Abstract
Migraine is a complex neurovascular disorder with substantial evidence supporting a genetic contribution. Prior attempts to localize susceptibility loci for common forms of migraine have not produced conclusive evidence of linkage or association. To date, no genomewide screen for migraine has been published. We report results from a genomewide screen of 50 multigenerational, clinically well-defined Finnish families showing intergenerational transmission of migraine with aura (MA). The families were screened using 350 polymorphic microsatellite markers, with an average intermarker distance of 11 cM. Significant evidence of linkage was found between the MA phenotype and marker D4S1647 on 4q24. Using parametric two-point linkage analysis and assuming a dominant mode of inheritance, we found for this marker a maximum LOD score of 4.20 under locus homogeneity (P=.000006) or locus heterogeneity (P=.000011). Multipoint parametric (HLOD = 4.45; P=.0000058) and nonparametric (NPLall = 3.43; P=.0007) analyses support linkage in this region. Statistically significant linkage was not observed in any other chromosomal region.
Introduction
Migraine (MIM 157300) is a highly prevalent primary headache disorder, which affects ∼10%–12% of the white population (Henry et al. 1992; Rasmussen and Olesen 1992; Stewart et al. 1992; O'Brien et al. 1994; Hagen et al. 2000). It is more prevalent among women, and, for both sexes, its prevalence peaks during middle age and declines thereafter (Lipton et al. 1994, 2001). The most common form of migraine is migraine without aura (MO), which, according to the International Headache Society’s (IHS) standardized guidelines, is characterized by unilateral pulsating pain of moderate to severe intensity, aggravated by physical activity and lasting 4–72 h. The attacks are associated with nausea, vomiting, photophobia, and phonophobia. Migraine with aura (MA) shares the same headache qualities, but the headache is usually preceded by aura—attacks of focal neurological symptoms, usually visual, that develop gradually within 5–20 min and that persist for <60 min (Headache Classification Committee of the International Headache Society 1988). Approximately one-third of individuals with migraine experience both types of migraine during their lifetimes.
Twin studies indicate that migraine has a significant genetic component, with heritability estimates of 40%–65% (Honkasalo et al. 1995; Larsson et al. 1995; Gervil et al. 1999b; Ulrich et al. 1999b). Approximately 50% of migraine sufferers have an affected first-degree relative (Bille 1997). However, the mode of transmission of the disorder is not clear (Mochi et al. 1993; Russell and Olesen 1993; Russell et al. 1995; Ulrich et al. 1999a). Several population-based family studies have suggested that both genetic and environmental factors are involved in migraine but that genetic factors are more influential in MA than in MO (Russell and Olesen 1995; Ziegler et al. 1998; Gervil et al. 1999a; Ulrich et al. 1999a, 1999b).
None of the numerous studies performed to date have led to identification of a gene responsible for the more common forms of migraine—that is, MO or MA—or even to specification of their mode of transmission. Only familial hemiplegic migraine (FHM1 [MIM 141500]), a rare autosomal dominant subtype of migraine, has had a gene identified for it, on 19p13 (Joutel et al. 1993; Ophoff et al. 1996). Mutations in this gene, CACNA1A (MIM 601011), a brain-specific P/Q-type calcium-channel gene, account for only a small fraction of all patients with migraine and for 50% of families with the FHM subtype (Ducros et al. 2001). Interestingly, mutations in this same gene also can cause either episodic ataxia type 2 (EA-2 [MIM 108500]) or spinocerebellar ataxia type 6 (SCA6 [MIM 183086]) (Ophoff et al. 1996; Jodice et al. 1997); on the other hand, 20% of families with FHM show linkage to 1q21-31 (FHM2 [MIM 602481), providing evidence that FHM is heterogeneous (Ducros et al. 1997; Gardner et al. 1997). Several studies have suggested that the 19p13 CACNA1A locus may be involved in nonhemiplegic migraine as well (May et al. 1995; Ophoff et al. 1997; Nyholt et al. 1998b; Terwindt et al. 2001), although contradictory data also have been reported (Hovatta et al. 1994; White et al. 2001).
Thus far, 1q has not been linked to either MA or MO; on the other hand, Xq has been implicated in typical familial migraine (MIM 300125) (Nyholt et al. 1998a , 2000; Oterino et al. 2001). In addition, a number of association studies have shown linkage between various gene loci and either MA or MO (Pardo et al. 1995; Peroutka et al. 1997; Del Zompo et al. 1998; Ogilvie et al. 1998; Kowa et al. 2000; Lea et al. 2000; Tzourio et al. 2001). Unfortunately, most of these studies have remained single reports and await confirmatory replication studies. No genomewide screens for susceptibility loci for migraine have been published thus far.
Identification of genes and allelic variants predisposing to common traits such as migraine has been complicated by inconsistencies in clinical diagnosis, a variable age at onset, an unknown mode of inheritance, locus heterogeneity, and the poorly understood role of environmental factors. However, the small number of founders, along with environmental and cultural homogeneity, can potentially make the Finnish population useful in genetic studies of complex diseases (de la Chapelle 1993; Lander and Schork 1994; Peltonen et al. 2000). This population, with well-established genealogical registries and well-kept population records, has been used successfully in past genetic studies, and most identified loci have been replicated also in mixed populations, suggesting their general significance (Mahtani et al. 1996; Kuokkanen et al. 1997; Hovatta et al. 1999; Leppävuori et al. 1999; Pajukanta et al. 1999; Ekelund et al. 2000; for a review, see Peltonen et al. 2000; Laitinen et al. 2001). We report here the results of a genomewide screen of 50 Finnish families with MA, implicating a susceptibility locus on 4q24.
Subjects and Methods
Study Design
The present study focuses on families ascertained for MA. This restriction was imposed because data suggest that the genetic component of MA may be stronger than that of MO (Honkasalo et al. 1995; Russell et al. 1995; Ulrich et al. 1999a). Also, by mandating that an aura be one of the symptoms, we diminish the risk that other types of headache will be included. Several studies also have indicated that MA and MO might be distinct disorders (Russell et al. 1996, 2001). Genetic analysis was performed only on individuals who gave their informed consent to the study. The ethics committees of the Helsinki University Central Hospital and the University of California, Los Angeles, approved the study protocol.
