Abstract
Multiple sclerosis (MS) is a chronic inflammatory disorder of the central nervous system, with a complex etiology that includes a strong genetic component. The contribution of the major histocompatibility complex (MHC) has been established in numerous genetic linkage and association studies. In addition to the MHC, the chromosome 19q13 region surrounding the apolipoprotein E (APOE) gene has shown consistent evidence of involvement in MS when family-based analyses were conducted. Furthermore, several clinical reports have suggested that the APOE-4 allele may be associated with more-severe disease and faster progression of disability. To thoroughly examine the role of APOE in MS, we genotyped its functional alleles, as well as seven single-nucleotide polymorphisms (SNPs) located primarily within 13 kb of APOE, in a data set of 398 families. Using family-based association analysis, we found statistically significant evidence that an SNP haplotype near APOE is associated with MS susceptibility (P=.005). An analysis of disease progression in 614 patients with MS from 379 families indicated that APOE-4 carriers are more likely to be affected with severe disease (P=.03), whereas a higher proportion of APOE-2 carriers exhibit a mild disease course (P=.02).
Introduction
Multiple sclerosis (MS [MIM 126200]) is a chronic inflammatory disorder of the CNS characterized by destruction of the myelin sheath, gliosis, varying degrees of axonal pathology, and progressive neurological dysfunction (Hauser and Goodkin 2001). MS is a major cause of morbidity and disability in young adults, with a prevalence of 0.1% in individuals of northern European origin (Kurtzke 1983; Rosati 1994). Genetic factors have been implicated by numerous studies, with an estimated sibling-recurrence risk of 20%–40% and a greatly increased concordance rate in MZ, compared with DZ, twins (Sadovnick et al. 1993; Ebers et al. 1995). Earlier clinical studies and recent genomic screens (Ebers et al. 1996; Sawcer et al. 1996; Haines et al. 1996; Kuokkanen et al. 1997) have found the strongest and most widely replicated evidence for an MS susceptibility gene at the major histocompatibility complex (MHC) region on chromosome 6p21.3. Here, the class II DR2 haplotype of the human leukocyte antigen (HLA) system (HLA-DRB1*1501-DQA1*0102-DQB1*0602) has been consistently associated with predisposition to MS in white populations of northern European descent. HLA has been estimated to account for 17%–62% of the genetic component of MS susceptibility (Haines et al. 1998), but the exact mechanism by which it increases disease risk has not yet been determined.
One of the first non-MHC regions of interest identified through linkage analysis was chromosome 19q13, near the apolipoprotein E (APOE [MIM 107741]) locus (Haines et al. 1993, 1996). Three other genomic screens (Ebers et al. 1996; Sawcer et al. 1996; Kuokkanen et al. 1997) have shown some support for linkage to this region, and a meta-analysis of all four genomic screens (Wise et al. 1999) identified 19q13 as the second-most-significant region after the MHC. Additional evidence for this region came from allelic association studies (Barcellos et al. 1997; Zouali et al. 1999; D'Alfonso et al. 2000) and, more recently, from follow-up analyses by the Multiple Sclerosis Genetics Group (MSGG) in both North American (Pericak-Vance et al. 2001) and San Marinese populations (Haines et al. 2000). As with most complex diseases, the data are not entirely consistent; not all studies have shown evidence of linkage (Chataway et al. 1998; D'Alfonso et al. 1999), and association results were based on polymorphisms in different markers.
One of several candidate genes in the 19q13 region is the APOE gene, which codes for a major lipid carrier protein (apoE) in the brain. The apoE protein has long been associated with regeneration of axons and myelin after lesions of central and peripheral nervous tissue (Ignatius et al. 1986), and its isoforms have been shown to have differential effects on neuronal growth (Nathan et al. 1994). Because decreased apoE concentrations have been observed in cerebrospinal fluid in patients with MS, some investigators have proposed that a corresponding decrease in intrathecal apoE synthesis influences the degree of MS exacerbation (Rifai et al. 1987; Gaillard et al. 1998). Immunomodulatory and antioxidant effects of APOE have also been reported (Laskowitz et al. 1998). Therefore, both coding-region polymorphisms and promoter-region variants that modify APOE expression levels are plausible MS candidate genes. In clinical and epidemiologic studies, the role of APOE in MS has been somewhat controversial because of reports of both the presence (Dousset et al. 1998; Evangelou et al. 1999; Lucotte et al. 2000) and the absence (Ferri et al. 1999; Pirttilä et al. 2000; Weatherby et al. 2000a, 2000b) of an association of the APOE-4 allele with susceptibility to or severity of the disease. The limited sample sizes of many of these earlier studies may partly explain the inconsistent findings. Recently, two larger studies reported statistically significant evidence of a more severe clinical course among APOE-4 carriers with MS. One of these studies (Fazekas et al. 2001) employed a disease activity score (progression index [Weinshenker et al. 1997]) to measure MS severity, whereas the other study (Chapman et al. 2001) examined latency to severe disease, by use of a Cox-regression model.
Given the strong evidence of a potential role of APOE in MS, the goal of the present study was to test the functional APOE polymorphism (alleles ε2, ε3, and ε4) and several nearby single-nucleotide polymorphisms (SNPs) for association with MS susceptibility or severity. The SNPs used were a subset of those previously found to be in linkage disequilibrium (LD) with the APOE polymorphism and to be associated with late-onset Alzheimer disease (Martin et al. 2000a), in which the APOE-4 allele is known to increase disease susceptibility (Corder et al. 1993; Saunders et al. 1993). To extract as much transmission information as possible, statistical methods included both single- and multilocus family-based association analyses. Genotype-phenotype association analyses, based on clinical variables among patients with MS, were also performed.
