contrast, we and others have found no alterations in LIT1, p57KIP2, or KvLQT1 in embryonal tumors (Reid et al. 1996; Mitsuya et al. 1999; M. P. Lee and A. P. Feinberg, unpublished data), suggesting specificity of the H19/IGF2 subdomain in embryonal tumors. In preliminary data, we described the association between abnormal H19 methylation and increased cancer risk in BWS (Niemitz et al. 2001). In addition, Bliek et al. (2001) documented an association between aberrant H19 methylation and cancer; however, in that study, it was unclear whether cancer was specific for abnormal H19 methylation or UPD, as there were only two cancer patients with abnormal H19 methylation in the absence of UPD. Additionally, Engel et al. (2000) described a 69% (20 of 29) frequency of midline abdominal-wall defects in patients with abnormal methylation of LIT1, but they did not compare this frequency to all patients with BWS without abnormal LIT1 methylation, so it cannot be determined whether this frequency is elevated. Because of small numbers of patients in both studies, discrimination of phenotypic association for UPD versus the individual affected genes could not be done. Both studies most likely had strong ascertainment bias, since children with either cancer or midline abdominal-wall defects who required surgery were probably referred to tertiary care centers. The current study addressed a large non–tertiary-care-center–based registry of children with BWS, and it represents the first report of BWS to examine multiple phenotypes associated with a given epigenotype, allowing the definition of epigenotype-phenotype relationships.
Patients and Methods
Patients
We established the BWS Registry in 1994 at the Genetic Epidemiology Branch of the National Cancer Institute to understand the natural history of this syndrome and to determine whether genotype-phenotype relationships exist. For those families who agreed to participate, a comprehensive questionnaire was completed, and informed consent and medical-information–release documents were obtained, as approved by the respective institutional review boards. For the purposes of this study, we defined a patient as having BWS if a clinical diagnosis of BWS had been made by a physician and the patient had at least two of the five most common features associated with BWS (Pettenati et al. 1986): (1) macroglossia, (2) birth weight and length >90th percentile, (3) hypoglycemia in the first month of life, (4) ear creases or ear pits, and (5) midline abdominal-wall defects (omphalocele, diastasis recti, and umbilical hernia). All positive phenotypic features were diagnosed by a physician and were documented in the questionnaire. The local physician defined idiopathic hemihypertrophy. In four patients, birth weight and length were unknown, and, thus, macrosomia could not be scored in those four patients. If a cancer was reported, the diagnosis was confirmed by review of the final pathology report and medical records. Among the patients studied, there were 16 embryonal tumors, including 13 Wilms tumors, 2 hepatoblastomas, and 1 rhabdomyosarcoma. The genotypes of all patients were not known prior to entering the registry. All molecular analyses were performed blinded for knowledge of the phenotype.
Isolation of DNA
Genomic DNA was prepared from peripheral-blood lymphocytes (2/3 of patients) or cultured skin fibroblasts (1/3 of patients) by standard proteinase K digestion and phenol extraction (Cui et al. 1998). This material is suitable for methylation assays involving DNA.
Analysis of H19 Methylation
A 1.8-kb PstI fragment of the H19 gene (nucleotides 1–1803 of the human H19 probe [GenBank accession number M32053]) was analyzed for DNA methylation by digestion with PstI and SmaI. Ten micrograms of DNA were digested at 25°C overnight with 40 U of SmaI, followed by an additional incubation at 37°C for 4 h with 40 U of PstI. The digested DNA was electrophoresed on a 1% agarose gel, was transferred to Hybond-N+ (Amersham), and was hybridized with the 1-kb PstI + SmaI fragment isolated from an H19 genomic clone and oligolabeled with α-[32P]-dATP. Signals were quantified using a PhosphorImager (Molecular Dynamics). Abnormal methylation in lymphocytes was defined as a methylation index of >0.63 (mean + 2 SD of 15 normal individuals). DNA samples were available from blood or fibroblasts but not both from any given patient. Fibroblasts from normal individuals (n=5) showed a methylation index of ∼0.33. All of the BWS fibroblast samples showed a methylation index of ∼0.33 (scored as normal) or ∼0.66 (scored as abnormal). There was no difference in the frequency of increased methylation when comparing BWS samples from lymphocytes or BWS samples from fibroblasts. Furthermore, the association between abnormal H19 methylation and cancer was unchanged when the fibroblast samples were excluded.
