Abstract
Several traits associated with asthma phenotypes, such as high total serum immunoglobulin E and bronchial hyperresponsiveness, have been linked by numerous genome-screen studies and linkage analyses to markers on human chromosome 5q31-q34. In the present article, we describe UGRP1 (encoding uteroglobin-related protein 1) as one of asthma-susceptibility genes that is located on chromosome 5q31-q32. UGRP1 is a homodimeric secretory protein of 17 kDa and is expressed only in lung and trachea. The G→A polymorphism was identified at −112 bp in the human UGRP1 gene promoter. The −112A allele is responsible for a 24% reduction in the promoter activity in relation to the −112G allele, as examined by transfection analysis. Electrophoretic mobility-shift analysis revealed that an unknown nuclear factor binds to the region around −112 bp. The binding affinity with the −112A oligonucleotide was reduced by approximately one half, as compared with the −112G oligonucleotide. In a case-control study using 169 Japanese individuals (84 patients with asthma and 85 healthy control individuals), those with a −112A allele (G/A or A/A) were 4.1 times more likely to have asthma than were those with the wild-type allele (G/G).
Introduction
Bronchial asthma is a complex, inheritable inflammatory disorder of the airways, characterized by reversible airflow obstruction, airway inflammation, and bronchial hyperresponsiveness (BHR) (McFadden and Gilbert 1992). A number of genetic studies have identified several chromosomal regions that contribute to the development of asthma and asthma-associated phenotypes such as BHR and atopy (Postma et al. 1995; The Collaborative Study on the Genetics of Asthma 1997; Ruffilli and Bonini 1997; Bleecker 1998; Cookson and Moffatt 2000; Ober and Moffatt 2000). Among these regions are human chromosomes 5q31-q34, 6p21-p23, 11q13, 12q15-q24, 14q12, and 16p12. Chromosome 5q31-q34 contains numerous gene candidates that potentially may play a role in the airway inflammation associated with atopic asthma, including a number of proinflammatory cytokines such as interleukin (IL)-3, -4, -5, -9, and -13, granulocyte macrophage colony-stimulating factor (CSF), and the β2-adrenergic receptor (ADRB2) (Postma et al. 1995; Ruffilli and Bonini 1997; Bleecker 1998; Cookson and Moffatt 2000; Ober and Moffatt 2000).
Our previous study on the mouse uteroglobin-related protein 1 (Ugrp 1) gene and its encoded protein (Niimi et al. 2001) suggested that the human UGRP1 gene (MIM 606531) is a plausible asthma candidate gene, for the following reasons: First, the mouse Ugrp1 gene is located on chromosome 18C-D, a region that is syntenic with human chromosome 5q31-q34, which has been assigned as one of the asthma-susceptibility gene loci (Niimi et al. 2001). Second, UGRP1 exhibits an amino acid sequence similar to that of Clara cell secretory protein (CCSP/CC16) (Niimi et al. 2001), which is thought to function as an anti-inflammatory agent by inhibiting chemotaxis and phagocytosis of monocytes and neutrophils and which modulates production and/or activity of various mediators of the inflammatory response, including phospholipase A2 (PLA2), interferon-γ, and tumor-necrosis factor α (Broeckaert and Bernard 2000; Mukherjee and Chilton 2000). Third, UGRP1 is mainly expressed in pulmonary airways, and a high level of expression was found in the epithelial cells of the trachea, bronchus, and bronchioles (Niimi et al. 2001). Lastly, the level of Ugrp1 mRNA decreased in the lungs of the antigen-induced inflammation model mouse (Niimi et al. 2001), in a way similar to that observed for CCSP/CC16; CCSP/CC16 protein expression was decreased in humans with asthma (Shijubo et al. 1999) and after induction of airway inflammation in humans (Lensmar et al. 2000) and animals (Arsalane et al. 2000; Hayashida et al. 2000).
UGRP1 is a homodimeric secretory protein of unknown function (Niimi et al. 2001). The mouse and human UGRP1 cDNAs encode a protein of 91 and 93 amino acids, respectively, of which the N-terminal 21 amino acid residues exhibit a characteristic signal sequence that may function to target the protein to a secretory pathway (Niimi et al. 2001). The mouse Ugrp1 gene is ∼2.9 kb in length and is composed of three exons (Niimi et al. 2001). Homologous UGRP2 genes have been identified in both mouse and human and, together with the mouse and human UGRP1 genes, constitute a new gene family (Niimi et al. 2001). The human UGRP1, human UGRP2, and mouse UGRP2 share 81%, 41%, and 33% amino acid sequence identity, respectively, with mouse UGRP1 (Niimi et al. 2001).