Consecutively identified families with at least three first-degree family members affected with MA were considered for the study. The families were recruited on the basis of patients attending headache clinics in three Finnish cities (Helsinki, Kemi, and Jyväskylä). Three neurologists (M.K., H.H., and M.I.) were in charge of the recruitment. Once a member of the family, the index case, had been clinically diagnosed, by a neurologist, as suffering from migraine, the index case was asked to contact all other members of the family who were believed to suffer from migraine and to ask if they would be willing to participate in the study. If at least three possible migraine sufferers were willing to participate, the validated Finnish Migraine Specific Questionnaire for Family Studies (FMSQFS) (Kallela et al. 2001) was mailed to each of them, to their parents, and to their siblings. The FMSQFS has been shown to be both specific and sensitive in both diagnosing MA and MO in a family setting (Kallela et al. 2001) and differentiating MA from MO. From among the first 300 participating families, 50 independent, multigenerational families including a total 646 family members were selected for genotyping. These families were chosen because they displayed a seemingly autosomal dominant mode of inheritance, with the MA phenotype passed from one generation to the next. In rare cases, an unaffected individual (i.e., a subject without MA) had affected offspring; this suggests that there is reduced penetrance for the disease gene or that phenocopies may exist. In several instances, there appeared to have been assorted mating within the pedigrees—that is, an offspring was the result of mating between an individual with the common MA phenotype and an individual who either (a) displayed either the MA or MO phenotype or (b) came from a family with a history of either MA or MO. In these instances, the spouse and his or her offspring were excluded from the genetic analysis. In addition to completing the FMSQFS, the participants were asked to provide a sample of their blood, for genotyping; blood samples were received from 430 individuals.
Diagnoses
Clinical data were available on 646 individuals (368 women and 278 men), including 246 individuals (174 women and 72 men) with MA and 53 individuals (36 women and 17 men) with MO. Table 1 describes the family sample used in our genomewide screen. Diagnoses of all index cases and family members were made according to the IHS criteria (Headache Classification Committee of the International Headache Society 1988), on the basis of the FMSQFS (Kallela et al. 2001). Table 2 describes the diagnosis of the 430 individuals from whom a blood sample was available for genotyping. Table 3 describes the clinical characteristics of headache, and table 4 describes the aura symptoms in the 246 subjects with MA. Families with FHM (IHS classification 1.2.3) were not included in our study.
Table 1.
Families with Migraine Who Were Used for Genomewide Screen
| Category | No. (Range) |
| Pedigree size: | |
| One generation | 5 |
| Two generations | 19 |
| Three generations | 24 |
| Four generations | 2 |
| Total | 50 |
| Subjects: | |
| Total | 646 |
| Total genotyped | 430 |
| Per-pedigree mean: | |
| Total | 13.0 (3–34) |
| Genotyped | 8.6 (3–20) |
| Affected (with either MA or MO) | 5.9 (3–17) |
Table 2.
Diagnosis of 430 Genotyped Subjects with Migraine
| Diagnosis (IHS Classification) | No. of Subjects |
| MA (1.2a) | 252 |
| MO (1.1) | 53 |
| HA | 10 |
| NoHA | 91 |
| DU | 24 |
Includes EQV (IHS classification 1.2.5), in 6 subjects.
Table 3.
Clinical Characteristics of Headache
| Symptom | Proportionof Subjects(%) |
| Unilateral headache | 76 |
| Pulsating headache | 75 |
| Headache intensity: | |
| Unbearable | 39 |
| Severe | 42 |
| Moderate | 17 |
| Mild | 2 |
| Aggravated by physical activity | 76 |
| Associated symptoms: | |
| Nausea | 85 |
| Vomiting | 57 |
| Photophobia | 87 |
| Phonophobia | 79 |
Table 4.
Aura Symptoms in 246 Subjects with MA
| Type of Aura | Proportionof Subjects(%) |
| Any visual aura | 94 |
| Hemianopia | 40 |
| Scintillating scotoma | 56 |
| Photopsia | 49 |
| Blurring of vision | 43 |
| Any sensory aura | 36 |
| Any motor aura | 14 |
| Speech disturbance | 29 |
Genotyping
Linkage analysis was conducted for the 22 autosomes and the X chromosome. In all, 350 polymorphic microsatellite markers were genotyped in the 430 subjects from the 50 families that provided blood. The markers were from The Human MapPairs Genome-Wide Screening Set (LI-COR). This compilation is the ninth version of the Weber-lab screening set (Broman et al. 1998), with a few modifications. The order and distances between the markers were determined on the basis of the genome database of the Center for Medical Genetics, Marshfield Medical Research Foundation. The markers are approximately evenly spaced throughout the genome, with an average of 11 cM between loci. The markers that failed to genotype well were replaced by microsatellite markers from The Genome Database and from the Marshfield genome database. Genomic DNA was extracted from peripheral blood samples, by standard techniques. DNA was amplified by multiplex PCR assays with fluorescent primers designed to detect the microsatellite loci. Pipetting of the reactions was performed by a Hydra Microdispenser (Robbins Scientific). The amplification reactions were run in microtiter 96-well plates, by Tetrad thermal cyclers (MJ Research). The resulting PCR fragments, along with a size-standard ladder and fragments from known control individuals from Fondation Jean Dausset CEPH, were separated on 6% acrylamide gels by electrophoresis using a LI-COR DNA 4200 Genetic Analyzer (LI-COR). The bands on the gels were automatically interpreted into genotypes by the Saga1.0 software package (University of Washington and LI-COR). Saga takes digital gel images, tracks the lanes, and calls the alleles on the basis of a combination of size, peak shape, and marker characteristics (e.g., di-, tri-, or tetranucleotide repeat). Allele sizes were standardized to those of CEPH control individuals. All genotypes were verified by human inspection. The PedCheck1.1 (O'Connell and Weeks 1998) and SimWalk2 2.82 (Sobel and Lange 1996) computer programs were used to detect genotyping errors. If the mistyping was not resolved by review of the gel images, the suspected genotypes were set to be unknown.
Linkage Analyses
All analyses were performed with the disease-gene frequency set at 0.001, under the assumption of autosomal dominant inheritance and a phenocopy rate of 2.4% (Hovatta et al. 1994). Allele frequencies were calculated from the genotypes of all individuals genotyped in the analysis. Initially, subjects with MA (IHS classification 1.2), with the exception of the subjects with migraine aura without headache (EQV [IHS classification 1.2.5]), were classified as affected. All other subjects, with MO (IHS classification 1.1), headache other than migraine (HA), no headache (NoHA), or diagnosis unavailable (DU), were classified as unknown. Their genotypes are useful in the reconstruction of missing parental genotypes and phase information. The initial linkage analysis was performed by means of a two-point approach—that is, by using a single marker and the trait. Additionally, two-point linkage analyses were also performed with a broader phenotype, in which both individuals with MA and individuals with MO were considered to be affected. Two-point analysis is known to be less sensitive to map and genotyping errors than are multipoint methods (Risch and Giuffra 1992). Two-point parametric linkage analysis was performed both under locus homogeneity and under locus heterogeneity, by the LINKAGE (Lathrop and Lalouel 1984) and HOMOG (Ott 1983) computer programs. To investigate the possibility that a putative migraine gene may act in a recessive fashion, we employed an affected-sib-pair (ASP) analysis. The identity-by-descent (IBD) status of each ASP was estimated by the SIBPAIR program. Within the ASP analysis, we anticipate that some power is lost because pedigrees have been split into nuclear families, which have been analyzed as though they had been ascertained independently; however, this is not expected to bias the linkage results in any manner (Göring and Terwilliger 2000b). The program ANALYZE (Göring and Terwilliger 2000a) was used to conduct these analyses.