Analysis of LIT1 Methylation
Ten micrograms of DNA were digested with BamHI and NotI at 37°C overnight. The digested DNA was electrophoresed on a 1% agarose gel, was transferred to Hybond-N+ (Amersham), and was hybridized with an expressed-sequence–tag probe (GenBank accession number AA155639) oligolabeled with α-[32P]-dATP. Signals were quantified using a PhosphorImager (Molecular Dynamics). Abnormal methylation was defined as a methylation index of <0.39 (mean − 2 SD of 15 normal individuals). The methylation index of LIT1 was unchanged in cultured skin fibroblasts.
Analysis of UPD
Patients who were determined to have epigenetic alterations at both BWS loci tested (i.e., to have hypermethylation of H19 and hypomethylation of LIT1) were evaluated for paternal UPD of 11p15.5. Patient and maternal DNA samples were genotyped with a panel of three microsatellite markers: D11S1318, D11S988, and D11S922. PCR primer sequences were obtained from the Genome Database. PCR was performed as recommended by the Genome Database by use of FAM-labeled primer pairs. PCR products were then analyzed by electrophoresis on a model 377 automated fluorescent DNA sequencer (Applied Biosystems). The raw data were analyzed using Genescan software (Applied Biosystems). The peak data generated by Genescan were then analyzed using the Genotyper program (Applied Biosystems). Patients in whose DNA the Genescan software indicated that the maternal allele(s) was absent were considered to have UPD; however, examination of the chromatograms for those patients revealed a small peak (<20% of the scored allele), corresponding to one of the maternal alleles and thereby indicating mosaicism for UPD for each patient.
Statistical Analysis
A χ2 statistic and Fisher's exact test were used to determine whether specific epigenotypes (aberrant methylation of either H19 or LIT1 or UPD of 11p15) were associated with the presence of specific phenotypes in children with BWS. When the expected value in a given cell was <5, Fisher's exact test was used (Glantz 1992). A P value <.05 was considered statistically significant. In addition, the odds ratio for cancer was calculated and was considered to be significantly increased if it was >1.0 and if the 95% confidence intervals (95%CIs) did not include 1.0.
Results
Comparison of Patients with BWS, with and without Molecular Analysis
A total of 279 children have met the criteria to be included in the BWS Registry, of whom material could be obtained from 92 for analysis of both LIT1 and H19 methylation and UPD. The average age of patients who provided biological specimens was 5.6 years, and the age range was 1 mo–30 years. Of the patients, 52% were male, and, of the parents, 87% identified themselves as white, 2% identified themselves as African American, 7% identified themselves as Hispanic, 2% identified themselves as Asian, and 2% identified themselves as Native American. No significant phenotypic differences were identified between patients who were molecularly analyzed and those who were not, including frequency of cancer, macroglossia, hypoglycemia, abdominal-wall defects, macrosomia, ear pits or creases, and hemihypertrophy (table 1). These findings indicate that the cohort of children with biological specimens in the BWS Registry were representative of the entire BWS Registry.
Table 1.
Comparison of Patients with BWS, with and without Molecular Analysis
| % With Phenotype |
|||
| Phenotype | WithMolecularAnalysis(n = 92) | WithoutMolecularAnalysis(n = 187) | Pa |
| Macrosomia | 88 | 53 | .25 |
| Ear grooves/pits | 63 | 71 | .17 |
| Abdominal-wall defects | 68 | 76 | .15 |
| Hypoglycemia | 53 | 52 | .83 |
| Macroglossia | 97 | 92 | .16 |
| Hemihypertrophy | 36 | 32 | .47 |
| Embryonal cancer | 16 | 19 | .08 |
Calculated by χ2 analysis.
