Abstract
Rhizomelic chondrodysplasia punctata (RCDP) is a genetically heterogeneous, autosomal recessive disorder of peroxisomal metabolism that is clinically characterized by symmetrical shortening of the proximal long bones, cataracts, periarticular calcifications, multiple joint contractures, and psychomotor retardation. Most patients with RCDP have mutations in the PEX7 gene encoding peroxin 7, the cytosolic PTS2-receptor protein required for targeting a subset of enzymes to peroxisomes. These enzymes are deficient in cells of patients with RCDP, because of their mislocalization to the cytoplasm. We report the mutational spectrum in the PEX7 gene of 78 patients (including five pairs of sibs) clinically and biochemically diagnosed with RCDP type I. We found 22 different mutations, including 18 novel ones. Furthermore, we show by functional analysis that disease severity correlates with PEX7 allele activity: expression of eight different alleles from patients with severe RCDP failed to restore the targeting defect in RCDP fibroblasts, whereas two alleles found only in patients with mild disease complemented the targeting defect upon overexpression. Surprisingly, one of the mild alleles comprises a duplication of nucleotides 45–52, which is predicted to lead to a frameshift at codon 17 and an absence of functional peroxin 7. The ability of this allele to complement the targeting defect in RCDP cells suggests that frame restoration occurs, resulting in full-length functional peroxin 7, which leads to amelioration of the predicted severe phenotype. This was confirmed in vitro by expression of the eight-nucleotide duplication–containing sequence fused in different reading frames to the coding sequence of firefly luciferase in COS cells.
Introduction
Rhizomelic chondrodysplasia punctata (RCDP) is an autosomal recessive peroxisomal disorder with a distinct clinical phenotype consisting of dwarfism due to symmetrical shortening of the proximal long bones (i.e., rhizomelia), cataracts, periarticular calcifications, multiple joint contractures, specific radiological abnormalities, and psychomotor retardation. The rhizomelia distinguishes RCDP clinically from other bone dysplasias. The disorder is genetically heterogeneous, consisting of three groups of patients with defects in different genes. By far the most common of these is RCDP type 1 (MIM 215100), which results from an inability to target proteins that contain a peroxisomal targeting signal type 2 (PTS2) to peroxisomes, because of mutations in PEX7 (GenBank accession numbers AF180806–AF180814), which encodes the cytosolic PTS2-receptor protein peroxin 7 (Braverman et al. 1997; Motley et al. 1997; Purdue et al. 1997). RCDP type 2 (MIM 222765) and type 3 (MIM 600121) are clinically indistinguishable from type 1 but are caused by mutations in the genes encoding the first and second enzyme of ether-phospholipid biosynthesis, respectively. Patients with RCDP type 2 have mutations in the gene that encodes peroxisomal dihydroxyacetonephosphate acyltransferase (Thai et al. 1997; Ofman et al. 1998), and patients with RCDP type 3 have mutations in the gene that encodes peroxisomal alkyl-dihydroxyacetonephosphate synthase (Wanders et al. 1994; de Vet et al. 1998).
Two well-defined targeting signals for directing proteins to the peroxisomal matrix have been identified. Most peroxisomal matrix proteins contain a PTS1 (peroxisome targeting signal type 1), which is a loosely conserved C-terminal tripeptide (Gould et al. 1989; Mullen et al. 1997; Sacksteder and Gould 2000; Subramani et al. 2000). PTS2 is found in only a few peroxisomal proteins and is a bipartite amino acid motif (located on the N terminus), the consensus of which comprises R[L/V/I]X5[H/Q][L/A] (Swinkels et al. 1991; Tsukamoto et al. 1994; Sacksteder and Gould 2000; Subramani et al. 2000). Two different receptor proteins have been identified that recognize these two PTSs in the cytoplasm and deliver the PTS-containing proteins to the peroxisomal membrane for import (for review, see Sacksteder and Gould 2000; Subramani et al. 2000). The inability to import PTS-containing proteins into peroxisomes renders most of the peroxisomal enzymes unstable or inactive in the cytoplasm of mammalian cells. The enzymatic deficiencies that result from an inability to import PTS-containing proteins are manifested as the severe disorders of peroxisome biogenesis, including Zellweger syndrome and RCDP (Wanders et al. 1995).
The biochemical deficiencies caused by the defective peroxin 7 in patients with RCDP type 1 reflect its function in PTS2-mediated protein transport: the PTS2-containing peroxisomal 3-ketoacyl-CoA thiolase remains unprocessed in the cytosol, and the PTS2-containing enzymes alkyl-dihydroxyacetonephosphate synthase and phytanoyl-CoA hydroxylase are both deficient (Heymans et al. 1985; Hoefler et al. 1988). The finding that patients with RCDP type 2 and type 3 have single-enzyme deficiencies in the ether-phospholipid biosynthetic pathway that result in the same clinical presentation as patients with RCDP type 1, indicates that the phenotype of RCDP is caused predominantly by a deficiency of ether phospholipids.
Few patients have been identified who have a mild form of RCDP type 1 displaying the same set of biochemical abnormalities as are observed in patients with classical type 1 but with a milder clinical presentation in that they lack the rhizomelia and have a much longer life expectancy (Poll-The et al. 1991; Smeitink et al. 1992; Nuoffer et al. 1994). In the patients with mild RCDP, ether-phospholipid biosynthesis is only moderately deficient, and residual enzyme activities are intermediate between those of patients with severe type 1 disease and those of normal controls (Smeitink et al. 1992; Nuoffer et al. 1994; Barth et al. 1996; Baumgartner et al. 1998). We have hypothesized previously that mutations in the PEX7 gene of these patients only partially affect the function of the PTS2 receptor (Barth et al. 1996; Motley et al. 1996). The finding that patients with a mild clinical course of the disease have higher residual levels of ether phospholipids confirms the importance of ether phospholipids in the pathogenesis of RCDP. Until now, however, this biochemical correlation with phenotype had not been linked to PEX7 genotype.
Here, we report the mutational spectrum in the PEX7 gene of 78 patients clinically diagnosed with RCDP type 1 and biochemically confirmed in our laboratory. In addition, we functionally analyzed 10 of the 22 different mutant alleles by assessing the ability of the encoded proteins to target a PTS2-tagged green fluorescent protein (GFP) to peroxisomes after overexpression in PTS2-mediated import-deficient cells. We found that the clinical severity of RCDP correlates with the residual activity of the PEX7 allele. Interestingly, two of the patients analyzed were homozygotes for a frameshift introducing an 8-nt duplication in the 5′ region of the PEX7 coding sequence, yet the disease was mild in both patients. Functional analysis of this PEX7 allele indicates that the duplication sequence leads to partial restoration of the reading frame, resulting in amelioration of a predicted severe phenotype.