Subjects and Methods
Subjects
A total of 398 white families that included one or more affected members were ascertained by the University of California at San Francisco and a network of collaborating clinical sites throughout the United States. Ascertainment methods included physician referrals, self-referrals, and responses to advertisements, as described elsewhere (Haines et al. 1996). Family ascertainment focused on probands with relapsing-remitting MS, thus enriching our overall patient sample for this disease type, relative to primary progressive MS. Of the 398 families, 148 had two or more sampled affected relatives. This includes most of the original 52 multiplex families genotyped in our genome screen (Haines et al. 1996) and the 46 families ascertained for follow-up analysis (Haines et al. 1998). An effort was made to extend the multiplex families through all affected first-degree relatives. The 250 additional families had one sampled affected member and no record of affected relatives. To include these families in our association analysis, one or more unaffected siblings and/or both parents of the patient were sampled as well. The collection of subjects and all experiments were performed under the approval of the committee of human research of the University of California at San Francisco. Institutional review board approval at all study sites and informed consent from all study participants were obtained. All affected family members were examined or had their medical records reviewed by one of the authors (S.L.H.), according to criteria described elsewhere (Goodkin et al. 1991). Only individuals with definite MS were considered as affected in the analyses.
The 398 families included a total of 633 patients with MS. For 614 of them (from 379 families), additional clinical variables were available for analysis of disease progression. A detailed definition of clinical features for these patients has been reported elsewhere (Barcellos et al. 2001). In brief, age at onset was defined as the first episode of neurological dysfunction suggestive of demyelinating disease. This information was obtained via patient recall and verified through review of medical records. Disability was assessed at entry by use of the Expanded Disability Status Scale (EDSS) (Kurtzke 1983) and was recorded in five categories (<3, 3 to <6, 6, 6.5, and ⩾7). EDSS scores were used in conjunction with disease duration to define mild (ability to walk normally or only mild gait disability) and severe (need for bilateral assistance to walk or wheelchair dependency) forms of MS (Barcellos et al. 2001). Finally, the number of years from disease onset until a patient reached an EDSS score ⩾7 (wheelchair dependency) was recorded whenever possible. The 614 patients included in the analysis of disease progression consisted of 379 probands, 178 siblings, 6 parents, 10 offspring, 18 avuncular relatives, 16 cousins, and 7 more distant relatives of the probands.
DNA Analysis and Genotyping
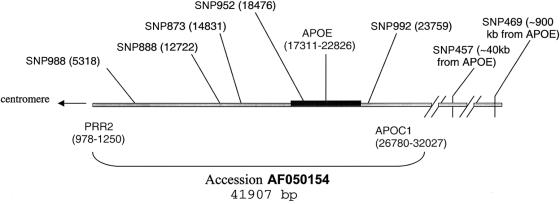
Genomic DNA was extracted from whole blood after isolation of blood lymphocytes according to established protocols (Vance 1998). APOE and HLA genotyping were performed as described elsewhere (Saunders et al. 1993; Barcellos et al. 2001). The generation and mapping of the SNPs used in the present study have been reported elsewhere (Lai et al. 1998; Martin et al. 2000a). SNP genotyping was performed by an oligonucleotide ligation assay (Eggerding et al. 1995), as described elsewhere (Martin et al. 2001b). A map of the SNPs, most of which are located in GenBank cosmid F19374 (accession number AF050154), is shown in figure 1. Allele designations and frequencies of all SNPs are given, for reference, in table 1. Note that allele frequencies were estimated from genotyped founders (individuals at the top of the pedigree and unrelated spouses) with unknown or unaffected phenotypes but were not relevant to the association analyses described below.
Figure 1.
Physical map of SNPs on cosmid F19374 containing APOE. snp888 and snp988 are within the TOM40 gene (LocusLink 10452). snp952 is in the 5′ UTR of APOE. A functional relationship with any particular APOE allele or other effect on APOE expression is not known.
Table 1.
Marker Characterization for Panel of SNPs Genotyped in 398 Families of Patients with MS
| SNP | Distancefrom APOE (kb) | Change | Minor Allelea | Frequencyb |
| 469 | 900 | A→C | A (1) | .13 |
| 457 | 40 | G→T | T (2) | .44 |
| 992 | 1.0 | T→C | T (1) | .38 |
| APOE | 0 | … | … | .12c |
| 952 | 5′ UTR | C→G | G (2) | .37 |
| 873 | 2.5 | G→C | C (2) | .41 |
| 888 | 4.6 | T→C | T (1) | .46 |
| 988 | 12.0 | A→G | G (2) | .20 |
Numeric labels (in parentheses) match those used in the text.
Estimated from genotyped founders with unknown or unaffected MS phenotype (individuals at the top of the pedigree and unrelated spouses, n=602–656 chromosomes).
Frequency for APOE ε4.
In accordance with quality-control (QC) procedures described elsewhere for fine-mapping (Rimmler et al. 1998), samples for six individuals were duplicated for each 96-well plate, and samples from the Centre d'Etude du Polymorphisme Humain were also included on all plates, with the laboratory technicians blinded to their location. Genotypes for the QC samples within and across plates were compared, to detect potential loading and reading errors and thus to minimize the actual genotype error rate. After the genotypes had passed the QC checks, they were uploaded into the PEDIGENE database and merged into the Lapis management system for creating analysis-input files (Haynes et al. 1995).
Statistical Analysis
Family genotypes were examined for Mendelian inconsistencies, using PedCheck (O'Connell and Weeks 1998). Samples yielding inconsistent results were sent back to the laboratory technician, without family-identifying information, for rereading. Given the small distances between markers (see fig. 1), families with obligate recombinants, as determined by haplotype analysis with SIMWALK2 (Sobel and Lange 1996), were reread and, if necessary, regenotyped.
Tests for Hardy-Weinberg equilibrium (HWE) at each marker and for LD between pairs of markers were conducted separately in a sample of unrelated affected individuals (one from each family) and a sample of unrelated unaffected individuals (one from each family, if available). Although individuals are independent within samples, there is clearly dependence between the two samples; however, we were interested only in estimating the extent of HWE and LD between markers, without formally testing whether it is different in affected versus unaffected individuals. An exact test of pairwise LD implemented in the Genetic Data Analysis program (Zaykin et al. 1995) was used.