Frequency of Genotypes
The most frequent molecular alteration observed in children with BWS was altered LIT1 methylation with normal H19 methylation (42% [39/92 patients]). The frequency of altered H19 methylation with normal LIT1 methylation was 11% (10/92 patients). Twelve patients (13%) exhibited altered methylation of both H19 and LIT1. We expected that these patients would show paternal UPD, since, in UPD, both maternal copies are replaced with a paternal copy that alters the methylation pattern of both genes. Unexpectedly, of these 12 patients, 9 showed UPD, and the remaining 3 exhibited altered methylation of both H19 and LIT1 in the absence of UPD. These patients therefore had an imprinting disturbance extending throughout the 11p15 imprinted-gene domain, which has not been described elsewhere.
Association of Cancer with Altered H19 Methylation
Abnormal H19 methylation was associated with an elevated risk for embryonal cancer (table 2). The frequency of altered H19 methylation in patients with embryonal cancer was significantly greater than that in patients without cancer—56% (9/16) versus 17% (13/76), respectively (P=.002). The odds ratio for cancer in children with altered H19 methylation was 6.2 (95%CI 1.7–19.8). In contrast, altered LIT1 methylation was not associated with an elevated risk for embryonal cancer (table 2). We also determined whether the increased frequency of altered H19 methylation in patients with cancer was specific for H19 or was simply due to UPD or an imprinting disturbance extending throughout the 11p15 imprinted-gene domain. To address this possibility, we excluded patients with UPD (n=9) or with aberrant methylation of both H19 and LIT1 (n=3) from the analysis; with the exclusion of these 12 patients, the frequency of altered H19 methylation among patients with embryonal cancer was still significantly higher than that in patients without cancer—36% (4/11) versus 9% (6/69), respectively (P=.01). The odds ratio for cancer in children with altered H19 methylation—6.0 (95%CI 1.3–26.5)—also did not change significantly when these 12 patients were excluded, providing strong evidence that the risk of cancer was specific for abnormal H19 methylation and was not simply due to UPD. Therefore, abnormal H19 methylation appears to be specifically associated with increased cancer risk in BWS.
Table 2.
Significant Relationships between Phenotype and Molecular Abnormalities in BWS
|
Abnormal H19 |
Abnormal LIT1 |
UPDa |
||||
| Phenotype | Frequency | P | Frequency | P | Frequency | P |
| Cancer: | ||||||
| Present | 9/16 (56%) | 6/16 (38%) | 5/16 (31%) | |||
| Absent | 13/76 (17%) | .002b | 45/76 (59%) | .19 | 7/76 (9%) | .03b |
| Macrosomia: | ||||||
| Present | 18/77 (23%) | 46/77 (60%) | 11/77 (14%) | |||
| Absent | 2/11 (18%) | 1.0b | 2/11 (18%) | .02 | 0/11 (0%) | .3b |
| Hypoglycemia: | ||||||
| Present | 14/49 (29%) | 28/49 (57%) | 10/49 (20%) | |||
| Absent | 8/43 (19%) | .4 | 23/43 (53%) | .9 | 2/43 (5%) | .05b |
| Hemihypertrophy: | ||||||
| Present | 12/33 (36%) | 19/33 (58%) | 10/33 (30%) | |||
| Absent | 10/59 (17%) | .07c | 32/59 (54%) | .9 | 2/49 (4%) | .003b |
| Midline defects: | ||||||
| Present | 14/63 (22%) | 41/63 (65%) | 8/63 (13%) | |||
| Absent | 8/29 (28%) | .76 | 10/29 (34%) | .012 | 4/29 (14%) | 1.0b |
Plus cases of abnormal methylation of both H19 and LIT1.