Subjects and Methods
Patients
All patients analyzed for PEX7 mutations in the present study showed the clinical characteristics described for RCDP. After we obtained informed consent, samples were collected from patients and, when indicated, from their parents and were sent to our laboratory for the biochemical and molecular diagnosis of RCDP. For the majority of patients, the biochemical diagnosis of RCDP type 1 was substantiated by detailed studies in primary skin fibroblasts, which included the following analyses: (1) de novo plasmalogen synthesis, (2) dihydroxyacetonephosphate acyltransferase and alkyl-dihydroxyacetonephosphate synthase activity measurements, (3) analysis of very-long-chain fatty-acid levels, (4) phytanic acid alpha-oxidation, (5) catalase immunofluorescence, and (6) immunoblot analysis of peroxisomal thiolase (Wanders et al. 1995).
PEX7 Mutation Analysis
PEX7 mutation analysis in the patients and their parents was performed at the cDNA and/or the genomic DNA level. Total RNA and genomic DNA were isolated from primary skin fibroblasts or lymphocytes of patients and, when available, their parents, using the Wizard RNA purification kit and the Wizard genomic DNA purification kit, respectively (Promega). For mutation analysis at the cDNA level, the coding region of PEX7 cDNA was amplified by PCR in three overlapping fragments from first-strand cDNA prepared from total RNA, as described elsewhere (Ijlst et al. 1994). The sequences of the cDNA primer sets are shown in table 1. The amplification of the PEX7 cDNA fragments was performed with our standard PCR program, which started with 2 min of denaturation at 96°C, followed by 30 cycles of 30 s at 94°C, 30 s at 50°C, and 1 min at 72°C, followed by a final extension step of 5 min at 72°C. For mutation analysis at the genomic level, the protein-encoding portions of exons 1 and 10 and the entire exons 2–9, plus flanking intron sequences from the PEX7 gene, were amplified by PCR, using the primer sets shown in table 1. The genomic primer sets were designed on the basis of the recently published PEX7 gene structure (Braverman et al. 2000; GenBank). Exons 4/5, 7, 8, 9, and 10 were amplified using a PCR program that started with a denaturation step at 96°C for 2 min, followed by 4 cycles of 30 s at 96°C, 30 s at 55°C (50°C for exon 8), and 1 min at 72°C, and 24 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, followed by a final extension step of 10 min at 72°C. Exons 1, 2, 3, and 6 were amplified using a “touchdown” amplification program consisting of a denaturation step of 2 min at 96°C, followed by 10 cycles of 30 s at 96°C, 30 s at 65–55°C, and 1 min at 72°C, during which the annealing temperature was lowered 1°C per cycle, followed by 24 cycles of 30 s at 96°C and 1 min at 72°C and a final extension step of 10 min at 72°C.
Table 1.
Primer Sets Used for PEX7 Mutation Analysis
| Analysis Type, Amplicon Name(Size in bp), and Primer Name | Sequencea |
| cDNA: | |
| Fragment 1 (373) | |
| PEX7−81 to −61 | 5′-[−21M13]-TCTCTCTAACCGCGCCAGTG-3′ |
| PEX7 256–236 | 5′-[M13Rev]-AGGTGATGAGGACATGTTCGT-3′ |
| Fragment 2 (494) | |
| PEX7 148–168 | 5′-[−21M13]-ATATTGGATCCAGATGAAGCT-3′ |
| PEX7 605– 586 | 5′-[M13Rev]-AAGATTTCTGCCTGATGTGC-3′ |
| Fragment 3 (697) | |
| PEX7 341–362 | 5′-[−21M13]-TGTATAGTGTTGATTGGAGCCA-3′ |
| PEX7 1001–981 | 5′-[M13Rev]-ATCTTCTGTTTCTGACCAAAG-3′ |
| gDNA: | |
| Exon 1 (633) | |
| PEX7 −245 to −226 | 5′-[−21M13]-GATCACTCCCCTGATAGATC-3′ |
| PEX7-IVS1-rev | 5′-[M13Rev]-ATGCACTTGCACACAACTGG-3′ |
| Exon 2 (406) | |
| PEX7-IVS1-fw | 5′-[−21M13]-CATTTGGTATTCAAGGTCCC-3′ |
| PEX7-IVS2-rev | 5′-[M13Rev]-TATGCAAACGCCAAGGTTCC-3′ |
| Exon 3 (357) | |
| PEX7-IVS2-fw | 5′-[−21M13]-TTGTGTAGCTGCCTATGTAAg-3′ |
| PEX7-IVS3-rev | 5′-[M13Rev]-ATGCTACGTTAACTTGTCCC-3′ |
| Exon 4 + 5 (704) | |
| PEX7-IVS3-fw | 5′-[−21M13]-TGCAATGTTGAACTTGATGG-3′ |
| PEX7-IVS5-rev | 5′-[M13Rev]-CCATTCACTACAAGTAAGGC-3′ |
| Exon 6 (374) | |
| PEX7-IVS5-fw | 5′-[−21M13]-AGGTGGCAATATCCTAACAC-3′ |
| PEX7-IVS6-rev | 5′-[M13Rev]-TTAATGGTCCAGGAAGCACC-3′ |
| Exon 7 (324) | |
| PEX7-IVS6-fw | 5′-[−21M13]-TTTCTAGTAGGAAAGCCTGC-3′ |
| PEX7-IVS7-rev | 5′-[M13Rev]-TAACTCCAATCCCTAAACCC-3′ |
| Exon 8 (381) | |
| PEX7-IVS7-fw | 5′-[−21M13]-CTCAAATTATAGCATATATGCC-3′ |
| PEX7-IVS8-rev | 5′-[M13Rev]-TTTAAATGTACCCATCTCAG-3′ |
| Exon 9 (416) | |
| PEX7-IVS8-fw | 5′-[−21M13]-ACGTAGGGCTTAATAGTGGG-3′ |
| PEX7-IVS9-rev | 5′-[M13Rev]-GTTTAATGCTCAAACGCTCC-3′ |
| Exon 10 (322) | |
| PEX7-IVS9-fw | 5′-[−21M13]-GAATTTTGTATGTCTAAATACG-3′ |
| PEX71138–1120 | 5′-[M13Rev]-TAAAGTCTTTATCAGCTCC-3′ |
−21M13 = 5′-TGTAAAACGACGGCCAGT-3′; M13Rev = 5′-CAGGAAACAGCTATGACC-3′.