For regions showing evidence of linkage in the parametric two-point analysis, multipoint parametric and nonparametric analyses were performed by the GENEHUNTER program, version 2.1_r2beta (Kruglyak et al. 1996). Parametric linkage analysis was performed using the model presented above, while allowing for locus heterogeneity. The nonparametric statistic, NPLall, which estimates the statistical significance of alleles shared IBD between all affected family members, was calculated also, together with an estimated P value.
P values for LOD scores detected under homogeneity were estimated according to the method proposed by Nyholt (2000); P values for LOD scores detected under heterogeneity were estimated according to the method proposed by Chiano and Yates (1995). To aid in the interpretation of linkage results, the thresholds were set according to the recommendation made by Lander and Kruglyak (1995). For LOD scores detected under homogeneity, the thresholds are as follows: a pointwise threshold of P=.000049 (which is equivalent to a LOD score of 3.3), for significant linkage; a pointwise threshold of P=.0017 (LOD score 1.9), for suggestive linkage, and a pointwise threshold of P=.05 (LOD score 0.59), for nominal linkage.
Results
Parametric and Nonparametric Two-Point Linkage Analyses
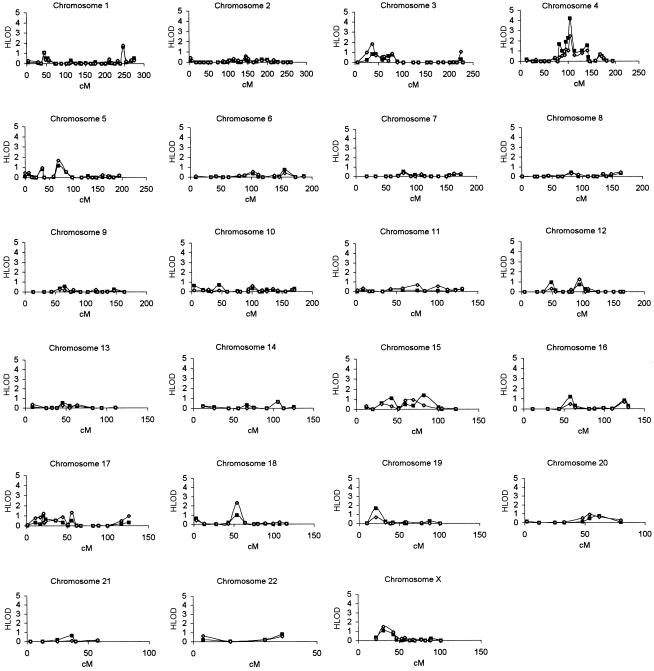
Parametric and nonparametric LOD scores were calculated using an affecteds-only strategy, for 350 markers. Initially, family members with MA (IHS classification 1.2.1) were classified as affected, and all others were classified as unknown (see the “Subjects and Methods” section); all results based on this phenotype classification are presented first. For parametric analyses, a dominant mode of inheritance and locus heterogeneity were employed. Plots of the two-point parametric LOD scores for each chromosome in the genomewide screen are shown in figure 1. In the genome screen, marker D4S1647 gave the highest LOD score, 4.20 (under homogeneity, P=.000006; under heterogeneity, P=.000011), at a recombination fraction (θ) of 0.18. This marker also gave an ASP LOD score of 2.25 (P=.0064) (table 5). Marker D4S2380, ∼4 cM proximal to marker D4S1647, had a two-point maximum LOD score of 2.26 (under heterogeneity; θ=0.08 and P=.0011), and an ASP LOD score of 1.30 (P=.0073). Marker D4S3240, ∼9 cM distal to marker D4S1647, had a two-point LOD score of 1.01 (under heterogeneity; θ=0.16 and P=.023) and an ASP LOD score of 0.63 (P=.041). Five other markers adjacent to marker D4S1647, spanning a region of 59 cM (D4S1517–D4S1520), showed two-point maximum LOD scores from 1.01 (P=.023) to 1.89 (P=.0026). Detailed results are presented in table 5.
Figure 1.
Two-point parametric linkage analyses (dominant model allowing for heterogeneity) for the 350-marker genomewide screen of 50 families with migraine. Blackened squares denote the two-point LOD scores when only individuals suffering from MA were considered to be affected; unblackened diamonds denote the two-point LOD scores for a broader phenotype, in which both individuals with MA and individuals with MO were considered to be affected. Analyses were performed by the LINKAGE program, as described in the text.
Table 5.