Determined by Fisher's exact test.
P=.5 if patients with UPD are excluded; thus, there is no independent association with H19 methylation.
Association of Macrosomia and Midline Abdominal-Wall Defects with Altered Methylation of LIT1
We examined the association between molecular abnormalities in BWS and macrosomia, hypoglycemia, macroglossia, ear pits or ear creases, hemihypertrophy, and midline abdominal-wall defects. The frequency of altered LIT1 methylation in patients with macrosomia was significantly higher than that in patients without macrosomia—60% (46/77) versus 18% (2/11), respectively (P=.02; table 2). Similarly, the frequency of altered LIT1 methylation in patients with midline abdominal-wall defects was significantly higher than that in patients without such defects—65% (41/63) versus 34% (10/29), respectively (P=.012; table 2). For both macrosomia and midline abdominal-wall defects, we also determined whether the increased frequency of altered LIT1 methylation was simply due to UPD or an imprinting disturbance extending throughout the 11p15 imprinted-gene domain. To address this possibility, we excluded patients with either UPD or aberrant methylation of both H19 and LIT1 from the analysis of macrosomia (n=11) and midline abdominal-wall defects (n=12). Excluding these patients, the frequency of altered LIT1 methylation among patients with macrosomia was still significantly higher than that in patients without macrosomia—53% (35/66) versus 18% (2/11), respectively (P=.03). Similarly, the frequency of altered LIT1 methylation among patients with midline abdominal-wall defects remained significantly higher than that in patients without such defects—60% (33/55) versus 24% (6/25), respectively (P=.003). Thus, the association between macrosomia and midline abdominal-wall defects was specific for abnormal LIT1 methylation and was not simply due to UPD. Therefore, abnormal LIT1 methylation appears to be specifically associated with macrosomia and midline abdominal-wall defects in BWS.
Association of Hemihypertrophy and Hypoglycemia with Altered Methylation of Both LIT1 and H19
We determined whether the 12 patients with aberrant methylation of both H19 and LIT1 (including the 9 patients with UPD) uniquely differed in phenotype in some way—that is, we determined whether there were any features specifically associated with such a domainwide alteration that were not found in association with H19 or LIT1 alone. We found that hemihypertrophy and hypoglycemia occurred significantly more frequently in these patients. The frequency of hemihypertrophy among patients with UPD or aberrant methylation of both H19 and LIT1 was significantly higher than that in patients without such defects—30% (10/33) versus 4% (2/49), respectively (P = .003). Similarly, the frequency of hypoglycemia was 20% (10/49) and 5% (2/43), respectively (P = .05). Thus, in BWS, abnormal methylation of both H19 and LIT1 appears to be specifically associated with hemihypertrophy and hypoglycemia. As expected, in these 12 patients, there was also a significantly increased frequency of cancer (table 2), since cancer is associated with abnormal methylation of only H19 (table 2). Somewhat surprisingly, that was not the case for macrosomia or midline abdominal-wall defects, which showed no significant increase in frequency in these 12 patients, even though these abnormalities are associated with abnormal methylation of only LIT1 (table 2).
Discussion
BWS is a model for the study of epigenetics in human disease, since it involves multiple imprinted genes, as well as both increased risk of cancer and a wide variety of birth defects. BWS offers a unique opportunity to understand the relationship between epigenotype and phenotype, because of its genetic heterogeneity and substantial variability in expressivity of the phenotype. The large number of patients in this study and their complete phenotypic assessment allowed for subgroup analysis to assess specific epigenotype-phenotype relationships. We focused this investigation on epigenetic changes in this disorder—specifically, altered methylations of H19 and LIT1, which account for most of the known alterations in these patients. There were three significant epigenotype-phenotype associations revealed by this analysis: (1) cancer is significantly associated with altered methylation of H19; (2) macrosomia and midline abdominal-wall defects are significantly associated with altered LIT1 methylation; and (3) hypoglycemia and hemihypertrophy are significantly associated with UPD or altered methylation of both LIT1 and H19.