Forward and reverse primers used for PEX7 mutation analysis were tagged with a −21M13 sequence and M13rev sequence, respectively. PCR fragments were sequenced in two directions using “−21M13” and “M13rev” fluorescent primers on an Applied Biosystems 377A automated DNA sequencer, according to the manufacturer’s protocol (PE Biosystems).
Functional Analysis of PEX7 Alleles
Selected PEX7 mutations identified in the patients were introduced in control PEX7 cDNA cloned into the EcoRI-XbaI sites of pUC19 (New England BioLabs). The PEX7 alleles encoding the amino acid substitutions L70W (209T→G), W95G (283T→G), D134N (400G→A) and the allele harboring the 370-396del27nt deletion were amplified from the corresponding patient’s cDNA samples, using the primers PEX7148–168 and PEX7605–586. The PCR fragments were digested with HindIII and BclI and ligated into the corresponding restriction sites of the control PEX7 cDNA in pUC19. The PEX7 alleles encoding the amino acid substitutions G217R (649G→A), A218V (653C→T), S262L (785C→T), H285R (854A→G), and L292X (875T→A) were amplified from the corresponding patient’s cDNA samples, using the primers PEX7341–362 and PEX71001–981. The PCR fragments were digested with BstEII and BglII and ligated into the corresponding restriction sites of the control PEX7 cDNA in pUC19. The 8-nt–duplication allele was amplified from the corresponding patient cDNA sample, using an EcoRI-tagged PEX71-19 primer (5′-CGGGAATTCCGGATGAGTGCGGTGTGCGGTG-3′ [underline indicates EcoRI site]) and the PEX7256-236 primer. After digestion with EcoRI and HindIII, this fragment was ligated into the corresponding sites of the control PEX7 cDNA in pUC19.
The constructed PEX7 alleles were released from pUC19 as EcoRI-XbaI fragments and ligated into the EcoRI-XbaI sites of the pcDNA3 vector (Invitrogen) under the transcriptional control of the cytomegalovirus (CMV) promoter. To determine the consequence of the mutations for the function of peroxin 7, the expression plasmids were introduced by nuclear microinjection in primary skin fibroblasts from a patient with RCDP homozygous for the L292X mutation and were cultured in Dulbecco’s minimal essential medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. Microinjection of PTS2-tagged GFP (final concentration of 90 μg/ml) and the various PEX7 pcDNA3 expression plasmids (final concentration 270 μg/ml) was followed by immunofluorescence microscopy, using anti-GFP antiserum (Clontech) and anti-rabbit Cy3 conjugates to assess the import of PTS2-tagged GFP into peroxisomes, as described elsewhere (Motley et al. 1994, 1997).
Luciferase Reading Frame Restoration Assay
The luciferase coding region lacking its initiation codon was fused in three different reading frames behind a short (8-nt) duplication containing a PEX7 cDNA sequence comprising 30 nucleotides and extending from the A at position 30 to the C at position 60. The fusion constructs were created by means of PCR, using control firefly luciferase cDNA in vector pcDNA1 as template, an SP6 primer as the reverse, and the following primers as the forward primers: frame 0, 5′-CCCGGATCCATGCTGCGGACGCCGGGACGCCGGGACGCCGAAGACGCCAAAAACATAAAGAAAG-3′; frame +1, 5′-CCCGGATCCATGCTGCGGACGCCGGGACGCCGGGACGCCAGAAGACGCCAAAAACATAAAGAAAG-3′; and frame +2, 5′-CCCGGATCCATGCTGCGGACGCCGGGACGCCGGGACGCCACGAAGACGCCAAAAACATAAAGAAAG-3′ (underlined sequence derived from PEX7, boldfaced italic sequence derived from firefly luciferase). The PCR products were cloned into the BamHI-XhoI sites of pcDNA3 and verified by sequencing to exclude errors introduced by PCR. The different constructs were transfected into COS cells by calcium phosphate precipitation (Chen and Okayama 1987). As a control for transfection efficiency, an expression plasmid encoding LacZ (Promega) was cotransfected with the PEX7-luciferase fusion constructs. Cells were harvested after 48 h, and luciferase was measured using the luciferase assay system of Promega according to the manufacturer’s instructions. Luciferase activities were corrected for transfection efficiency by normalization with β-galactosidase activity (Rosenthal 1987).
In Vitro Expression of PEX7 Alleles
The various PEX7-pcDNA3 plasmids used for expression in fibroblasts (from the CMV promoter) were also used for expression in vitro (from the T7 promoter), using the TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s instructions. The labeled proteins were tested on 7.5% SDS polyacrylamide gels and were visualized by autoradiography.
Secondary Structure Predictions of Peroxin 7
The human peroxin 7 amino acid sequence was analyzed by use of the PHD program for protein structure prediction (Rost 1996). Predictions were re-evaluated and confirmed by analysis in the ExPasy Secondary Structure Prediction Package and by aligning the peroxin 7 sequence with the sequence of the β-subunit of heterotrimeric G protein, a WD-repeat–containing protein for which the crystal structure has been resolved (Sondek et al. 1996). The combined data allowed the construction of the topology folding model of peroxin 7.
Results
PEX7 Mutations in Patients with RCDP Type 1
Sequence analysis of the PEX7 cDNA and/or gene of 73 patients (excluding the five sib pairs) in whom RCDP type 1 was clinically and biochemically diagnosed revealed 22 different mutations, 18 of which have not been reported before. The mutations are detailed in table 2 and involve three deletions and one insertion and nine missense, six nonsense, and three splice-site mutations.
Table 2.