Locations and Two-Point LOD Scores for Markers with P<.05 in the Genomewide Screen[Note]
|
Results under Locus Homogeneity |
Results under Locus Heterogeneity |
Results of ASP Analysis |
|||||||
| Marker (Location) | Positiona(cM) | LOD Score | P | θb | HLOD Score | P | θb | LOD Score | P |
| D1S552 (1p36.13) | 45 | (.36) | (.099) | .28 | 1.08 | .020 | .00 | (.00) | (.50) |
| D1S3462 (1q42.2) | 247 | .70 | .036 | .30 | 1.66 | .0046 | .34 | (.00) | (.50) |
| D3S1259 (3p25.2) | 37 | .80 | .026 | .30 | .82 | .038 | .26 | .71 | .036 |
| D3S3038 (3p24.3) | 45 | .82 | .026 | .30 | .82 | .038 | .30 | (.52) | (.062) |
| D3S2409 (3p21.31) | 71 | .64 | .043 | .36 | (.64) | (.061) | .36 | (.13) | (.22) |
| D3S1766 (3p14.2) | 79 | .77 | .030 | .30 | .77 | .045 | .00 | (.12) | (.23) |
| D4S1517 (4q13.3) | 82 | 1.66 | .0029 | .26 | 1.66 | .0046 | .26 | 1.02 | .015 |
| D4S3243 (4q) | 88 | 1.01 | .016 | .26 | 1.01 | .023 | .26 | (.17) | (.19) |
| D4S2361 (4q21.23) | 93 | .61 | .047 | .30 | (.68) | (.055) | .22 | (.12) | (.23) |
| D4S2409 (4q22.1) | 96 | 1.85 | .0018 | .20 | 1.89 | .0026 | .16 | 1.00 | .016 |
| D4S2380 | 101 | 1.75 | .0023 | .22 | 2.26 | .0011 | .08 | 1.30 | .0073 |
| D4S1647 (4q24) |
105 | 4.20 | .000006 |
.18 | 4.20 | .000011 |
.18 | 2.25 | .0064 |
| D4S3240 (4q25) | 114 | .76 | .031 | .30 | 1.01 | .023 | .16 | .63 | .041 |
| D4S2394 (4q28.2) | 130 | .98 | .017 | .28 | 1.05 | .021 | .22 | (.02) | (.38) |
| D4S1520 (4q31.1) | 141 | 1.55 | .0038 | .20 | 1.55 | .0061 | .20 | (.47) | (.071) |
| D5S2845 (5p14.3) | 36 | .82 | .026 | .24 | .81 | .039 | .24 | (.36) | (.10) |
| D5S2500 (5q12.1) | 69 | 1.11 | .012 | .26 | 1.16 | .016 | .20 | (.36) | (.099) |
| D6S2436 (6q25.2) | 155 | .75 | .032 | .34 | .75 | .046 | .34 | .61 | .047 |
| D12S1042 (12p11.23) | 49 | .94 | .019 | .32 | .94 | .028 | .32 | (.35) | (.10) |
| D12S1064 (12q21.33) | 95 | (.42) | (.082) | .36 | .74 | .047 | .00 | (.00) | (.50) |
| D15S659 (15q21.1) | 43 | (.05) | (.32) | .42 | 1.09 | .019 | .06 | (.00) | (.50) |
| D15S655 (15q25.3) | 83 | .70 | .036 | .24 | 1.38 | .0092 | .04 | (.31) | (.12) |
| D16S753 (16p12.3) | 58 | (.004) | (.45) | .40 | 1.22 | .014 | .00 | (.07) | (.29) |
| D16S539 (16q24.1) | 125 | .84 | .025 | .34 | .84 | .036 | .34 | (.00) | (.5) |
| D17S945 (17p13.1) | 21 | .96 | .018 | .30 | 1.01 | .023 | .24 | .61 | .047 |
| D18S877 (18q12.1) | 54 | (.18) | (.18) | .40 | 1.00 | .024 | .00 | (.08) | (.27) |
| D19S427 (19p13.2) | 21 | 1.70 | .0026 | .22 | 1.70 | .0042 | .22 | (.08) | (.27) |
| D22S683 (22q12.3) | 36 | (.41) | (.085) | .34 | .82 | .038 | .08 | (.44) | (.077) |
| DXS9896 (Xp) | 31 | 1.08 | .013 | .30 | 1.08 | .020 | .30 | (.13) | (.22) |
| DXS6810 (Xp11.4) | 43 | .65 | .042 | .34 | (.65) | (.060) | .34 | (.31) | (.12) |
Note.— Results are based on analyses of individuals with MA; parentheses denote entries for which P>.05; the significant P value (and the marker for it) is underlined, and the suggestive P value (and the marker for it) is in boldface italic.
Distance from pter, according to the map published by the Center for Medical Genetics, Marshfield Medical Research Foundation.
Values are those at which the maximum LOD score was found.
Statistically significant or suggestive linkage was not observed in any other chromosomal region. However, nominal linkage with parametric two-point LOD scores giving P values ⩽.05 were found at 21 additional loci, in 1p, 1q, 3p, 5p, 5q, 6q, 12p, 12q, 15q, 16p, 16q, 17p, 18q, 19p, 22q, and Xp. These results are presented in detail in table 5. Of these regions, two—1q and 19p—are of specific interest because they previously have been linked to either FHM (Joutel et al. 1993; Ducros et al. 1997; Gardner et al. 1997) or MA and MO (May et al. 1995; Ophoff et al. 1997; Nyholt et al. 1998b; Terwindt et al. 2001). Therefore, results regarding these regions are presented here in more detail. Also, results regarding the X chromosome will be presented in more detail because several previous and recent studies have provided evidence of linkage of the X chromosome to common forms of migraine (Nyholt et al. 1998a, 2000; Oterino et al. 2001; Wieser et al. 2001).
Ten markers were genotyped on chromosome 19, from 10 to 101 cM (where pter is at 0 cM and qter is at 105 cM). Marker D19S1150, at a distance of ∼38 cM, is an intragenic marker of CACNA1A and was included in the screen. This marker showed no evidence of linkage in our families (HLOD 0.00). However, marker D19S427, located ∼20 cM distal to marker D19S1150, showed nominal evidence of linkage, with a maximum LOD score of 1.70 (θ=0.22; under homogeneity, P=.0026; under heterogeneity, P=.0042).
Thirty-one markers were genotyped on chromosome 1, from 4 to 275 cM (where pter is at 0 cM and qter is at 290 cM). Marker D1S3462, on 1q42 (at 247 cM), and marker D1S522, on 1p36 (at 45 cM), showed nominal evidence of linkage, with HLOD values of 1.66 (θ=0.34; P=.0046) and 1.08 (θ=0.00; P=.02), respectively.
Fourteen markers were genotyped on the X chromosome from 22 to 101 cM (where pter is at 0 cM and qter is at 103 cM); of these, marker DXS9896, at 31 cM, showed a two-point maximum LOD score of 1.08 (θ=0.30; under homogeneity, P=.013; under heterogeneity, P=.02), with nominal evidence of linkage. No evidence of linkage was observed on Xq.
In addition, markers D14S1426 (at 126 cM), D17S1830 (at 117 cM), and D18S535 (at 64 cM) gave LOD scores of 1.13 (P=.011), 1.44 (P=.005 ), and 1.41 (P=.005), respectively, in the nonparametric ASP analysis.
Parametric Three-Point Linkage Analysis
Parametric three-point linkage analysis—that is, analysis using two markers and the trait, on all pairs of adjacent markers—was performed in the region from marker D4S2409 (at 96 cM) to marker D4S3240 (at 114 cM). The maximum three-point HLOD, 3.68 (P=.0004), was observed with the D4S2380-D4S1647 pair, suggesting that the best evidence of linkage is obtained in this 4-cM region.