Family-Based Association Analysis
To test the markers for linkage and association with MS, the data were analyzed with the Pedigree Disequilibrium Test (PDT) (Martin et al. 2000b, 2001) and with the likelihood-ratio test implemented in TRANSMIT (Clayton 1999). PDT can analyze only one marker locus at a time, whereas TRANSMIT is able to consider the transmission of multiple marker haplotypes, even in the presence of phase uncertainty and missing parental genotypes. For this haplotype analysis, TRANSMIT assumes absence of recombination between markers; thus, this analysis was applied only to the interval between snp992 and snp988, where markers were spaced 1–7 kb apart and where no recombinations were expected to occur in our sample, given the observed number of meioses (∼2,080). Genotypes for families that did show evidence of recombinations in this interval were regenerated, and the discrepancy could be resolved for all families included in this analysis.
TRANSMIT two- and three-locus analyses were performed, using a sliding-window approach, to examine haplotypes for each successive set of adjacent SNPs. Four- and five-locus analyses were performed only for selected haplotypes, in which two- and three-locus analyses indicated a possible association with disease susceptibility. A formal correction for multiple testing was considered too conservative, especially because of the high level of pairwise LD between the SNPs.
Analysis of APOE and Disease Progression
Analyses that examined the relationship of APOE-4 and APOE-2 carrier status and clinical variables were also conducted. Methods that take into account the correlation of multiple affected members from the same families were used throughout. Specifically, a logistic-regression model with variance estimation based on generalized estimating equations (PROC GENMOD, SAS version 8.0; SAS Institute) was used to model a binary variable that quantifies disease severity (defined in the “Results” section) as a function of several covariates of interest. To model disease severity as an ordinal response variable, a proportional-odds model (Agresti 1990) was employed. Here, the association was measured by an odds ratio for being in a disease severity category ⩽j (j=1 for mild, j=2 for moderate, j=3 for severe MS, see the “Results” section) rather than >j for exposed versus unexposed patients, under the assumption that this odds ratio is independent of the particular categories used. Familial correlations in the multiplex data were accounted for by use of generalized estimating equations, using a publicly available SAS macro (Williamson et al. 1999). Finally, we analyzed a Cox-regression model based on latency to EDSS score ⩾7 as a censored response variable. This was similar to the analysis by Chapman et al. (2001), except that their study defined the event as EDSS ⩾4 and ⩾6, respectively. Familial correlations were accounted for by using the robust variance estimate of the Cox-regression parameters (Lin and Wei 1989), as implemented in the STATA package (version 7.0; StataCorp 2001).
Results
No evidence of deviations from HWE for any of the eight markers was found in the samples of affected (n=398) or unaffected (n=214) individuals (P⩾.09). In both affected and unaffected subjects, there was strong LD (P<.0001) between each pair of the snp992, APOE, snp952, snp873, snp888, and snp988 markers.
Family-Based Association Analysis
The results of the PDT and TRANSMIT single-locus analysis, as well as TRANSMIT multilocus analyses, are shown in table 2. The single-locus analyses did not show any evidence of association, but significant results were observed for several multilocus haplotypes within this region. Both the global and haplotype-specific multilocus results indicated that the 21112 haplotype of markers APOE, snp952, snp873, snp888, and snp988 was associated with increased MS susceptibility (P=.02 for five-marker haplotype, and P=.005 for four-marker haplotype without snp988; both are uncorrected P values from the TRANSMIT global test). The first “2” allele in this haplotype is the APOE-2 polymorphism. The other (lower-order) significant multilocus tests shown in table 2 result from analyzing subsets of these five alleles and illustrate that the haplotype effect is seen consistently, regardless of the number of loci jointly analyzed. When families were stratified by HLA-DR2 status (216 families with presence and 149 families with absence of DR2 in all affected individuals), similar single-locus and multilocus haplotype associations were observed in both subgroups (data not shown).
Table 2.
Probabilities Based on PDT and TRANSMIT (TRM) Analysis of 398 Families of Patients with MS[Note]
|
Global Test |
Haplotype-Specific Test |
Transmissions to Affected Offspring |
|||||||
| Marker(s) | df | PDTa | TRMb | OvertransmittedHaplotype | df | TRMb | Total No.c | Observed(%) | Expected(%) |
| snp469 | 1 | .51 | .22 | ||||||
| snp457 | 1 | .86 | .80 | ||||||
| snp992 | 1 | .38 | .71 | ||||||
| APOE | 2 | .99 | .70 | ||||||
| snp952 | 1 | .59 | .72 | ||||||
| snp873 | 1 | .41 | .07 | ||||||
| snp888 | 1 | .53 | 1.0 | ||||||
| snp988 | 1 | .32 | .33 | ||||||
| 873-888 | 3 | … | .006 | 11 | 1 | .06 | 1,192 | 6.6 | 5.8 |
| 952-873-888 | 4 | … | .01 | 111 | 1 | .03 | 1,192 | 6.3 | 5.4 |
| 873-888-988 | 4 | … | .02 | 112 | 1 | .02 | 1,192 | 6.0 | 5.1 |
| APOE-952-873-888 | 5 | … | .005 | 2111d | 1 | .02 | 1,230 | 4.5 | 3.8 |
| 952-873-888-988 | 6 | … | .01 | 1112 | 1 | .02 | 1,192 | 6.1 | 5.1 |
| APOE-952-873-888-988 | 6 | … | .02 | 21112d | 1 | .01 | 1,202 | 4.5 | 3.6 |
Note.— Only significant P values (uncorrected for multiple comparisons) are shown for multilocus analyses, for which a sliding-window approach was used (see “Subjects and Methods” section).
P value (uncorrected) for sum version of PDT (Martin et al. 2001a); P values for the average version of PDT were similar (data not shown). Sample comprised 228–284 discordant sib pairs and 371–447 parent-offspring triads.
P value (uncorrected) for TRANSMIT. Alleles/haplotypes with <3% frequency were combined to ensure that variance of observed minus expected transmission counts was ⩾5. The robust variance estimator was used throughout. P values were essentially identical when extended multiplex pedigrees were analyzed and when only a single nuclear family from such pedigrees was included. Sample comprised 551–615 affected offspring with scorable transmissions.
Number of transmissions is twice the number of affected offspring.
The first “2” allele is the APOE-2 allele.