PEX7 Mutations Identified in 78 Patients with RCDP Type 1
|
No. of Patients Identified by |
||||||||
| cDNA Analysis |
gDNA Analysis |
|||||||
| NucleotideChangea | Exon | Predicted Effecton CodingSequence | +/+ | +/− | +/+ | +/− | AlleleFrequencyb (n = 146) | Ethnic/Geographic Originof Index Patients (No.) |
| Missense: | ||||||||
| 209T→G | 3 | L70W | 1 | 1.4 | Turkish (1) | |||
| 257G→A | 3 | C86Y | 1 | .7 | Dutch (1) | |||
| 283T→G | 3 | W95G | 3 | 4.1 | Israelic (3) | |||
| 400G→A | 4 | D134N | 1 | 1.4 | Italian (1) | |||
| 649G→Ac |
7 | G217R | 3 | 2.0 | British (2), Dutch (1) | |||
| 653C→Tc,d,e |
7 | A218V | 6 | 1 | 1 | 3 | 12.3 | Spanish (5), French (3), German (1), American (1), Chilean (1) |
| 722A→T | 7 | H241L | 1 | .7 | Spanish (1) | |||
| 785C→T | 8 | S262L | 1 | .7 | Dutch (1) | |||
| 854A→G | 9 | H285R | 1 | .7 | Dutch (1) | |||
| Nonsense: | ||||||||
| 60C→A | 1 | Y20X | 1 | .7 | Turkish (1) | |||
| 120C→G | 1 | Y40X | 2 | 1.4 | British (1), German (1) | |||
| 345T→G | 4 | Y115X | 1 | .7 | Swedish (1) | |||
| 376C→T | 4 | Q126X | 1 | .7 | Italian (1) | |||
| 695C→Te |
7 | R232X | 1 | 1.4 | Japanese (1) | |||
| 875T→A c,d,e,f |
9 | L292X | 30g | 6 | 8h | 4 | 52.1 | Dutch (23), German (15), British (5), Danish (1), Belgian (1), Australian (1), Chilean (1) |
| Deletion: | ||||||||
| 195-196delCT | 3 | FDW64-66VALRi | 1 | .7 | Swedish (1) | |||
| 370-396del27nt | 4 | Del aa124-132 | 3 | 2 | 6.8 | Turkish (3), German (2) | ||
| 842delC | 9 | Frameshift | 1 | .7 | Turkish (1) | |||
| Insertion: | ||||||||
| 52ins GGGACGCC | 1 | Frameshift | 2 | 2.7 | Swiss (1), French (1) | |||
| Splicing: | ||||||||
| IVS1+1G→C | ? | 1 | .7 | British (1) | ||||
| IVS1+3G→C | ? | 1 | 1.4 | Dutch (1)? | ||||
| IVS9+1G→Cj | Frameshift | 1 | 5 | 4.8 | British (3), Italian (1), French (1), Dutch (1) | |||
| Unknownk | 2 | 1.4 | Dutch (2) | |||||
Mutations described elsewhere are underlined.
To calculate allele frequency, apparent homozygotes have been assumed to be homozygotes, and sibs have been excluded.
Braverman et al. (1997).
Motley et al. (1997).
Shimozawa et al. (1999).
Purdue et al (1997).
Includes four sib pairs; homozygosity was confirmed in parents of three patients.
Includes one sib pair; homozygosity was confirmed in parents of six patients..
Deletion of 2 nt in exon 3 results in a 3′ shift of the intron 2 splice-acceptor site from position 188 to position 183, with the predicted substitution of amino acid residues FDW at position 64–66 by VALR.
Splice donor site mutation resulting in aberrantly spliced mRNA lacking exon 9 (804-903del100nt).
Two patients were heterozygous for L292X at the genomic level but homozygous at the cDNA level. No second mutation was identified.
Of the 18 patients who appeared to be heterozygous for two mutations, 4 were confirmed as compound heterozygotes by analysis of parental PEX7 DNA or by finding the mutations on two different alleles (cDNA subcloning). Two patients appeared to be homozygous for the L292X mutation at the cDNA level but were heterozygotes at the genomic level; no second mutation could be detected in the PEX7 gene. Analysis of genomic PEX7 DNA identified 14 patients as apparent homozygotes; for 6 of these, the homozygosity was confirmed by analysis of parental PEX7 DNA. PEX7 cDNA analysis identified 41 apparent homozygotes, with homozygosity confirmed by parental PEX7 DNA analysis in 4 of them.
When we assumed that all apparent homozygotes were true homozygotes, we found the nonsense mutation L292X to be by far the most common mutation causing RCDP type 1; it had an allele frequency of ∼52% and was detected in 43 of the 73 patients (table 2). Other relatively common mutations are A218V (11 patients; allele frequency ∼12%), 370-369del27nt (5 patients; allele frequency ∼7 %), and IVS9+1G→C (6 patients; allele frequency ∼5%). Remaining mutations were identified in only one to three patients.
Three of the patients we analyzed showed a mild clinical presentation of the disease and no rhizomelia. Surprisingly, two of these patients were apparent homozygotes for an 8-nt duplication of nucleotides 45–52 in PEX7 cDNA (52insGGGACGCC), predicted to result in a frameshift at codon 17 and no functional peroxin 7. The homozygosity for this duplication was confirmed by PCR amplification of genomic DNA of both patients and is in line with the reported consanguinity of the respective parents (Poll-The et al. 1991, Nuoffer et al. 1994). It is not known, however, whether the two patients are related. The third patient with mild RCDP is a compound heterozygote for two mutations, L292X and H285R. As the common L292X nonsense mutation does not lead to functional peroxin 7, the H285R mutation must be the allele responsible for the mild presentation of the disease in this patient.
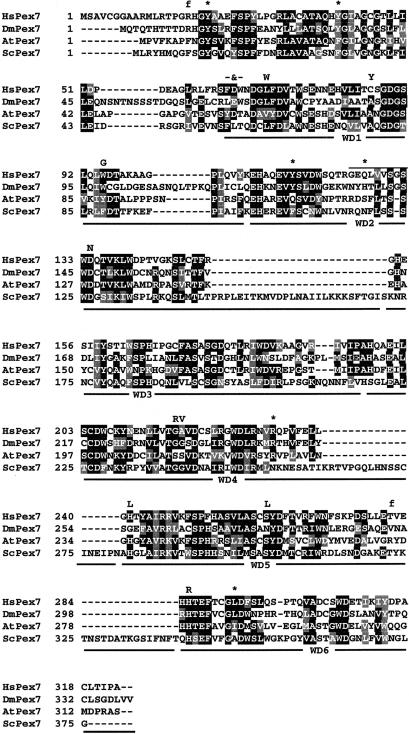
The positions of all the mutations identified in the coding region of PEX7 are shown in an alignment of human peroxin 7 with its orthologues from three evolutionarily distant phyla, represented by Drosophila melanogaster (Flybase), Arabidopsis thaliana, and Saccharomyces cerevisiae (fig. 1). Most missense mutations affect amino acids that are highly conserved among the orthologues and/or predicted to be essential for the functional integrity of one of the six WD repeats that are present in peroxin 7.
Figure 1.