Multipoint Analyses
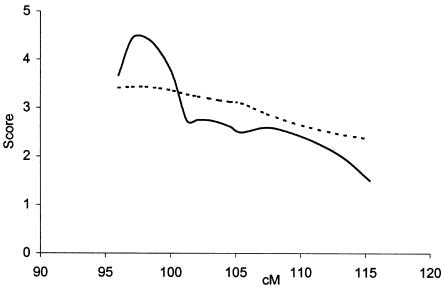
To pool information from multiple markers and to position the linked locus on 4q24, we performed parametric and nonparametric multipoint analyses, using the GENEHUNTER program. The parametric test gave the highest HLOD, 4.45 (P=.0000058), between markers D4S2409 and D4S2380 (fig. 2). The estimated fraction of linked pedigrees, α, at HLOD was 0.23–0.5. The results of the nonparametric analysis are consistent with those of the parametric result, giving a maximum NPLall of 3.43 (P=.0007) at the same position (fig. 2).
Figure 2.
Multipoint parametric and nonparametric linkage analyses of 4q, with markers D4S2409 (96 cM), D4S2380 (101 cM), D4S1647 (105 cM), and D4S3240 (114 cM). Analyses were performed by the GENEHUNTER program, as described in the text. The vertical axis presents both the parametric (i.e., HLOD [unbroken line]) and nonparametric (i.e., NPLall [dotted line]) values along a 20-cM region of chromosome 4.
Parametric Two-Point Linkage Analysis for a Broader Phenotype
Two-point linkage analyses were also performed with use of a broader phenotype, in which both individuals with MA and individuals with MO were considered to be affected. A plot of the two-point parametric LOD scores (under heterogeneity) for each chromosome is shown in figure 1. No significant linkage to any chromosomal region was detected. Marker D4S1647 showed only suggestive evidence of linkage, with a LOD score of 2.48 (θ=0.22), both under homogeneity (P=.00037) and under heterogeneity (P=.00064). Marker D18S877, at 31 cM, also showed suggestive evidence of linkage, with a two-point maximum LOD score of 2.32 (θ=0.00; under heterogeneity, P=.00093).
Discussion
Our results, based on the genotyping of 50 Finnish families with MA, provide strong evidence of a novel migraine-susceptibility locus on 4q24. Statistically significant evidence of linkage, both under locus homogeneity and under locus heterogeneity (LOD score 4.20; P=.000006 and P=.000011, respectively), between the common MA phenotype and marker D4S1647 was obtained with a parametric two-point linkage analysis, under a dominant mode of inheritance. Parametric and nonparametric multipoint analyses supported linkage to this chromosomal region. The multipoint analyses found the highest HLOD scores, 4.45 (P=.0000058), and NPLall = 3.43 (P=.0007), in the region between markers D4S2409 and D4S2380, 7 cM proximal of the D4S1647 locus.
Our successful identification of linkage to 4q24 was most probably due to the selection strategy used in our study sample, which emphasized clustering in families with a specific phenotype, MA. Additionally, to reduce heterogeneity, a single neurologist carefully analyzed all the clinical data. Only families with MA throughout the pedigree were considered for further analysis. On the basis of these carefully diagnosed families with predominant MA, we were able to select reasonably large, multigenerational families for our linkage study. The importance of careful family selection in linkage studies of complex traits has recently, elegantly been demonstrated by Brzustowicz et al. (2000), who were able to locate, on 1q21-q22, a major susceptibility locus for familial schizophrenia in 22 extended families with high rates of schizophrenia. In genetically homogeneous populations, such as the Finns, in which there are potentially only a few affected founders, linkage analysis of complex traits can be especially powerful, provided that the clinical phenotype is properly dissected. Finland is not the only country in which there is a genetically isolated population appropriate for genetic studies, but the additional features of well-kept population records and a high-quality, state-run health-care system make it a very reliable resource for the genetic studies of complex traits (Peltonen et al. 2000).
The strength of analysis of a more stringently defined migraine phenotype was reflected in the drop in the two-point LOD scores for 4q when a broader disease classification was included, in which both individuals with MA and individuals with MO were considered to be affected. Additionally, no other statistically significant locus was detected for this combined phenotype. It is likely that the increased heterogeneity and possible phenocopies introduced into the etiology by inclusion of the MO phenotypes are responsible for these differences. However, the current sample is not large enough to allow us to investigate whether MA and MO are genetically distinct entities.
To the best of our knowledge, no other report of a statistically significant linkage of common MA phenotypes to any autosomal region has been published. The present study's finding therefore suggests the existence of a novel MA locus and must be tested in independent studies. However, results of two other candidate-gene studies have indicated that two possible loci on chromosome 4 are associated with migraine. An association study by Pardo et al. (1995), of 112 unrelated patients with migraine (62 patients with MO and 50 patients with MA), provided evidence that the group-component locus (GC [MIM 139200]) on 4q12-13.3 is involved in migraine. GC codes for a multifunctional protein found in plasma, for example, where it carries the vitamin-D sterols and prevents polymerization of actin by binding its monomers. Tzourio et al. (2001) found an association between migraine and a polymorphism at the endothelin type A–receptor gene (ENDRA [MIM 131243]) on 4q31. Endothelin receptors mediate the biological effects of endothelin (ET-1 [MIM 131240]), a potential vasoconstrictor, which has been implicated in migraine, by recent studies showing increased plasma ET-1 levels during and between migraine attacks (Färkkilä et al. 1992; Kallela et al. 1998; Hasselblatt et al. 1999). GC and ENDRA are good candidate genes for our further studies, although they are located some distance from marker D4S1647 (GC is 30 Mb proximal, and ENDRA is 50 Mb distal). The ∼60-cM region around our most informative marker, D4S1647, contains ⩾170 known genes, including several obvious candidate genes (UCSC Human Genome Project Working Draft).