Analysis of APOE and Disease Progression
A summary of clinical variables among the patients with MS is shown in table 3. To analyze the relationship of APOE-4 and APOE-2 carrier status and clinical phenotypes, while adjusting for other relevant variables, a logistic-regression model was used, with age at onset, sex, DR2 status, and APOE-2/APOE-4 carrier status as covariates. Patients were classified as having “severe” or “mild” MS, with “severe” being defined as reaching EDSS >6 (combined categories 6.5 [need for bilateral assistance to walk] and 7 [wheelchair dependency]) ⩽10 years after disease onset (23 patients) and “mild” being defined as EDSS <3 that continued >10 years after disease onset (55 patients). The severe MS group included 8 patients with relapsing-remitting MS, 9 with secondary progressive, 3 with primary progressive, and 3 with progressive-relapsing MS; the mild MS group included 53 patients with relapsing-remitting disease and 2 with secondary progressive MS. A highly significant association between age at onset and disease severity was observed for both severe and mild MS patient groups, as reported elsewhere (Barcellos et al. 2001). The average age at onset was 36.8 years (severe), compared with 29.9 years (nonsevere, P=.0003), and 26.5 years (mild), compared with 30.5 years (nonmild, P=.0002).
Table 3.
Summary of Clinical/Demographic Characteristics of 614 Patients with MS from 379 Families
| Clinical/DemographicInformationa | Mean No. of Years (Range) ± SD |
| Age at onset | 30.1 (6–63) ± 8.8 |
| Disease duration | 15.1 (0–59) ± 10.3 |
| Years to EDSS ⩾7b | 15.8 (1–42) ± 8.3 |
| No. (%) of Patients |
|
| Female | 457 (74.4) |
| EDSS: | |
| <3 | 140 (22.8) |
| 3–<6 | 209 (34.0) |
| 6 | 96 (15.6) |
| 6.5 | 61 (9.9) |
| ⩾7 | 108 (17.6) |
| Disease course: | |
| Mild | 55 (9.0) |
| Nonmild | 559 (91.0) |
| Severe | 23 (3.8) |
| Nonsevere | 591 (96.2) |
| Disease type: | |
| Relapsing-remitting | 379 (61.7) |
| Primary progressive | 30 (4.9) |
| Secondary progressive | 182 (29.6) |
| Progressive-relapsing | 19 (3.1) |
| HLA-DR2: | |
| Positive | 390 (63.5) |
| Negative | 224 (36.5) |
Disease types and courses are defined in the text.
A total of 91 patients reached EDSS ⩾7 and had information for the corresponding disease duration.
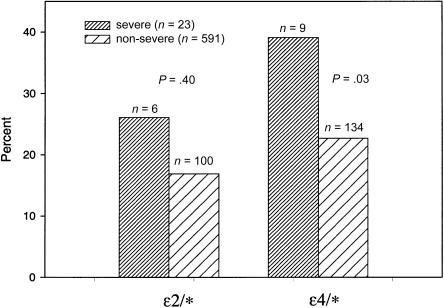
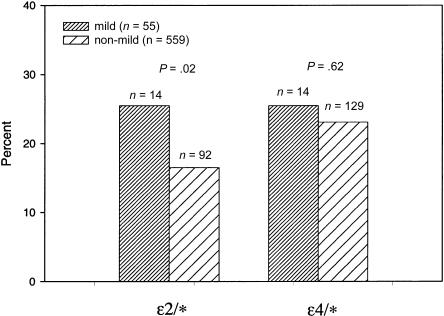
The proportion of APOE-4 carriers was significantly higher in the severe disease group (n=9, 39.1%) than the nonsevere group (n=134, 22.7%; odds ratio (OR) for severe MS = 2.67, 95% confidence interval (CI) 1.12–6.36, P=.03; fig. 2). On the other hand, the proportion of APOE-2 carriers was significantly higher in the mild disease group (n=14, 25.5%) than in the nonmild group (n=92, 16.5%; OR for mild MS = 2.10, 95% CI 1.11–3.95, P=.02; fig. 3). For these analyses, APOE-2 and APOE-4 carriers were compared with APOE-3/3 individuals. When nonsevere and nonmild patient groups were restricted to patients with disease duration ⩾10 years (n=374 with nonsevere MS; n=319 with nonmild MS), very similar results were observed (P=.03 for association of APOE-4 and severe disease; P=.01 for association of APOE-2 and mild disease). Exclusion of all primary-progressive patients from this analysis did not alter our results (data not shown).
Figure 2.
Comparison of APOE genotypes for severe and nonsevere MS. P values from PROC GENMOD, using logistic regression with correction for familial correlations and adjustment for age at onset, sex, and DR2 status. Odds ratio of severe MS for APOE-4 carriers = 2.67, 95% CI 1.12–6.36, P=.03; reference group APOE-3/3 genotype.
Figure 3.
Comparison of APOE genotypes for mild and nonmild MS. P values from PROC GENMOD, using logistic regression with correction for familial correlations and adjustment for age at onset, sex, and DR2 status. Odds ratio of mild MS for APOE-2 carriers = 2.10, 95% CI 1.11–3.95, P=.02; reference group APOE-3/3 genotype.
As an extension of the logistic-regression model, a proportional-odds model (see “Subjects and Methods” section) with the same covariates was also applied. Here, three ordered categories of disease severity were used as the response variable: patients with mild, moderate, or severe MS; “mild” and “severe” were defined as above, and all other patients were considered to have “moderate” disease. Results were similar to those from the logistic-regression model but did not reach statistical significance: APOE-2 carriers were more likely to have mild disease (OR 1.54, 95% CI 0.77–3.08, P=.22), and APOE-4 carriers were less likely to have mild disease (OR 0.79, 95% CI 0.42–1.49, P=.47), when compared with APOE-3/3 individuals.