Alignment of peroxin 7 orthologues from four different phyla, represented by human, Drosophila melanogaster, Arabidopsis thaliana, and Saccharomyces cerevisiae. Alignment was determined using the Clustal W program (Thompson et al. 1994). Amino acids that are identical and conserved in at least three sequences are indicated in blackened and shaded boxes, respectively. The horizontal black lines underneath the alignment indicate the positions of the six WD repeats. Mutations identified in the patients and affecting the human peroxin 7 sequence are indicated above the alignment as amino acid substitutions (one-letter code), nonsense mutations (*), and frameshift mutations (f). The FDW64–66VALR mutation is indicated (-&-), and the overlined residues represent the 370-369del27nt mutation (deletion of amino acid residues 124–132). For details of these mutations, see table 2.
Functional Analysis of PEX7 Alleles
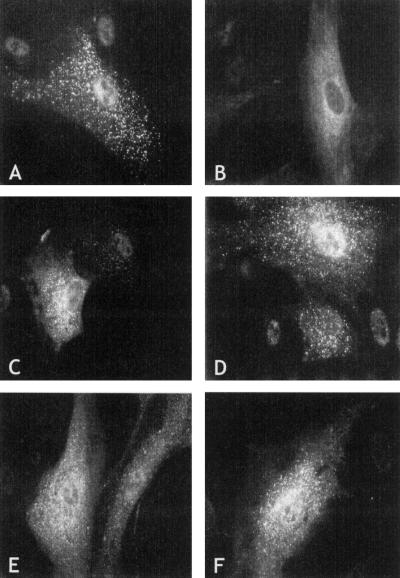
To test the effect of mutations on the function of peroxin 7, we expressed 10 different patient PEX7 alleles, under transcriptional control of the CMV promoter, in cultured skin fibroblasts from a confirmed L292X homozygote, together with a PTS2-tagged GFP (as reporter protein for PTS2-mediated protein import into peroxisomes [fig. 2]). Expression of a control PEX7 allele resulted in restoration of peroxisomal PTS2-mediated protein import, as indicated by the appearance of punctate peroxisomal fluorescence (fig. 2A). In contrast, none of the eight different PEX7 alleles derived from patients with severe RCDP were able to restore PTS2-mediated protein import, and the fluorescence observed remained invariably diffuse, indicative of a cytosolic localization of PTS2-tagged GFP (shown only for the L70W allele, in fig. 2B). Expression of the two alleles found in the patients with mild RCDP, however, resulted in restoration of PTS2-mediated peroxisomal protein import, although to varying degrees (fig. 2C–F). Import restoration by the 8-nt–duplication allele was rather efficient, with many GFP-expressing cells showing punctate peroxisomal fluorescence without background cytosolic labeling and with only few cells showing partial import restoration (fig. 2C and D). Complementation of the import defect by the second mild allele, H285R, was weaker and, although punctate peroxisomal fluorescence often could be detected, it was always against a background of cytosolic labeling (fig. 2E and F). These results indicate that the severity of the clinical phenotype correlates with the ability of a PEX7 allele to restore PTS2-mediated protein import into peroxisomes in RCDP cells.
Figure 2.
Functional complementation of PTS2-mediated peroxisomal protein import by PEX7 alleles. Ten different PEX7 alleles identified in the patients were coexpressed with PTS2-tagged GFP to test their ability to restore PTS2-mediated peroxisomal protein import in skin fibroblasts from a patient homozygous for the L292X mutation. A, Expression of control PEX7 resulted in punctate peroxisomal fluorescence in >90% of GFP-expressing cells, with 20%–40% of the cells showing cytosolic fluorescence in addition to punctate fluorescence. B, None of the eight alleles derived from patients with severe RCDP (see Subjects and Methods) were able to complement the PTS2-mediated protein import defect, and PTS2-tagged GFP fluorescence was invariably cytosolic (the L70W allele, which is representative for all other seven alleles, is shown). C and D, Expression of the 8-nt duplication PEX7 allele resulted in punctate peroxisomal fluorescence in 90% of the cells. In 40%–60% of the GFP-expressing cells, however, cytosolic fluorescence and punctate fluorescence were evident, indicating that complementation by this allele is less efficient than complementation by the control allele. E and F, Expression of the H285R PEX7 allele resulted in punctate peroxisomal fluorescence in 50%–70% of GFP-expressing cells, but this was always against a background of cytosolic fluorescence; no cells were found in which fluorescence was exclusively punctate.
In Vitro Expression of Patient PEX7 Alleles
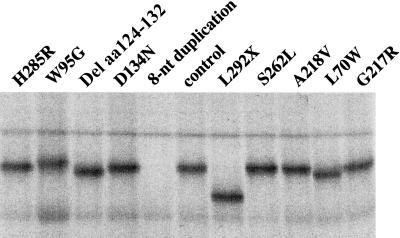
To determine whether PEX7 mutations have an effect on the synthesis and/or stability of peroxin 7, we also expressed the 10 different patient alleles in vitro, using a coupled transcription-translation system (fig. 3). For these studies, we used the same pcDNA3 constructs as described above for expression in fibroblasts, because pcDNA3 contains both a CMV promoter for expression in mammalian cells and a T7 promoter for in vitro expression. As shown infigure 3, most patient alleles produced a protein of approximately the predicted size. In the case of the 8-nt–duplication allele, however, no full-length peroxin 7 could be detected, not even after long exposures or when more lysate was layered.
Figure 3.
In vitro transcription and translation of PEX7 alleles. The same 10 constructs used for the complementation studies were used to express the PEX7 alleles in a T7-coupled reticulocyte lysate expression system. Each allele produced a polypeptide of approximately the expected size, with the exception of the 8-nt duplication allele, which gave no detectable product.
Restoration of Luciferase Reading Frame by the 8-nt–Duplication Allele
Because the 8-nt duplication occurs downstream of two potential in-frame translational start codons and because no other in-frame AUG codons are present in PEX7 mRNA, we hypothesized that the duplication sequence leads to restoration of the reading frame and the production of functional peroxin 7. To further analyze this, we studied the ability of the 8-nt–duplication containing sequence to restore luciferase activity by fusing the coding sequence of luciferase in three different reading frames behind a short 8-nt-duplication-containing PEX7 sequence followed by expression of these constructs in COS cells (table 3). As translation initiation codon for the 8-nt–duplication sequence in the fusion constructs, the second ATG of PEX7 cDNA has been chosen, and the first ATG of luciferase has been removed to prevent internal initiation. The fusion constructs were transfected into COS cells, and transfection efficiency was determined by cotransfection with a plasmid encoding β-galactosidase. Luciferase activity was normalized by correcting for β-galactosidase activity. As can be seen from table 3, luciferase activity was observed for both out-of-frame fusions, although to varying degrees, with frame restoration being slightly more efficient for the frame +2 fusion. As a control, a construct lacking both the PEX7-derived sequence and the luciferase start codon showed no luciferase activity, indicating that the observed luciferase activity does not result from internal translational initiation within the luciferase coding sequence. These results indicate that molecular misreading must occur during expression of these PEX7-luciferase fusion sequences, leading to partial correction of the introduced frameshifts and resulting in low-level luciferase activity.