The association, in BWS, between paternal UPD of 11p15 and cancer was initially suggested by Henry et al. (1993), in a small case series in which three of six patients with UPD had cancer. Similarly, because of the small number of patients with cancer (n=7), Bliek et al. (2001) could not perform a statistical analysis distinguishing the risk of cancer with UPD from the risk of cancer with abnormal H19 methylation. Here, we show that this risk is specific for abnormal H19 methylation.
Given that the epigenetic alteration associated with cancer risk in BWS is altered methylation of the H19 DMR, what might be the mechanism for cancer in these patients? The only known function of H19 is the regulation of IGF2 expression by the H19 DMR, since knockout of the H19 RNA itself has no effect in mice (Schmidt et al. 1999). We and others previously showed that sporadically occurring embryonal tumors (i.e., those not occurring in the context of BWS) undergo LOI of IGF2 (Ogawa et al. 1993; Rainier et al. 1993). We found that LOI of IGF2, which involves activation of the normally silent maternal allele, is often coupled, in these tumors, to abnormal methylation of the maternal allele of the H19 DMR (Moulton et al. 1994; Steenman et al. 1994). This methylation appears to inhibit binding of CTCF or of other insulator proteins to the normally unmethylated maternal H19 DMR, thus allowing access by a shared enhancer to the IGF2 promoter and thereby causing biallelic expression of IGF2 (reviewed by Feinberg [2001]). We hypothesized that a similar mechanism may be at work in patients with constitutional epigenetic alterations (i.e., in patients with BWS). Our study supports a role for the H19 DMR in the pathogenesis of human cancer. The human H19 DMR extends from the promoter to 6 kb upstream from H19, and the region examined here is the same (promoter region) as that identified earlier by us and others as showing abnormal methylation in Wilms tumor (Moulton et al. 1994; Steenman et al. 1994). We have found that altered methylation in Wilms tumor also includes CTCF-binding sites 5 kb upstream and within the DMR (Cui et al. 2001), and it will be of interest in further studies to map these sites precisely in BWS.
IGF2 imprinting could not be directly analyzed in most patients, since it would require examination of expressing tissues as well as informativeness for the requisite polymorphic markers to distinguish individual alleles. One problem with examining IGF2 in cultured fibroblasts is that we have found IGF2 to undergo loss of imprinting in cultured normal fibroblasts (E. L. Niemitz and A. P. Feinberg, unpublished data), which may explain the apparent discordance of IGF2 imprinting and H19 methylation reported by Joyce et al. (1997). In the study by Bliek et al. (2001), the source of DNA was not given. We also found that H19 methylation, but not LIT1 methylation, is reduced in cultured skin fibroblasts; however, in the present study, there was no difference in the frequency of increased H19 methylation among samples from patients with BWS derived from blood or from cultured skin fibroblasts, and thus altered imprinting of IGF2 may be methylation independent—at least at the H19 DMR. Note that the source of DNA did not affect the results, since the association between increased H19 methylation and cancer was seen even if the patients with fibroblast samples were excluded.
Given that, in BWS, the epigenetic alteration associated with midline abdominal-wall defects and macrosomia is altered LIT1 methylation, what is the mechanism for such congenital malformations in these patients? The function of LIT1 is unknown, although we have previously hypothesized that abnormal hypomethylation of LIT1 might cause the silencing of the nearby p57KIP2 gene, by a mechanism similar to the silencing of IGF2 by the H19 DMR (Lee et al. 1999). In support of this hypothesis, p57KIP2 is mutated in a small fraction of patients with BWS (Lee et al. 1997a; O'Keefe et al. 1997). Furthermore, knockout mice lacking a functional p57KIP2 gene show similar midline abdominal-wall defects (Zhang et al. 1997). As with IGF2, direct examination of p57KIP2 imprinting would require analysis of expressing tissues, such as kidney and tongue, which are not readily available in a large cohort of patients.