Survival Analysis
Latency (in years) to EDSS ⩾7 was analyzed as a censored response variable, using the Cox-regression model (n=597 patients). The same covariates were included, and disease duration was used for patients who had not reached EDSS ⩾7. No significant effect of APOE genotypes on rate of progression to disability was detected (hazard ratio for APOE-4 carriers = 0.84, 95% CI 0.51–1.39, P=.50; for APOE-2 carriers = 0.84, 95% CI 0.43–1.64, P=.60). The mean latency to EDSS ⩾7 for APOE-4 carriers was 16.3 years, compared with 15.7 years for noncarriers. Because of the limited number of individuals with EDSS ⩾7, this patient group included only 13 and 18 carriers of the APOE-2 and APOE-4 alleles, respectively.
Discussion
We have used a well-characterized panel of informative SNPs to analyze the APOE region on chromosome 19q13 for an association with MS. Family-based association analysis identified a disease-associated SNP haplotype, suggesting that an allele in LD with this haplotype may confer an increased risk of MS. This result confirms and extends our previous reports of linkage and association of the 19q13 region with MS (Pericak-Vance et al. 2001). From our previous analysis, the effect of the 19q13 locus is likely to be small, with an estimated locus-specific λs=1.5 that accounts for 4%–6% of the overall genetic component in MS (Pericak-Vance et al. 2001). We did not find evidence of an interaction between the established DR2 effect and the 19q13 loci examined in this study. Our results also support the previously suggested hypothesis that APOE may be involved in modulation of clinical expression. Specifically, we observed significant associations between APOE-4 and a more severe form of MS and between APOE-2 and mild disease. In a survival analysis similar to that employed by a previous study (Chapman et al. 2001), we failed to find evidence of faster progression to very severe disease (EDSS ⩾7) in APOE-4 carriers, possibly because of a lack of statistical power.
We examined whether the significant haplotype effect was more likely a result of the presence of a non-APOE gene that influences MS susceptibility or whether it could also be explained by the putative APOE association with disease severity. Using TRANSMIT output, we analyzed which families contributed positively and negatively, respectively, to the score for excesstransmission of the 21112 haplotype (APOE-snp952-snp873-snp888-snp988). If the first group of families predominantly included patients with mild MS and the second those with severe MS, this would suggest that the haplotype effect could be the result of an association of the APOE-2 allele with mild MS. However, of the 30 affected offspring whose transmitted haplotype contributed positively to the TRANSMIT score, only 6 had mild MS; and, of the 10 affected offspring whose transmitted haplotype had a score contribution of −0.5 or less, none had severe MS. This argues against the hypothesis that the APOE-2 association with mild MS found in our patient population accounts for the haplotype association in the family data set. Thus, there may be a non-APOE MS susceptibility locus on 19q13 that is in LD with an SNP haplotype that includes the APOE-2 allele.
Several studies have reported an association of the APOE-4 allele and more severe disease, as measured by older age at onset or by disease activity/progression index (Dousset et al. 1998; Evangelou et al. 1999; Lucotte et al. 2000; Fazekas et al. 2001) or by faster rate of disease progression (Chapman et al. 2001). Fewer studies have reported APOE effects on MS susceptibility, although one case-control study reported a higher risk of developing MS for APOE-4/4 homozygotes (Høgh et al. 2000). Some biological support for the APOE-4 association with disease progression was suggested by an MRI study of brain lesions among patients with MS, which revealed more-extensive tissue destruction or less-efficient repair in APOE-4 carriers (Fazekas et al. 2000). However, several studies have not detected any APOE-4 association with MS (Ferri et al. 1999; Pirttilä et al. 2000; Weatherby et al. 2000a, 2000b). In addition to the obvious influence of sample size, other factors that may contribute to the lack of consistency across studies include differences between study populations with respect to proportion of mild and severe MS, proportion of disease types (relapsing-remitting vs. primary progressive), or distribution of disease duration. The present study confirmed both the strong association of delayed disease onset and more severe disease, and the higher risk of severe MS for APOE-4 carriers. Chapman et al. (2001) found that the influence of APOE-4 on disease progression is more pronounced after the first 5 years of disease duration. If APOE genotypes indeed influence the extent of neuronal repair, it is plausible that an effect of APOE-4 becomes apparent only in later disease stages when axonal damage is incurred. This might explain why we observed an APOE-4 effect only on the probability of severe disease (defined as progression to EDSS >6 within the first 10 years of disease) but no effect on the probability of mild disease.
Fewer studies have examined the association of the APOE-2 allele and MS phenotypes. One study (Carlin et al. 2000) suggested that remyelination may be impaired in individuals with the APOE-2 allele, since the apoE-2 isoform was shown to have reduced affinity for receptors in glial cells. Another study (Ballerini et al. 2000) reported some evidence of a protective effect of the APOE-2 allele, observing that the time to reach secondary progression for patients whose initial disease type was relapsing-remitting was significantly longer for those with APOE-2/3 genotypes than for those with APOE-3/3 and 3/4 genotypes. Our results, based on a substantially larger patient population than those of most studies published to date, provide additional support for a protective effect of the APOE-2 allele, in terms of disease severity. However, it would be useful to refine the clinical definition of different disease phenotypes beyond a binary classification of mild and severe MS, perhaps on the basis of a combination of neurological examinations with longitudinal follow-up and MRI scans.
Methodologically, survival analysis may be a more sensitive instrument for measuring effects on disease progression, compared with a variable such as progression index/disease activity score (raw EDSS score divided by disease duration). A difficulty with this approach is the determination of which endpoint (EDSS ⩾4, 6, or 7) is clinically most meaningful for assessing a patient’s disease course. A limitation of our study is the fact that the only latency variable ascertained so far is duration to EDSS ⩾7, which is a rather rigorous definition of severe disease. Such a stringent definition makes it more difficult to obtain a large enough sample of patients who experience the event of interest, thus reducing statistical power to detect small-to-moderate effects of genotypes on disease progression.