Table 3.
Reading Frame–Dependent Restoration of Luciferase Activity by the 8-nt Duplication
| Frame | Constructa | Luciferase Activityb(%) |
| 0 | PCMV- GAA …c | 0 |
| 0 | PCMV-ATG CTG CGG ACG CCG GGA CGC CGG GAC GCC GAA … |
100 |
| +1 | PCMV-ATG CTG CGG ACG CCG GGA CGC CGG GAC GCC AGA A… |
1.1 |
| +2 | PCMV-ATG CTG CGG ACG CCG GGA CGC CGG GAC GCC ACG AA… |
4.3 |
ATG codon following CMV promoter (PCMV) is second ATG of PEX7 (nt 31–33 of cDNA). Open reading frame of firefly luciferase starts at the second codon and is shown in bold. Underlined sequence denotes the 8-nt duplication
100% is 3.106 units luciferase activity normalized for β-galactosidase activity
Control construct without PEX7 sequence and without the luciferase ATG.
Discussion
We analyzed the PEX7 genotypes in a large cohort of patients with clinical and biochemical diagnosis of RCDP type 1. Initially, our genetic analysis involved sequence analysis of patient PEX7 cDNAs prepared from total RNA isolated from primary skin fibroblasts or lymphocytes. After the recent elucidation of the PEX7 gene structure (Braverman et al. 2000), we introduced a PCR-based method for analysis of the PEX7 gene at the genomic level, which involves sequencing of all coding exons plus flanking intron-exon boundaries. In our patients we found 22 different mutations, including 3 deletions, 1 insertion, and 9 missense, 6 nonsense, and three splice-site mutations. In two patients, we could find only one mutation in heterozygous form after analysis of the PEX7 gene at the genomic level. Subsequent PEX7 cDNA analysis of these patients showed this mutation in homozygous form, indicating that the second, undetected mutation affects mRNA expression and/or stability and, for example, might be located in the PEX7 promoter. Of the 22 mutations, only 4 had been reported previously, as indicated in table 1. Of a fifth mutation, IVS9+1G→C, only its consequence on mRNA splicing has been reported (i.e., skipping of exon 9, comprising nucleotides 804–903 [Purdue et al. 1997]). The mutation underlying this aberrant splicing could be resolved only by analysis of the PEX7 gene at the genomic level.
We found the nonsense mutation L292X to be by far the most common mutation causing RCDP type 1, followed by the A218V missense mutation. In our cohort, the frequencies of the L292X and A218V mutations were ∼52% and ∼12%, respectively, which is similar to the frequencies of 49% and 6% that have been reported by Braverman and colleagues (2000), who analyzed 36 patients with RCDP type 1 exclusively for three previously reported mutations, including these two. The third mutation reported by Braverman and colleagues (2000)—the G217R mutation, with a frequency of 7%—was observed in heterozygous form in only three of our patients. The high frequency of the L292X mutation has been shown to result from a founder effect (Braverman et al. 2000).
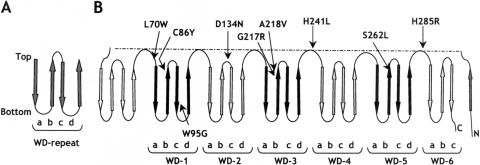
Most of the mutations detected in our patients affect amino acids conserved among peroxin 7 orthologues from different phyla and/or amino acids that are predicted to have an important function in the conformation of the protein. Peroxin 7 is a 323–amino acid protein containing six WD repeats. Proteins with such WD repeats have been implicated in diverse cellular processes, and the WD repeats have been postulated to be involved in establishing protein-protein interactions (Smith et al. 1999). The crystal structures of two WD repeat–containing proteins, the β-subunit of heterotrimeric G proteins and Tup1 (Sondek et al. 1996; Sprague et al. 2000), revealed that each WD repeat folds into four β-strands (named “a,” “b,” “c,” and “d”), which are arranged in an antiparallel fashion (fig. 4A). As a result, the overall appearance of a WD-repeat protein resembles that of a propeller, with as many blades as the number of WD repeats (Garcia-Higuera et al. 1998). Secondary structure predictions revealed that peroxin 7 is composed solely of β-strands, indicating that it probably displays a propeller-like structure with seven blades. On the basis of these predictions and of comparison with the crystallized WD repeat–containing proteins, we created a folding topology diagram of peroxin 7 (fig. 4B). Interestingly, mapping the various missense mutations in the diagram revealed that, except for the W95G mutation, they all affect amino acid residues located in the exposed top surface of the propeller-like structure, either in the connecting loops or at the ends of the β-strands (fig. 4B). Since peroxin 7 has been shown to interact with both the PTS2 sequence of peroxisomal matrix proteins and the PTS1 receptor peroxin 5 (Otera et al. 2000), this clustering suggests that the mutations interfere with PTS2 and/or peroxin 5 binding. A similar clustering of mutations has been reported elsewhere for the RAG2 gene in patients who have severe combined immune deficiency syndrome (Corneo et al. 2000), and the clustering has been demonstrated to affect the interaction of RAG2 with RAG1 (Villa et al. 1998).
Figure 4.
Predicted folding topology of peroxin 7 based on WD-repeat secondary structure features and locations of PEX7 mutations. A, Folding of an individual WD repeat. Each WD repeat is composed of four β-strands named “a,” “b,” “c,” and “d” (arrows) separated by loops and turns (lines), which together make up one blade of a propeller. The top region of a folded WD repeat is defined by the tight turn between β-strands b and c. B, Topology model showing the structural arrangement of the six WD repeats predicted for peroxin 7 and mapping of the various amino acid residues affected by the missense mutations identified in the patients with RCDP. The WD repeats (indicated as WD-1–WD-6) are alternately shaded and blackened and are separated by connecting loops.