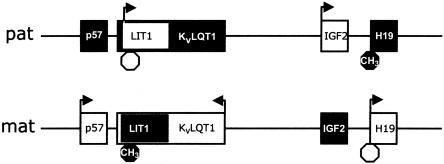
These observations fit very well a two-domain hypothesis for BWS we have proposed elsewhere (Lee et al. 1999). By this model, 11p15 contains two separate imprinted subdomains (fig. 1). The more telomeric subdomain includes IGF2 and H19, and the more centromeric subdomain includes p57KIP2, LIT1, and KvLQT1. BWS arises by the disrupted imprinting of either subdomain. The likely target gene for BWS is IGF2 in the more telomeric subdomain and p57KIP2 in the more centromeric subdomain. By this model, a DMR lies between a given BWS target gene and its enhancer, and abnormal hypermethylation permits access of this enhancer to the maternal allele of IGF2 on the maternal allele, while abnormal hypomethylation inhibits access of this enhancer to the maternal allele of p57KIP2. The chromosomal rearrangements in KvLQT1 would have the same effect (Lee et al. 1997b). Support for our model comes from a comparative genomic analysis showing a similar organization in the mouse (Onyango et al. 2001) and from a study indicating that the p57KIP2 enhancer is in the location our model predicts, although the enhancer itself was not identified in that study (Cleary et al. 2001). The data in the current study would suggest that the more centromeric subdomain is involved primarily in growth regulation and that the more telomeric subdomain is involved primarily in tumor suppression.
Figure 1.
One-megabase region of chromosome 11p15.5 containing two imprinted-gene subdomains. The more centromeric subdomain includes p57KIP2, LIT1, and KvLQT1, and the more telomeric subdomain includes IGF2 and H19. Unblackened boxes represent transcriptionally active genes, and arrows indicate the transcriptional orientation; octagons represent the DMRs analyzed here, and blackened octagons indicate methylation. Paternal chromosome is designated by “pat”; maternal chromosome is designated by “mat.” (Not all genes are listed.)
An additional conclusion of this study is that hypoglycemia and hemihypertrophy are associated specifically with UPD or with methylation abnormalities affecting both H19 and LIT1. We address each of these separately:
-
1.
Hemihypertrophy. This is an asymmetric enlargement of one half of the body or a portion thereof. We suggest that hemihypertrophy in BWS is simply the mosaic form of macrosomia, which was associated with altered methylation of LIT1. To date, all of the patients with UPD reported—including those studied here—are mosaic. We predict that there is an asymmetric spatial distribution of UPD on the two sides of the body in these patients.
-
2.
Hypoglycemia. The association between hypoglycemia and UPD (or abnormal methylation of both genes) and the lack of association between hypoglycemia and either methylation of only H19 or only LIT1 suggest that another gene on 11p15 may contribute to this phenotype. Such a gene would also undergo paternal duplication in patients with UPD. What gene could this be? There are several additional imprinted genes on 11p15 (reviewed by Feinberg [1999]). One intriguing possibility is that the insulin gene (INS [MIM 176730]), although not generally thought to be imprinted, may show imprinting in the yolk sac (Moore et al. 2001). It will be of interest to examine the imprinting of the insulin gene in the developing pancreas.
As with any case-cohort study, our analysis is associated with several limitations. We did not obtain biological samples from all patients in the BWS Registry; however, the frequency of cancer and other phenotypic features did not differ significantly between those with and without molecular analysis. Another limitation is that we were not able to assess the relationship between phenotype and p57KIP2 mutations or KvLQT1 rearrangements, because of the low frequency of these alterations; in our previous work and work by others, the frequency of p57KIP2 mutations is 5% (Lee et al. 1997a; O’Keefe et al. 1997), and the frequency of KvLQT1 rearrangements is <1% (reviewed by Elias et al. [1998]). The focus of the present study has been epigenetic alterations, rather than mutations.