In summary, our results provide further evidence of a role of APOE in MS progression. It is important to remember that the statistical associations we found cannot establish whether APOE itself is the etiologically relevant polymorphism. As observed elsewhere (Martin et al. 2000a) for associations of SNPs near APOE with risk of late-onset Alzheimer disease, lack of association, particularly when SNPs in noncoding regions are used, may not necessarily rule out a candidate gene. On the other hand, positive association may exist with nonetiologic polymorphisms that are merely in LD with the functional allele. Having said this, our haplotype analysis indicates that APOE itself is unlikely to be the 19q13 susceptibility gene, unless it plays a role in susceptibility for only a subset of clinically distinct patients with MS, a role that has yet to be identified. Despite the positive association of APOE-4 and severe MS, disease progression may actually be influenced by a different allele that is in LD with APOE-4. For example, polymorphisms in the APOE promoter region may play a role in determining disease severity. A previous study indicated that the AA homozygous state of the –491 A/T polymorphism in the APOE regulatory region may be associated with cognitive impairment in patients with MS (Oliveri et al. 1999). For Alzheimer disease, several promoter polymorphisms were shown to regulate transcriptional activity of the APOE gene and to correlate with disease risk independently of the APOE-2 and APOE-4 effects (Artiga et al. 1998; Lambert et al. 1998). These regulatory region variants, as well as other candidate genes in 19q13, will be the subject of future MS studies. Since the potential APOE effect on MS progression is likely to be moderate, it will be crucial to determine the precise clinical phenotype that is most strongly influenced by this polymorphism. Therefore, further studies that include detailed clinical measurements of an even larger number of patients and their families than those provided by this data set are needed to dissect the role of the 19q13 region in MS.
Acknowledgments
We thank the patients with MS and their families for making this study possible. This research was funded by National Multiple Sclerosis Society (NMSS) grants RG2901 (to J.R.O.) and RG2542 (to S.L.H.), by National Institutes of Health grants NS26799 (to S.L.H. and J.R.O.) and NS32830 (to J.L.H., MAP-V), and by the Nancy Davis Foundation.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Human Genetics, http://wwwchg.mc.duke.edu/software/pdt.html (for Pedigree Disequilibrium Test computer program)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession number AF050154)
- Lewis Labs, http://lewis.eeb.uconn.edu/lewishome/software.html (for Genetic Data Analysis computer program)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MS [MIM 126200] and APOE [MIM 107741])
- MRC Biostatistics Unit, http://www.mrc-bsu.cam.ac.uk/ (for TRANSMIT computer program)
References
- Agresti A (1990) Categorical data analysis. John Wiley & Sons, New York, p 322 ff [Google Scholar]
- Artiga MJ, Bullido MJ, Frank A, Sastre I, Recuero M, García MA, Lendon CL, Han SW, Morris JC, Vázquez J, Goate A, Valdivieso F (1998) Risk for Alzheimer's disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet 7:1887–1892 [DOI] [PubMed] [Google Scholar]
- Ballerini C, Campani D, Rombola G, Gran B, Macmias B, Pia Amato M, Siracusa G, Bartolozzi L, Sorbi S, Massacesi L (2000) Association of apolipoprotein E polymorphism to clinical heterogeneity of multiple sclerosis. Neurosci Let 296:174–176 [DOI] [PubMed] [Google Scholar]
- Barcellos LF, Oksenberg JR, Green AJ, Bucher P, Rimmler JB, Schmidt S, GarciaME, Lincoln RR, Pericak-Van MA, Haines JL, Hauser SL (2002) Genetic basis for clinical expression in multiple sclerosis. Brain 125:150–158 [DOI] [PubMed] [Google Scholar]
- Barcellos LF, Thomson G, Carrington M, Schafer J, Begovich AB, Lin P, Xu XH, Min BQ, Marti D, Klitz W (1997) Chromosome 19 single-locus and multilocus haplotype associations with multiple sclerosis: evidence of a new susceptibility locus in Caucasian and Chinese patients. JAMA 278:1256–1261 [PubMed] [Google Scholar]
- Carlin C, Murray L, Graham D, Doyle D, Nicoll J (2000) Involvement of apolipoprotein E in multiple sclerosis: absence of remyelination associated with possession of the APOE ε2 allele. J Neuropathol Exp Neurol 59:361–367 [DOI] [PubMed] [Google Scholar]
- Chapman J, Vinokurov S, Achiron A, Karussis DM, Mitosek-Szewczyk K, Birnbaum M, Michaelson DM, Korczyn AD (2001) APOE genotype is a major predictor of long-term progression of disability in MS. Neurology 56:312–316 [DOI] [PubMed] [Google Scholar]
- Chataway J, Feakes R, Coraddu F, Gray J, Deans J, Fraser M, Robertson N, Broadley S, Jones H, Clayton D, Goodfellow P, Sawcer S, Compston A (1998) The genetics of multiple sclerosis: principles, background and updated results of the United Kingdom systematic genome screen. Brain 121:1869–1887 [DOI] [PubMed] [Google Scholar]
- Clayton D (1999) A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet 65:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923 [DOI] [PubMed] [Google Scholar]
- D'Alfonso S, Nistico L, Bocchio D, Bomprezzi R, Marrosu MG, Murru MR, Lai M, Massacesi L, Ballerini C, Repice A, Salvetti M, Montesperelli C, Ristore G, Trojano M, Liguori M, Gambi D, Quattrone A, Tosi R, Momigliano-Richiardi P (2000) An attempt of identifying MS-associated loci as a follow-up of a genomic linkage study in the Italian population. J Neurovirol Suppl 6:S18–S22 [PubMed] [Google Scholar]