Using a PTS2-tagged GFP as reporter protein for PTS2-mediated peroxisomal targeting, we were able to distinguish between PEX7 alleles from patients with severe disease and those from patients with mild disease: whereas eight alleles identified in patients with severe RCDP did not restore the PTS2-mediated targeting defect in vitro, both alleles from patients with mild disease did, although to varying degrees and only after CMV-promoter–driven overexpression. In this respect, it should be noted that previously we had been unable to distinguish between fibroblasts from the patients with mild disease and those with severe disease when we analyzed these cells for the ability to import endogenous peroxisomal thiolase and a PTS2-chloramphenicol acyltransferase reporter protein expressed from the RSV promoter; none of the cells were able to import either of these PTS2-containing proteins, although import of a PTS1-containing reporter protein was normal (Motley et al. 1996). The partial restoration upon overexpression of the H258R mutation, in conjunction with the fact that this mutation is not located in the main cluster of mutations in the topology model, suggests that the histidine at position 258 is located in a region less important for protein-protein interaction. On the other hand, in contrast to the other missense mutations, this mutation involves the conserved substitution of an amino acid, which may not completely abolish peroxin 7 activity.
The 8-nt–duplication allele found in two of the three patients with mild disease is particularly interesting. As the 8-nt duplication occurred just downstream of the two potential start codons of PEX7 and no other AUG codons are present in the PEX7 mRNA, we hypothesized that the frameshift caused by the 8-nt duplication is corrected with a low frequency at either the transcriptional or the translational level. Indeed, our luciferase reading frame restoration assay confirmed the occurrence of frame restoration within the duplication-containing sequence. This result also makes a possible internal initiation event at non-AUG codons of PEX7, as has been reported for other cellular mRNAs, unlikely (Ronsin et al. 1999; Arnaud et al. 1999). At this moment, it is not clear whether the frame restoration of the 8-nt duplication PEX7 allele results from molecular misreading at the transcriptional level, the translational level, or both. Unfortunately, this could not be resolved using an in vitro coupled transcription-translation system, because, in this system, no detectable peroxin 7 protein was synthesized. A possible explanation for this could be that frame restoration at the transcriptional level may be mediated only by RNA polymerase II but not by the phage T7 polymerase used in the in vitro system. Alternatively, the frame restoration could result from ribosomal shifting, which may occur only in intact cells but not in reticulocyte lysates.
The repeat-containing sequence in PEX7 that undergoes frame restoration does not resemble any previously described sequence involved in transcriptional (van Leeuwen et al. 1998; Linton et al. 1997; van den Hurk et al. 2001) or translational (Giedroc et al. 2000) misreading. Indeed, searching of the sequence databases indicates that this repeat-containing sequence is unique to PEX7. To our knowledge, this is only the third example of a frame restoration event resulting in amelioration of a predicted severe phenotype reported to date. The other two examples involve frame restoration due to transcriptional and/or translational errors occurring in a stretch of 10 adenines created by the deletion of a thymidine in the factor VIII gene, resulting in unexpectedly mild hemophilia A (Young et al. 1997), and the insertion of an additional adenine into a stretch of eight adenines created by the deletion of a cytosine in the apolipoprotein B gene, resulting in hypobetalipoproteinemia (Linton et al. 1992).
Acknowledgments
We thank the many physicians and investigators who made the initial diagnosis of RCDP in the patients and/or contributed skin fibroblasts, blood samples, or DNA samples from their patients. This work was supported by fellowships from the Netherlands Organization for Scientific Research (to A.M.M.), the Calouste Gulbenkian Foundation with Program Praxis XXI-FCT (BD9805/96) (to P.B.), and the Royal Netherlands Academy of Arts and Sciences (to H.R.W.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- ExPasy Secondary Structure Prediction Package, http://www.expasy.ch/#secondary
- Flybase, http://hedgehog.lbl.gov:7081 (for Drosophila melanogaster PEX7 orthologue [annotation number Fban0006486])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human PEX7 gene [accession numbers AF180806–AF180814], for PEX7 cDNA [accession number U69171], Arabidopsis thaliana PEX7 orthologue [accession number AF130973], and Saccharomyces cerevisiae PEX7 orthologue [accession number X81424])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RCDP type 1 [MIM 215100], RCDP type 2 [MIM 222765], and RCDP type 3 [MIM 600121])
- PHD Program for Protein Structure Prediction, http://www.embl-heidelberg.de/predictprotein/
References
- Arnaud E, Touriol C, Boutonnet C, Gensac MC, Vagner S, Prats H, Prats AC (1999) A new 34 kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol 19:505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth PG, Wanders RJA, Schutgens RBH, Staalman C (1996) Variant RCDP with normal phytanic acid: clinico-biochemical delineation of a subtype and complementation studies. Am J Med Genet 62:164–168 [DOI] [PubMed] [Google Scholar]
- Baumgartner MR, Poll-The BT, Verhoeven NM, Jakobs C, Espeel M, Roels F, Rabier D, Levade T, Rolland MO, Martinez M, Wanders RJA, Saudubray JM (1998) Clinical approach to inherited peroxisomal disorders: a series of 27 patients. Ann Neurol 44:720–730 [DOI] [PubMed] [Google Scholar]
- Braverman N, Steel G, Lin P, Moser A, Moser H, Valle D (2000) PEX7 gene structure, alternative transcripts and evidence for a founder haplotype for the frequent RCDP allele L292ter. Genomics 63:181–192 [DOI] [PubMed] [Google Scholar]
- Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D (1997) Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet 15:369–376 [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H (1987) High efficiency of transfection of mammalian cells by plasmid DNA. Mol Cell Biol 7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B, Moshous D, Callebaut I, de Chasseval R, Fischer A, de Villartay JP (2000) Three-dimensional clustering of human RAG2 gene mutations in severe combined immune deficiency. J Biol Chem 275:12672–12675 [DOI] [PubMed] [Google Scholar]
- de Vet EC, Ijlst L, Oostheim W, Wanders RJA, van den Bosch H (1998) Alkyl-dihydroxyacetonephosphate synthase: fate in peroxisome biogenesis disorders and identification of the point mutation underlying a single enzyme deficiency. J Biol Chem 273:10296–10301 [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Gaitatzes C, Smith TF, Neer EJ (1998) Folding a WD repeat propeller. J Biol Chem 273:9041–9049 [DOI] [PubMed] [Google Scholar]