Finally, we have here reported the first examples of abnormal methylation that affects both LIT1 and H19, in the absence of UPD. These results suggest that these three patients exhibit disrupted methylation throughout the 11p15 imprinted-gene domain. We postulate that there is a hierarchical epigenetic organization of this domain, with some patients showing epigenetic disruption including that of LIT1, with others showing epigenetic disruption including that of H19, and with a final group showing epigenetic disruption affecting the domain as a whole. Such patients may have “imprinting center” mutations analogous to those seen in some patients with Prader-Willi syndrome (MIM 176270) or Angelman syndrome (MIM 105830) with microdeletions (Buiting et al. 2000).
Acknowledgments
This work was supported by grant CA54358 from the National Institutes of Health (A.P.F.) and by the Robert Wood Johnson Minority Faculty Development Program and the Doris Duke Clinical Scientist Foundation (M.R.D.). We thank T. Litzi for technical assistance.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human H19 probe [accession number M32053] and human LIT1 probe [accession number AA155639])
- Genome Database, The, http://gdbwww.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BWS [MIM 130650], IGF2 [MIM 147470], H19 [MIM 103280], p57KIP2 [MIM 600856], KvLQT1 [MIM 192500], and LIT1 [MIM 604115])
References
- Bliek J, Maas SM, Ruijter JM, Hennekam RCM, Alders M, Westerveld A, Mannens MMAM (2001) Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Hum Mol Genet 10:467–476 [DOI] [PubMed] [Google Scholar]
- Buiting K, Farber C, Kroisel P, Wagner K, Brueton L, Robertson ME, Lich C, Horsthemke B (2000) Imprinting centre deletions in two PWS families: implications for diagnostic testing and genetic counseling. Clin Genet 58:284–290 [DOI] [PubMed] [Google Scholar]
- Cleary MA, van Raamsdonk CD, Levorse J, Zheng B, Bradley A, Tilghman SM (2001) Disruption of an imprinted gene cluster by a targeted chromosomal translocation in mice. Nat Genet 29:78–82 [DOI] [PubMed] [Google Scholar]
- Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP (1998) Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med 4:1276–1280 [DOI] [PubMed] [Google Scholar]
- Cui H, Niemitz EL, Ravenel JD, Onyango P, Brandenburg SA, Lobanenkov VV, Feinberg AP (2001) Loss of imprinting of insulin-like growth factor-II in Wilms' tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res 61:4947–4950 [PubMed] [Google Scholar]
- DeBaun MR, Tucker MA (1998) Risk of cancer during the first four years of life in children from the Beckwith-Wiedemann Syndrome Registry. J Pediatr 132:398–400 [DOI] [PubMed] [Google Scholar]
- Elias ER, DeBaun MR, Feinberg AP (1998) Beckwith-Wiedemann syndrome. In: Jameson JL (ed) Principles of molecular medicine. Humana, Totowa, New Jersey, pp 1047–1052 [Google Scholar]
- Engel JR, Smallwood A, Harper A, Higgins MJ, Oshimura M, Reik W, Schofield PN, Maher ER (2000) Epigenotype-phenotype correlations in Beckwith-Wiedemann syndrome. J Med Genet 37:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP (1999) Imprinting of a genomic domain of 11p15 and loss of imprinting in cancer: an introduction. Cancer Res 59 Suppl:1743s–1746s [PubMed] [Google Scholar]
- Feinberg AP (2001) Cancer epigenetics takes center stage. Proc Natl Acad Sci USA 98:392–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz S (1992) Primer of biostatistics: the program. McGraw-Hill, New York [Google Scholar]
- Henry I, Puech A, Riesewijk A, Ahnine L, Mannens M, Beldjord C, Bitoun P, Tournade MI, Junien C (1993) Somatic mosaicism for partial paternal isodisomy in Wiedemann-Beckwith fertilization event. Eur J Hum Genet 1:19–29 [DOI] [PubMed] [Google Scholar]