In addition to the significant finding with regard to 4q, the present study has revealed 21 markers, on 12 different chromosomes, that provide nominal evidence of linkage. However, in light of the LOD scores for them, the risk of false-positive signals is considerable. Nonetheless, of special interest are regions on chromosomes 1q, 19p, and X, where several earlier studies found evidence of linkage or association with migraine. The CACNA1A locus on 19p13, which Joutel et al. (1993) reported as being linked to FHM, has also been indicated as being involved in nonhemiplegic migraine. May et al. (1995) studied how CACNA1A markers are involved with migraine in 28 German families, and Terwindt et al. (2001) do so later in 36 extended Dutch families. Both groups concluded that the increased allele sharing around the CACNA1A gene on 19p13 is consistent with an important involvement of this region in migraine, especially MA. Nyholt et al. (1998b) reported significant cosegregation and allele sharing for markers situated within or adjacent to the CACNA1A locus in one of the four large Australian families that they analyzed. On the other hand, our two-point linkage analysis of Finnish families showed no evidence of linkage of the MA phenotype to the intragenic marker D19S1150 of CACNA1A. This finding is in agreement with our previous results excluding this region as a site for a migraine locus in Finnish families with migraine (Hovatta et al. 1994); however, two of the four families in that previous study (Hovatta et al. 1994) also were included in the present study. It should be stressed that our data are from families collected from a genetically homogeneous population and that, when other ethnic groups are analyzed, locus heterogeneity is likely to exist; however, we have found nominal linkage to marker D19S427 (LOD score 1.70; P=.0042), located in the proximity of the insulin-receptor gene (INSR [MIM 147670]) that White et al. (2001) recently have associated with migraine. Both the findings by White et al. (2001) and those of our group support the notion that genes other than CACNA1A might contribute to the linkage and association findings on 19p13. These regions obviously require further study with other samples and, possibly, with further phenotypic stratification.
We have found nominal evidence of linkage at marker D1S3462 (LOD score 1.70; P=.0046) on 1q42. There is no previous linkage evidence that would indicate that this region is involved in migraine. One locus—or, possibly, two loci—for FHM have been mapped to 1q21-31 (Ducros et al. 1997; Gardner et al. 1997). Interestingly, the 1q21-42 region harbors several ion-channel genes that are possible candidates for both FHM and MA.
Our finding of nominal linkage to the X chromosome, at marker DXS9896 (LOD score 1.08; P=.02) is noteworthy, since several other studies have provided evidence of linkage of the X chromosome to migraine. Studies by Nyholt et al. (1998a, 2000) have implicated a locus on Xq24-28 in two Australian families with migraine. Interestingly, Oterino et al. (2001) have suggested the connexin 32 gene (GJB1 [MIM 304040]) on Xq13 as a possible candidate for MA in a family with both Chargot-Marie-Tooth disease (MIM 302800) and MA. Furthermore, Wieser et al. (2001) have reported 80 families (from Spain, Germany, and the United States) with migraine and possible X-linked dominant inheritance, providing evidence of allele sharing and linkage to the X chromosome. Further studies are necessary to confirm the role of the X-chromosomal loci in genetic predisposition to migraine.
In conclusion, the present study provides strong evidence of an MA-susceptibility locus on 4q24. Further studies are needed to delineate the role of the other potential loci involved in MA in the families analyzed in the present study. However, the statistically significant evidence of the linkage to 4q24 should facilitate efforts to identify an underlying migraine-susceptibility gene. The detection of underlying mutation(s) will provide clues to the further elucidation of the complex molecular pathways of migraine and, finally, will help in the development of rational treatment strategies.
Acknowledgments
Financial support by National Institutes of Health grants RO1 NS37675-02 (to A.P.) and R01 HG00008 (to J.O.) and by the Oxnard Foundation (support to A.P), the Research Funds of the Helsinki University Hospital (support to A.P., M.F, M.K, and M.A.K.), the Academy of Finland (support to M.W. and P.M.), the Paulo Foundation (support to M.W. and P.M.), the Finnish Society of Neurology (support to M.K.), the Finnish Cultural Foundation (support to M.A.K. and P.M.), the Wihuri Research Foundation (support to P.M.), and the Emil Aaltonen Foundation (support to P.M.) is gratefully acknowledged. We especially thank the Finnish families with migraine, for their devoted participation in this study. Finally, we wish to thank our laboratory technicians—Jovita Sain, Maritta Putkiranta, Susanna Wikman, and Arja Korhonen—and our nurses—Liisa Hilden, Ms. Liisa Aaltoila, and Mari Kuutti—for their excellent work in this migraine project.
Electronic Data-Base Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for genetic-linkage distances)
- Fondation Jean Dausset CEPH, http://www.cephb.fr (for heterozygosity and allele sizes)
- Genome Database, The, http://www.gdb.org/ (for primers used for genotyping, heterozygosity, and allele sizes)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for migraine [MIM 157300], FHM1 [MIM 141500], CACNA1A [MIM 601011], EA-2 [MIM 108500], SCA6 [MIM 183086], FHM2 [MIM 602481], typical familial migraine [MIM 300125], GC [MIM 139200], ENDRA [MIM 131243], ET-1 [MIM 131240], INSR [MIM 147670], GJB1 [MIM 304040], and Chargot-Marie-Tooth disease [MIM 302800])
- UCSC Human Genome Project Working Draft, http://genome.ucsc.edu/ (for gene information)
References
- Bille B (1997) A 40-year follow-up of school children with migraine. Cephalalgia 17:488–491 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 288:678–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiano MN, Yates JRW (1995) Linkage detection under heterogeneity and the mixture problem. Ann Hum Genet 59:83–95 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A (1993) Disease gene mapping in isolated human populations: the example of Finland. J Med Genet 30:857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Zompo M, Cherchi A, Palmas MA, Ponti M, Bocchetta A, Gessa GL, Piccardi MP (1998) Association between dopamine receptor genes and migraine without aura in a Sardinian sample. Neurology 51:781–786 [DOI] [PubMed] [Google Scholar]