- Giedroc DP, Theimer CA, Nixon PL (2000) Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J Mol Biol 298:167–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S (1989) A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol 108:1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymans HSA, Oorthuijs JWE, Nelck G, Wanders RJA, Schutgens RBH (1985) Rhizomelic chondrodysplasia punctata: another peroxisomal disorder. N Engl J Med 313:187–188 [PubMed] [Google Scholar]
- Hoefler G, Hoefler S, Watkins PA, Chen WW, Moser A, Baldwin V, McGillivary B, Charrow J, Friedman JM, Rutledge L, Hashimoto T, Moser HW (1988) Biochemical abnormalities in rhizomelic chondrodysplasia punctata. J Pediatr 112:726–733 [DOI] [PubMed] [Google Scholar]
- Ijlst L, Wanders RJA, Ushikubo S, Kamijo T, Hashimoto T (1994) Molecular basis of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency: identification of the major disease-causing mutation in the α-subunit of the mitochondrial trifunctional protein. Biochim Biophys Acta 1215:347–350 [DOI] [PubMed] [Google Scholar]
- Linton MF, Pierotti V, Young SG (1992) Reading-frame restoration with an apolipoprotein B gene frame shift mutation. Proc Natl Acad Sci USA 89:11431–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A, Hettema E, Distel B, Tabak HF (1994) Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol 125:755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Hettema EH, Hogenhout EM, Brites P, ten Asbroek ALMA, Wijburg FA, Baas F, Heijmans HS, Tabak HF, Wanders RJA, Distel B (1997) Rhizomelic chondrodysplasia punctata is a peroxisomal protein targetting disease caused by a non-functional PTS-2 receptor. Nat Genet 15:377–380 [DOI] [PubMed] [Google Scholar]
- Motley AM, Tabak HF, Smeitink JAM, Poll-The BT, Barth PG, Wanders RJA (1996) Non-rhizomelic and rhizomelic chondrodysplasia punctata within a single complementation group. Biochim Biophys Acta 1315:153–158 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Flynn CR, Trelease RN (1997) Diverse amino acid residues function within the type 1 peroxisomal targetting signal. Plant Physiol 115:881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer JM, Pfammatter JP, Spahr A, Toplak H, Wanders RJA, Schutgens RBH, Wiesmann UN (1994) Chondrodysplasia punctata with mild clinical course. J Inher Metab Dis 17:60–66 [DOI] [PubMed] [Google Scholar]
- Ofman R, Hettema EH, Hogenhout EM, Caruso U, Muijsers AO, Wanders RJA (1998) Acyl-CoA: dihydroxyacetonephosphate acyltransferase: cloning of human cDNA and resolution of the molecular basis in rhizomelic chondrodysplasia punctata type 2. Hum Mol Genet 7:847–853 [DOI] [PubMed] [Google Scholar]
- Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, Fujiki Y (2000) The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type I transporter, translocates the Pex7p-PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J Biol Chem 275:21703–21714 [DOI] [PubMed] [Google Scholar]
- Poll-The BT, Maroteaux P, Narcy C, Quentin P, Guesnu M, Wanders RJA, Schutgens RBH, Saudubray JM (1991) A new type of chondrodysplasia punctata associated with peroxisomal dysfunction. J Inher Metab Dis 14:361–363 [DOI] [PubMed] [Google Scholar]
- Purdue PE, Zhang JW, Skoneczny M, Lazarow PB (1997) Rhizomelic chondrodysplasia punctata is caused by deficiency of human PEX7, a homologue of the yeast PTS2 receptor. Nat Genet 15:381–384 [DOI] [PubMed] [Google Scholar]
- Ronsin C, Chung-Scott V, Poullion I, Aknouche N, Gaudin C, Triebel F (1999) A non-AUG-defined alternative ORF of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ. J Immunol 163:483–490 [PubMed] [Google Scholar]
- Rosenthal N (1987) Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol 152:704–720 [DOI] [PubMed] [Google Scholar]
- Rost B (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol 266:525–539 [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Gould SJ (2000) The genetics of peroxisome biogenesis. Annu Rev Genet 34:623–652 [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Suzuki Y, Zhang Z, Miura K, Matsumoto A, Nagaya M, Castillo-Taucher S, Kondo N (1999) A novel nonsense mutation of the PEX7 gene in a patient with rhizomelic chondrodysplasia punctata. J Hum Genet 44:123–125 [DOI] [PubMed] [Google Scholar]
- Smeitink JAM, Beemer FA, Espeel M, Donckerwolcke RAMG, Jakobs C, Wanders RJA, Schutgens RBH, Roels F, Duran M, Dorland L, Berger R, Poll-The BT (1992) Bone dysplasia associated with phytanic acid accumulation and deficient plasmalogen synthesis: a peroxisomal entity amenable to plasmapheresis. J Inher Metab Dis 15:377–380 [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24:181–185 [DOI] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB (1996) Crystal structure of a G-protein βγ dimer at 21A resolution. Nature 379:369–374 [DOI] [PubMed] [Google Scholar]
- Sprague ER, Redd MJ, Johnson AD, Wolberger C (2000) Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J 19:3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Koller A, Snyder WB (2000) Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem 69:399–418 [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10:3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TP, Heid H, Rackwitz HR, Hunziker A, Gorgas K, Just WW (1997) Ether lipid biosynthesis: isolation and molecular characterization of human dihydroxyacetonephosphate acyltransferase. FEBS Lett 420:205–211 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T (1994) Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem 269:6001–6010 [PubMed] [Google Scholar]
- Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta LB, Ochs HD, Schwarz K, Notarangelo LD, Vezzoni P, Spanopoulou E (1998) Partial V(D)J recombination activity leads to Omenn syndrome. Cell 93:885–896 [DOI] [PubMed] [Google Scholar]
- van den Hurk W, Willems HJJ, Bloemen M, Martens GJM (2001) Novel frameshift mutations near short simple repeats. J Biol Chem 276:11496–11498 [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, Grosveld FG, Peter J, Burbach H, Hol EM (1998) Frameshift mutants of β amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science 279:242–247 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Dekker C, Horvath VAP, Schutgens RBH, Tager JM (1994) Human alkyldihydroxyacetonephosphate synthase deficiency: a new peroxisomal disorder. J Inher Metab Dis 17:315–318 [DOI] [PubMed] [Google Scholar]
- Wanders RJA, Schutgens RBH, Barth PG (1995) Peroxisomal disorders: a review. J Neuropathol Exp Neurol 54:726–739 [DOI] [PubMed] [Google Scholar]
- Young M, Inaba H, Hoyer LW, Higuchi M, Kazazian HH, Antonarakis SE (1997) Partial correction of a severe molecular defect in hemophilia A, because of errors during expression of the factor VIII gene. Am J Hum Genet 60:565–573 [PMC free article] [PubMed] [Google Scholar]