- D'Alfonso S, Nistico L, Zavattari P, Marrosu MG, Murru R, Lai M, Massacesi L, Ballerini C, Gestri D, Salvetti M, Ristori G, Bomprezzi R, Trojano M, Liguori M, Gambi D, Quattrone A, Fruci D, Cucca F, Richiardi PM, Tosi R (1999) Linkage analysis of multiple sclerosis with candidate region markers in Sardinian and Continental Italian families. Eur J Hum Genet 7:377–385 [DOI] [PubMed] [Google Scholar]
- Dousset V, Gayou A, Brochet B, Iron A (1998) ApoE polymorphism in multiple sclerosis. Mult Scler 4:357 [Google Scholar]
- Ebers GC, Kukay K, Bulman DE, Sadovnick AD, Rice G, Anderson C, Armstrong H, Cousin K, Bell RB, Hader W, Paty DW, Hashimoto S, Oger J, Duquette P, Warren S, Gray T, O'Connor P, Nath A, Auty A, Metz L, Francis G, Paulseth JE, Murray TJ, Pryse-Phillips W, Risch N (1996) A full genome search in multiple sclerosis. Nat Genet 13:472–476 [DOI] [PubMed] [Google Scholar]
- Ebers GC, Sadovnick AD, Risch NJ (1995) A genetic basis for familial aggregation in multiple sclerosis. Nature 377:150–151 [DOI] [PubMed] [Google Scholar]
- Eggerding FA, Iovannisci DM, Brinson E, Grossman P, Winn-Deen ES (1995) Fluorescence-based oligonucleotide ligation assay for analysis of cystic fibrosis transmembrane conductance regulator gene mutations. Hum Mutat 5:153–165 [DOI] [PubMed] [Google Scholar]
- Evangelou N, Jackson M, Beeson D, Palace J (1999) Association of the APOE ε4 allele with disease activity in multiple sclerosis. J Neurol Neurosurg Psychiatry 67:203–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas F, Strasser-Fuchs S, Kollegger H, Berger T, Kristoferitsch W, Schmidt H, Enzinger C, Schiefermeier M, Schwarz C, Kornek B, Reindl M, Huber K, Grass R, Wimmer G, Vass K, Pfeiffer KH, Hartung HP, Schmidt R (2001) Apolipoprotein E epsilon4 is associated with rapid progression of multiple sclerosis. Neurology 57:853–857 [DOI] [PubMed] [Google Scholar]
- Fazekas F, Strasser-Fuchs S, Schmidt H, Enzinger C, Ropele S, Lechner A, Flooh E, Schmidt R, Hartung HP (2000) Apolipoprotein E genotype related differences in brain lesions of multiple sclerosis. J Neurol Neurosurg Psychiatry 69:25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri C, Sciacca FL, Veglia F, Martinelli F, Comi G, Canal N, Grimaldi LME (1999) APOE ε2–4 and –491 polymorphisms are not associated with MS. Neurology 53:888–889 [DOI] [PubMed] [Google Scholar]
- Gaillard O, Gervais A, Meillet D, Plassart E, Fontaine B, Lyon-Caen O, Delattre J, Schuller E (1998) Apolipoprotein E and multiple sclerosis: a biochemical and genetic investigation. J Neurol Sci 158:180–186 [DOI] [PubMed] [Google Scholar]
- Goodkin DE, Doolittle TH, Hauser SL, Ransohoff RM, Roses AD, Rudick RA (1991) Diagnostic criteria for multiple sclerosis research involving multiply affected families. Arch Neurol 48:805–807 [DOI] [PubMed] [Google Scholar]
- Haines JL, Ashley-Koch A, Jackson CE, Booze M, Ribble RC, Rimmler JB, Garcia ME, Vance JM, Barcellos L, Lincoln R, Hauser SL, Oksenberg JR, Pericak-Vance MA (2000) A genomic screen for multiple sclerosis loci in a San Marino population supports the presence of a locus on 19q. Am J Hum Genet Suppl 67:A66 [Google Scholar]
- Haines JL, Seboun E, Goodkin DE, Usuku K, Lincoln R, Rimmler J, Gusella JF, Roses AD, Pericak-Vance MA, Hauser SL (1993) Genetic dissection of the multiple sclerosis phenotype. Am J Hum Genet Suppl 53:A266 [Google Scholar]
- Haines JL, Ter-Minassian M, Bazyk A, Gusella JF, Kim DJ, Terwedow H, Pericak-Vance MA, Rimmler JB, Haynes CS, Roses AD, Lee A, Shaner B, Menold M, Seboun E, Fitoussi RP, Gartioux C, Reyes C, Ribierre F, Gyapay G, Weissenbach J, Hauser SL, Goodkin DE, Lincoln R, Usuku K, Oksenberg JR (1996) A complete genomic screen for multiple sclerosis underscores a role for the major histocompatability complex. Nat Genet 13:469–471 [DOI] [PubMed] [Google Scholar]
- Haines JL, Terwedow HA, Burgess K, Pericak-Vance MA, Rimmler JB, Martin ER, Oksenberg JR, Lincoln R, Zhang DY, Banatao DR, Gatto N, Goodkin DE, Hauser SL (1998) Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. The Multiple Sclerosis Genetics Group. Hum Mol Genet 7:1229–1234 [DOI] [PubMed] [Google Scholar]
- Hauser SL, Goodkin SL (2001) Multiple Sclerosis and other demyelinating diseases. In: Braunwald E, Fauci AD, Kasper DL, Hauser SL, Longo DL, Jameson JL (eds) Harrison's principles of internal medicine. McGraw-Hill, New York, pp 2452–2461 [Google Scholar]
- Haynes C, Speer MC, Peedin M, Roses AD, Haines JL, Vance JM, Pericak-Vance MA (1995) PEDIGENE: a comprehensive data management system to facilitate efficient and rapid disease gene mapping. Am J Hum Genet Suppl 57:A193 [Google Scholar]
- Høgh P, Oturai A, Schreiber K, Blinkenberg M, Jorgensen OS, Ryder L, Paulson OB, Sorensen PS, Knudsen GM (2000) Apolipoprotein E and multiple sclerosis: impact of the ε-4 allele on susceptibility, clinical type and progression rate. Mult Scler 6:226–230 [DOI] [PubMed] [Google Scholar]
- Ignatius MJ, Gebicke-Harter PJ, Skene JH, Schilling JW, Weisgraber KH, Mahley RW, Shooter EM (1986) Expression of apolipoprotein E during nerve degeneration and regeneration. Proc Natl Acad Sci USA 83:1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S, Gschwend M, Rioux JD, Daly MJ, Terwilliger JD, Tienari PJ, Wikstrom J, Palo J, Stein LD, Hudson TJ, Lander ES, Peltonen L (1997) Genomewide scan of multiple sclerosis in Finnish multiplex families. Am J Hum Genet 61:1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzke JF (1983) Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurol 33:1444–1452 [DOI] [PubMed] [Google Scholar]