- Ducros A, Denier C, Joutel A, Cecillon M, Lescoat C, Vahedi K, Darcel F, Vicaut E, Bousser MG, Tournier-Lasserve E (2001) The clinical spectrum of familial hemiplegic migraine associated with mutations in a neuronal calcium channel. N Engl J Med 345:17–24 [DOI] [PubMed] [Google Scholar]
- Ducros A, Joutel A, Vahedi K, Cecillon M, Ferreira A, Bernard E, Verier A, Echenne B, Lopez de Manain A, Boussier MG, Tournier-Lasserve E (1997) Mapping of a second locus for familial hemiplegic migraine to 1q21-q23 and evidence of further heterogeneity. Ann Neurol 42:885–890 [DOI] [PubMed] [Google Scholar]
- Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, Juvonen H, Varilo T, Arajarvi R, Kokko-Sahin ML, Lönnqvist J, Peltonen L (2000) Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet 9:1049–1057 [DOI] [PubMed] [Google Scholar]
- Färkkilä M, Palo J, Saijonmaa O, Fyhrquist F (1992) Raised plasma endothelin during acute migraine attack. Cephalalgia 12:383–384 [DOI] [PubMed] [Google Scholar]
- Gardner K, Barmada MM, Ptacek LJ, Hoffman EP (1997) A new locus for hemiplegic migraine maps to chromosome 1q31. Neurology 49:1231–1238 [DOI] [PubMed] [Google Scholar]
- Gervil M, Ulrich V, Kaprio J, Olesen J, Russell MB (1999a) Genetic and environmental factors in migraine. Neurology 53:995–999 [DOI] [PubMed] [Google Scholar]
- Gervil M, Ulrich V, Kyvik KO, Olesen J, Russell MB (1999b) Migraine without aura: a population-based twin study. Ann Neurol 46:606–611 [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD (2000a) Linkage analysis in the presence of errors III: marker loci and their map as nuisance parameters. Am J Hum Genet 66:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2000b) Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen K, Zwart J, Vatten L, Stovner L, Bovim G (2000) Prevalence of migraine and non-migrainous headache--head-HUNT, a large population-based study. Cephalalgia 20:900–906 [DOI] [PubMed] [Google Scholar]
- Hasselblatt M, Kohler J, Volles E, Ehrenreich H (1999) Simultaneous monitoring of endothelin-1 and vasopressin plasma levels in migraine. Neuroreport 10:423–425 [DOI] [PubMed] [Google Scholar]
- Headache Classification Committee of the International Headache Society (1988) Classification and diagnostic criteria for headache disorders, cranial neuralgias and facial pain. Cephalalgia 8 Suppl 7:1–96 [PubMed] [Google Scholar]
- Henry P, Michel P, Brochet B, Dartigues JF, Tison S, Salamon R (1992) A nationwide survey of migraine in France: prevalence and clinical features in adults. GRIM. Cephalalgia 12:229–237 [DOI] [PubMed] [Google Scholar]
- Honkasalo ML, Kaprio J, Winter T, Heikkila K, Sillanpaa M, Koskenvuo M (1995) Migraine and concomitant symptoms among 8167 adult twin pairs. Headache 35:70–78 [DOI] [PubMed] [Google Scholar]
- Hovatta I, Kallela M, Färkkilä M, Peltonen L (1994) Familial migraine: exclusion of the susceptibility gene from the reported locus of familial hemiplegic migraine on 19p. Genomics 23:707–709 [DOI] [PubMed] [Google Scholar]
- Hovatta I, Varilo T, Suvisaari J, Terwilliger J, Ollikainen V, Arajärvi R, Juvonen H, Kokko-Sahin M, Väisänen L, Mannila H, Lönnqvist J, Peltonen L (1999) A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet 65:1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodice C, Mantuano E, Veneziano L, Trettel F, Sabbadini G, Calandriello L, Francia A, Spadaro M, Pierelli F, Salvi F, Ophoff RA, Frants RR, Frontali M (1997) Episodic ataxia type 2 (EA2) and spinocerebellar ataxia type 6 (SCA6) due to CAG repeat expansion in the CACNA1A gene on chromosome 19p. Hum Mol Genet 6:1973–1978 [DOI] [PubMed] [Google Scholar]
- Joutel A, Bousser M-G, Biousse V, Labauge P, Chabriat H, Nibbio A, Maciazek J, Meyer B, Bach M-A, Weissenbach J, Lathrop GM, Tournier-Lasserve E (1993) A gene for familial hemiplegic migraine maps to chromosome 19. Nat Genet 5:40–45 [DOI] [PubMed] [Google Scholar]
- Kallela M, Färkkilä M, Saijonmaa O, Fyhrquist F (1998) Endothelin in migraine patients. Cephalalgia 18:329–332 [DOI] [PubMed] [Google Scholar]
- Kallela M, Wessman M, Färkkilä M (2001) Validation of a migraine specific questionnaire for use in family studies. Eur J Neurol 8:61–66 [DOI] [PubMed] [Google Scholar]
- Kowa H, Yasui K, Takeshima T, Urakami K, Sakai F, Nakashima K (2000) The homozygous C677T mutation in the methylenetetrahydrofolate reductase gene is a genetic risk factor for migraine. Am J Med Genet 96:762–764 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S, Gschwend M, Rioux J, Daly M, Terwilligr J, Tienari P, Wikstrom J, Palo J, Stein L, Hudson T, Lander E, Peltonen L (1997) Genomewide scan of multiple sclerosis in Finnish multiplex families. Am J Hum Genet 61:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen T, Daly MJ, Rioux JD, Kauppi P, Laprise C, Petays T, Green T, Cargill M, Haahtela T, Lander ES, Laitinen LA, Hudson TJ, Kere J (2001) A susceptibility locus for asthma-related traits on chromosome 7 revealed by genome-wide scan in a founder population. Nat Genet 28:87–91 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guideline for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lander E, Schork N (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed] [Google Scholar]
- Larsson B, Bille B, Pederson NL (1995) Genetic influence in headaches: a Swedish twin study. Headache 35:513–519 [DOI] [PubMed] [Google Scholar]
- Lathrop G, Lalouel J (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lea RA, Dohy A, Jordan K, Quinlan S, Brimage PJ, Griffiths LR (2000) Evidence for allelic association of the dopamine β-hydroxylase gene (DBH) with susceptibility to typical migraine. Neurogenetics 3:35–40 [DOI] [PubMed] [Google Scholar]
- Leppävuori J, Kujala U, Kinnunen J, Kaprio J, Nissila M, Heliovaara M, Klinger N, Partanen J, Terwilliger JD, Peltonen L (1999) Genome scan for predisposing loci for distal interphalangeal joint osteoarthritis: evidence for a locus on 2q. Am J Hum Genet 65:1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton RB, Silberstein SD, Stewart WF (1994) An update on the epidemiology of migraine. Headache 34:319–328 [DOI] [PubMed] [Google Scholar]
- Lipton RB, Stewart WF, Diamond S, Diamond ML, Reed M (2001) Prevalence and burden of migraine in the United States: data from the American Migraine Study II. Headache 41:646–657 [DOI] [PubMed] [Google Scholar]