Mutation screening of asthma candidate genes aims to detect sequence variations that alter the concentration or activity of the protein that is coded by that gene and that ultimately contributes to the development of clinical asthma. To this end, the human UGRP1 gene was isolated and characterized. Screening for sequence variations in the exons and the 5′ promoter region of the gene among patients with asthma and healthy control individuals revealed the presence of a point mutation, in the promoter region, that appears to be responsible for reduced transcriptional activity of the gene and is associated with an increased risk of asthma.
Material and Methods
Cloning and DNA Sequencing
A human UGRP1 genomic DNA was isolated from a human BAC DNA library (Incyte Genomics) and Human GenomeWalker kit (Clontech). The genomic DNA fragments were sequenced using an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit and a model 377 DNA sequencer (PE Applied Biosystems). The transcription start site of the human UGRP1 transcript was determined by a SMART RACE cDNA amplification kit (Clontech), using 2 μg of human adult lung total RNA. DNA sequence analyses indicated the presence of multiple transcription start sites. Since the majority of clones (12/16) had exactly the same sequence, we refer to this site as the major transcription start site.
Chromosomal Mapping
A human UGRP1 probe of an entire BAC genomic clone labeled with biotin or digoxigenin was used for FISH of chromosomes derived from methotrexate-synchronized normal peripheral lymphocytes. Conditions of hybridization; detection of hybridization signals; digital-image acquisition, processing, and analysis; and direct fluorescent signal localization on banded chromosomes were performed as described elsewhere (Zimonjic et al. 1995).
Genotyping
Genomic DNA was extracted from peripheral leukocytes isolated from EDTA-anticoagulated blood through use of a commercially available DNA isolation kit (DnaQuick; Dainippon Pharmaceutical). Genotyping for the −112G/A polymorphism was performed using a PCR fragment amplified by the primers 5′-CCTCCAGATTGCTTTCACAACTGGG-3′ and 5′-CAAAGTGTGATGGCTGCTTTTGCAC-3′. PCR was performed, in a 20-μl reaction mixture containing 50 ng of genomic DNA, under the following conditions: denaturation at 94°C for 30 s and annealing and extension at 68°C for 1 min, for 35 cycles. Amplified DNA fragments were purified and sequenced.
Transfection and Reporter-Gene Assays
A 294-bp fragment (from −209 to +85) of the human UGRP1 gene promoter was prepared by PCR, using two forms (−112G/G and −112A/A) of genomic DNA as a template, each of which was separately subcloned into an NheI-XhoI site of the pGL3-Basic luciferase reporter vector (Promega) to generate the pGL3−112G and pGL3−112A plasmids.
Conditions used for the culturing of human lung adenocarcinoma NCI-H441 cells, as well as the method of transfection, are described elsewhere (Niimi et al. 2001). Luciferase activity was normalized to β-galactosidase activity, and the relative luciferase activity of the reporter constructs was expressed in relation to the activity of the pGL3-Basic plasmid, which was assigned a value of 1. Data are the mean values of four independent experiments, plus or minus the SD.
Electrophoretic Mobility–Shift Assays (EMSAs)
Single-stranded oligonucleotides were annealed to produce double-stranded DNA having either G or A at −112 bp (−112G or −112A oligonucleotide). Double-stranded DNA was end-labeled with α-[32P]dCTP and a DNA polymerase Klenow fragment (Life Technologies). Nuclear extracts were prepared from human lung NCI-H441 cells, and EMSA was performed as described elsewhere (Niimi et al. 2001). An oligonucleotide containing a CCAAT/enhancer binding protein (C/EBP) α/β consensus binding sequence (TTGCGCAAT) (Ryden and Beemon 1989) was from the C/EBP Gelshift kit (Geneka Biotechnology).
Subjects for the Case-Control Study
Initial screening for a possible variation of the UGRP1 gene sequence was performed using 51 subjects (26 males and 25 females) from various ethnic backgrounds, with and without asthma/allergy (rhinitis), who were volunteers living in the local Philadelphia area. For a case-control study, a total of 84 Japanese patients (40 males and 44 females) with bronchial asthma were recruited from the pulmonary clinic at the Hokkaido University Hospital, Sapporo, Japan, between 1990 and 1992. Asthma was diagnosed by the following three criteria: (1) presence of at least two symptoms (recurrent cough, wheezing, or dyspnea), (2) presence of reversible airflow limitation (15% variability in forced expiratory volume per second or in peak expiratory flow rate, either spontaneously or with an inhaled short-acting β2-agonist) or airway hyperresponsiveness to methacholine, and (3) absence of other pulmonary diseases, such as chronic obstructive pulmonary disease. Antigen-specific immunoglobulin E (IgE) was examined for 10 common inhaled antigens, including house-dust mite, molds, pollens, and animal dander (cat and dog). Of 84 patients, 57 (67.9%) were positive for at least one of these antigens.