- Joyce JA, Lam WK, Catchpoole DJ, Jenks P, Reik W, Maher ER, Schofield PN (1997) Imprinting of IGF2 and H19: lack of reciprocity in sporadic Beckwith-Wiedemann syndrome. Hum Mol Genet 6:1543–1548 [DOI] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP (1999) Loss of imprinting of a paternally expressed transcript, with antisense orientation to KvLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA 96:5203–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, DeBaun M, Richard BA, Elledge SJ, Feinberg AP (1997a) Low frequency of p57KIP2 mutation in Beckwith-Wiedemann syndrome. Am J Hum Genet 61:304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, Hu RJ, Johnson LA, Feinberg AP (1997b) Human KvLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet 15:181–185 [DOI] [PubMed] [Google Scholar]
- Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M (1999) LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet 8:1209–1217 [DOI] [PubMed] [Google Scholar]
- Moore GE, Abu-Amero SN, Bell G, Wakeling EL, Kingsnorth A, Stanier P, Jauniaux E, Bennett ST (2001) Evidence that insulin is imprinted in the human yolk sac. Diabetes 50:199–203 [DOI] [PubMed] [Google Scholar]
- Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T (1994) Epigenetic lesions at the H19 locus in Wilms' tumour patients. Nat Genet 7:440–447 [DOI] [PubMed] [Google Scholar]
- Niemitz EL, Feinberg AP, DeBaun MR (2001) Specific association of altered DNA methylation of the H19 gene with cancer risk in Beckwith-Wiedemann syndrome. In: Proceedings of the American Association of Cancer Research, Vol 42. p 3796 [Google Scholar]
- Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, Smith PJ, Reeve AE (1993) Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature 362:749–751 [DOI] [PubMed] [Google Scholar]
- O'Keefe D, Dao D, Zhao L, Sanderson R, Warburton D, Weiss L, Anyane-Yeboa K, Tycko B (1997) Coding mutations in p57KIP2 are present in some cases of Beckwith-Wiedemann syndrome but are rare or absent in Wilms tumors. Am J Hum Genet 61:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango P, Miller W, Lehoczky J, Leung CT, Birren B, Wheelan S, Dewar K, Feinberg AP (2001) Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res 10:1697–1710 [DOI] [PubMed] [Google Scholar]
- Pettenati MJ, Haines JL, Higgins RR, Wappner RS, Palmer CG, Weaver DD (1986) Wiedemann-Beckwith syndrome: presentation of clinical and cytogenetic data on 22 new cases and review of the literature. Hum Genet 74:143–154 [DOI] [PubMed] [Google Scholar]
- Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP (1993) Relaxation of imprinted genes in human cancer. Nature 362:747–749 [DOI] [PubMed] [Google Scholar]
- Reid LH, Crider-Miller SJ, West A, Lee MH, Massague J, Weissman BE (1996) Genomic organization of the human p57KIP2 gene and its analysis in the G401 Wilms' tumor assay. Cancer Res 56:1214–1218 [PubMed] [Google Scholar]
- Reik W, Brown KW, Schneid H, Le Bouc Y, Bickmore W, Maher ER (1995) Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum Mol Genet 4:2379–2385 [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Levorse JM, Tilghman SM (1999) Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc Natl Acad Sci USA 96:9733–9738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, et al (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA 96:8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenman M, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP (1994) Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumor. Nat Genet 7:433–439 [DOI] [PubMed] [Google Scholar]
- Weksberg R, Shen DR, Fei YL, Song QL, Squire J (1993) Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat Genet 5:143–150 [DOI] [PubMed] [Google Scholar]
- Zhang P, Liegeois NJ, Wong C, Finegold M, Hou H, Thompson JC, Silverman A, Harper JW, DePinho RA, Elledge SJ (1997) Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature 387:151–158 [DOI] [PubMed] [Google Scholar]