- Mahtani MM, Widen E, Lehto M, Thomas J, McCarthy M, Brayer J, Bryant B, Chan G, Daly M, Forsblom C, Kanninen T, Kirby A, Kruglyak L, Munnelly K, Parkkonen M, Reeve-Daly MP, Weaver A, Brettin T, Duyk G, Lander ES, Groop LC (1996) Mapping of a gene for type 2 diabetes associated with an insulin secretion defect by a genome scan in Finnish families. Nat Genet 14:90–94 [DOI] [PubMed] [Google Scholar]
- May A, Ophoff RA, Terwindt GM, Urban C, van Eijk R, Haan J, Diener HC, Lindhout D, Frants RR, Sandkuijl LA, Ferrari MD, van Eijk R (1995) Familial hemiplegic migraine locus on 19p13 is involved in the common forms of migraine with and without aura. Hum Genet 96:604–608 [DOI] [PubMed] [Google Scholar]
- Mochi M, Sangiorgi S, Cortelli P, Carelli V, Scapoli C, Crisci M, Monari L, Pierangeli G, Montagna P (1993) Testing models for genetic determination in migraine. Cephalalgia 13:389–394 [DOI] [PubMed] [Google Scholar]
- Nyholt DR (2000) All LODs are not created equal. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR, Curtain RP, Griffiths LR (2000) Familial typical migraine: significant linkage and localization of a gene to Xq24-28. Hum Genet 107:18–23 [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Dawkins JL, Brimage PJ, Goadsby PJ, Nicholson GA, Griffiths LR (1998a) Evidence for an X-linked genetic component in familial typical migraine. Hum Mol Genet 7:459–463 [DOI] [PubMed] [Google Scholar]
- Nyholt D, Lea R, Goadsby P, Brimage P, Griffiths L (1998b) Familial typical migraine: linkage to chromosome 19p13 and evidence for genetic heterogeneity. Neurology 50:1428–1432 [DOI] [PubMed] [Google Scholar]
- O'Brien B, Goeree R, Streiner D (1994) Prevalence of migraine headache in Canada: a population-based survey. Int J Epidemiol 23:1020–1026 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie AD, Russell MB, Dhall P, Battersby S, Ulrich V, Smith CA, Goodwin GM, Harmar AJ, Olesen J (1998) Altered allelic distributions of the serotonin transporter gene in migraine without aura and migraine with aura. Cephalalgia 18:23–26 [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouwe MN, Frants RR, Ferrari MD (1997) Involvement of a Ca2+channel gene in familial hemiplegic migraine and migraine with and without aura. Headache 37:479–485 [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouve MN, van Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW, Bulman DE, Ferrari M, Haan J, Lindhout D, van Ommen GJ, Hofker MH, Ferrari MD, Frants RR (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutation in the Ca2+ channel gene CACNL1A4. Cell 87:543–552 [DOI] [PubMed] [Google Scholar]
- Oterino A, Monton F, Cid C, Ruiz-Lavilla N, Gardner K, Barmada M, Pascual J (2001) A new locus for migraine with aura on Xq13. Cephalalgia 21:346 [Google Scholar]
- Ott J (1983) Linkage analysis and family classification under heterogeneity. Ann Hum Genet 47:311–320 [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Terwilliger JD, Perola M, Hiekkalinna T, Nuotio I, Ellonen P, Parkkonen M, Hartiala J, Ylitalo K, Pihlajamaki J, Porkka K, Laakso M, Viikari J, Ehnholm C, Taskinen MR, Peltonen L (1999) Genomewide scan for familial combined hyperlipidemia genes in Finnish families, suggesting multiple susceptibility loci influencing triglyceride, cholesterol, and apolipoprotein B levels. Am J Hum Genet 64:1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J, Carracedo A, Munoz I, Castillo J, Lema M, Noya M (1995) Genetic markers: association study in migraine. Cephalalgia 15:200–204 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Peroutka SJ, Wilhoit T, Jones K (1997) Clinical susceptibility to migraine with aura is modified by dopamine D2-receptor (DRD2) Nco I alleles. Neurology 49:201–206 [DOI] [PubMed] [Google Scholar]
- Rasmussen BK, Olesen J (1992) Migraine with aura and migraine without aura: an epidemiological study. Cephalalgia 12:221–228; discussion 186 [DOI] [PubMed] [Google Scholar]
- Risch N, Giuffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed] [Google Scholar]
- Russell MB, Iselius L, Olesen J (1995) Inheritance of migraine investigated by complex segregation analysis. Hum Genet 96:726–730 [DOI] [PubMed] [Google Scholar]
- Russell M, Olesen J (1993) The genetics of migraine without aura and migraine with aura. Cephalalgia 13:245–248 [DOI] [PubMed] [Google Scholar]
- ——— (1995) Increased familial risk and evidence of genetic factor in migraine. BMJ 311:541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell MB, Rasmussen BK, Fenger K, Olesen J (1996) Migraine without aura and migraine with aura are distinct clinical entities: a study of four hundred and eighty-four male and female migraineurs from the general population. Cephalalgia 16:239–245 [DOI] [PubMed] [Google Scholar]
- Russell MB, Ulrich V, Gervil M, Olesen J (2001) Migraine without aura and migraine with aura are distinct disorders: a population-based twin survey. Cephalalgia 21:346 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: application to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Stewart W, Lipton R, Celentano D, Reed M (1992) Prevalence of migraine headache in the United States. JAMA 267:64–69 [PubMed] [Google Scholar]
- Terwindt GM, Ophoff RA, van Eijk R, Vergouwe MN, Haan J, Frants RR, Sandkuijl LA, Ferrari MD (2001) Involvement of the CACNA1A gene containing region on 19p13 in migraine with and without aura. Neurology 56:1028–1032 [DOI] [PubMed] [Google Scholar]
- Tzourio C, El Amrani M, Poirier O, Nicaud V, Bousser MG, Alperovitch A (2001) Association between migraine and endothelin type A receptor (ETA -231 A/G) gene polymorphism. Neurology 56:1273–1277 [DOI] [PubMed] [Google Scholar]
- Ulrich V, Gervil M, Fenger K, Olesen J, Russell MB (1999a) The prevalence and characteristics of migraine in twins from general population. Headache 39:173–180 [DOI] [PubMed] [Google Scholar]
- Ulrich V, Gervil M, Kyvik K, Olesen J, Russell M (1999b) Evidence of a genetic factor in migraine with aura: a population-based Danish twin study. Ann Neurol 45:242–246 [DOI] [PubMed] [Google Scholar]
- White NJ, Hosford DA, Humprey PP, Boyd PR, Griffits LR, Peroutka S, Roses AD, Purvis IJ, McCarthy LC (2001) Single nucleotide polymorphism (SNP) alleles in the insulin receptor (INSR) gene are associated with migraine. Cephalalgia 21:280 [DOI] [PubMed] [Google Scholar]
- Wieser T, Pascual J, Barmada MM, Soso M, Onterino A, Gardner KL (2001) Genetic analysis of the X-chromosome in migraine families. Cephalalgia 21:280 [Google Scholar]
- Ziegler DK, Hur YM, Bouchard TJJ, Hassanein RS, Barter R (1998) Migraine in twins raised together and apart. Headache 38:417–422 [DOI] [PubMed] [Google Scholar]