- Lai E, Riley J, Purvis I, Roses A (1998) A 4-Mb high-density single nucleotide polymorphism-based map around human APOE. Genomics 54:31–38 [DOI] [PubMed] [Google Scholar]
- Lambert JC, Pasquier F, Cottel D, Frigard B, Amouyel P, Chartier-Harlin MC (1998) A new polymorphism in the APOE promoter associated with risk of developing Alzheimer's disease. Hum Mol Genet 7:533–540 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Horsburgh K, Roses AD (1998) Apolipoprotein E and the CNS response to injury. J Cereb Blood Flow Metab 18:465–471 [DOI] [PubMed] [Google Scholar]
- Lin DY, Wei LJ (1989) The robust inference for the Cox proportional hazards model. J Am Statist Assoc 84:1074–1078 [Google Scholar]
- Lucotte G, Mercier G, Clavel C (2000) Apolipoprotein E-e4 allele, the major risk factor for Alzheimer's disease, is also involved in the severity of multiple sclerosis. Alzheimer's Rep 3:151–154 [Google Scholar]
- Martin ER, Bass MP, Kaplan NL (2001a) Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet 68:1065–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari AJ, Riley J, Finch KL, Stevens JF, Livak KJ, Slotterbeck BD, Slifer SH, Warren LL, Conneally PM, Schmechel DE, Purvis I, Pericak-Vance MA, Roses AD, Vance JM (2000a) SNPing away at complex disease: analysis of SNPs around APOE in Alzheimer disease. Am J Hum Genet 67:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000b) The pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH Jr, Goetz CG, Small GW, Masterman D, Mastaglia F, Laing NG, Stajich JM, Zhang F, Gibson RA, Middleton LT, Roses AD, Haines JL, Scott BL, Pericak-Vance MA, Vance JM (2001) Association of single-nucleotide polymorphisms of the τ gene with late-onset Parkinson disease. JAMA 286:2245–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE (1994) Differential effects of apolipoprotein E3 and E4 on neural growth in vitro. Science 264:850–852 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri RL, Cittadella R, Sibilia G, Manna I, Valentino P, Gambardella A, Aguglia U, Zappia M, Romeo N, Andreoli V, Bono F, Caracciolo M, Quattrone A (1999) APOE and risk of cognitive impairment in multiple sclerosis. Acta Neurol Scand 100:290–295 [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Rimmler JB, Martin ER, Haines JL, Garcia ME, Oksenberg JR, Barcellos LF, Lincoln R, Goodkin DE, Hauser SL (2001) Linkage and association analysis of chromosome 19q13 in multiple sclerosis. Neurogenetics 3:195–201 [DOI] [PubMed] [Google Scholar]
- Pirttilä T, Haanpää M, Mehta PD, Lehtimäki T (2000) Apolipoprotein E (APOE) phenotype and APOE concentrations in multiple sclerosis and acute herpes zoster. Acta Neurol Scand 102:94–98 [DOI] [PubMed] [Google Scholar]
- Rifai N, Christenson RH, Gelman BB, Silverman LM (1987) Changes in cerebrospinal fluid IgG and apolipoprotein E indices in patients with multiple sclerosis during demyelination and remyelination. Clin Chem 33:1155–1157 [PubMed] [Google Scholar]
- Rimmler J, McDowell JG, Slotterback BD, Haynes CS, Menold MM, Rogala A, Speer MC, Gilbert JR, Hauser ER, Vance JM, Pericak-Vance MA (1998) Development of a data coordinating center (DCC): data quality control for complex disease studies. Am J Hum Genet Suppl 63:A240 [Google Scholar]
- Rosati G (1994) Descriptive epidemiology of multiple sclerosis in Europe in the 1980s: a critical overview. Ann Neurol Suppl 36:S164–S174 [DOI] [PubMed] [Google Scholar]
- Sadovnick AD, Armstrong H, Rice GP (1993) A population-based study of multiple sclerosis in twins: update. Ann Neurol 33:281–285 [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, Hulette C, Crain B, Goldgaber D, Roses AD (1993) Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology 43:1467–1472 [DOI] [PubMed] [Google Scholar]
- Sawcer S, Jones HB, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow PN, Compston A (1996) A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet 13:464–476 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Vance JM (1998) The collection of biological samples for DNA analysis. In: Haines JL, Pericak-Vance MA (eds) Approaches to gene mapping in complex human diseases. Wiley-Liss, New York, pp 201–211 [Google Scholar]
- Weatherby SJ, Mann CL, Davies MB, Carthy D, Fryer AA, Boggild MD, Young C, Strange RC, Ollier W, Hawkins CP (2000a) Polymorphisms of apolipoprotein E: outcome and susceptibility in multiple sclerosis. Mult Scler 6:32–36 [DOI] [PubMed] [Google Scholar]
- Weatherby SJ, Mann CL, Fryer AA, Strange RC, Hawkins CP, Stevenson VL, Leary SM, Thompson AJ (2000b) No association between the APOE ε4 allele and outcome and susceptibility in primary progressive multiple sclerosis. J Neurol Neurosurg Psychiatry 68:532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshenker BG, Wingerchuk DM, Liu Q, Bissonet AS, Schaid DJ, Sommer SS (1997) Genetic variation in the tumor necrosis factor alpha gene and the outcome of multiple sclerosis. Neurology 49:378–385 [DOI] [PubMed] [Google Scholar]
- Williamson JM, Lipsitz SR, Kim KM (1999) GEECAT and GEEGOR: computer programs for the analysis of correlated categorical response data. Comput Methods Programs Biomed 58:25–34 [DOI] [PubMed] [Google Scholar]
- Wise LH, Lanchbury JS, Lewis CM (1999) Meta-analysis of genome searches. Ann Hum Genet 63:263–272 [DOI] [PubMed] [Google Scholar]
- Zaykin D, Zhivotovsky L, Weir BS (1995) Exact tests for association between alleles at arbitrary numbers of loci. Genetica 96:169–178 [DOI] [PubMed] [Google Scholar]
- Zouali H, Faure-Delanef L, Lucotte G (1999) Chromosome 19 locus apolipoprotein C-II association with multiple sclerosis. Mult Scler 5:134–136 [DOI] [PubMed] [Google Scholar]