Eighty-five subjects were frequency matched by age and were selected from among healthy Japanese volunteers without history of bronchial asthma or other respiratory symptoms. For both patients with asthma and healthy control individuals, information about lifestyle, such as cigarette smoking, was obtained by standardized interview by a physician. The study was approved by the University Ethics Committee. Informed consent was obtained from all participants, according to the Helsinki Declaration.
Statistical Analyses
Frequencies of the −112A allele and of the G/A or A/A genotype were compared between patients with asthma and healthy control individuals, by the use of χ2 test. The mean age and IgE level were continuous variables and were compared between cases and controls by the nonparametric Kruskal-Wallis test. Smoking status was dichotomized as current smokers versus ex-/nonsmokers. The odds ratio (OR) and 95% confidence interval (CI) for the association of asthma diagnosis with the UGRP1 genotype were estimated by logistic regression (Breslow and Day 1980).
Results
Characterization of the Human UGRP1 Gene
The human UGRP1 gene is ⩾2,900 bp in length and consists of three exons; the structure resembles the orthologous mouse Ugrp1 gene (fig. 1) (Niimi et al. 2001). All the exon-intron boundaries demonstrated a consensus sequence for RNA splicing. Analysis of human UGRP1 mRNA through use of a human multiple tissue expression array (Clontech) revealed that it was only expressed in lung and trachea; fetal lung had the highest expression (fetal trachea was not tested), whereas, in adults, higher expression was found in trachea, as compared with very low expression in lung (fig. 2).
Figure 1.
Human UGRP1 gene structure. The structure of the gene and the sequences at the exon-intron boundaries are shown in uppercase (exon) and lowercase (intron) letters. Splicing donor and acceptor consensus sequences are shown in boldface.
Figure 2.
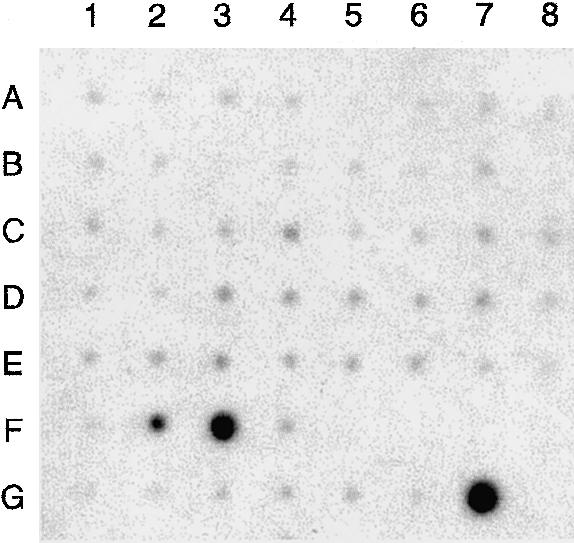
UGRP1 expression. Human multiple tissue expression array was hybridized with 32P-labeled human UGRP1 cDNA probe. RNA sources are as follows: A1 = whole brain; A2 = amygdala; A3 = caudate nucleus; A4 = cerebellum; A5 = cerebral cortex; A6 = frontal lobe; A7 = hippocampus; A8 = medulla oblongata; B1 = occipital lobe; B2 = putamen; B3 = substantia nigra; B4 = temporal lobe; B5 = thalamus; B6 = subthalamic nucleus; B7 = spinal cord; C1 = heart; C2 = aorta; C3 = skeletal muscle; C4 = colon; C5 = bladder; C6 = uterus; C7 = prostate; C8 = stomach; D1 = testis; D2 = ovary; D3 = pancreas; D4 = pituitary gland; D5 = adrenal gland; D6 = thyroid gland; D7 = salivary gland; D8 = mammary gland; E1 = kidney; E2 = liver; E3 = small intestine; E4 = spleen; E5 = thymus; E6 = peripheral leukocyte; E7 = lymph node; E8 = bone marrow; F1 = appendix; F2 = lung; F3 = trachea; F4 = placenta; G1 = fetal brain; G2 = fetal heart; G3 = fetal kidney; G4 = fetal liver; G5 = fetal spleen; G6 = fetal thymus; and G7 = fetal lung.
Chromosomal Localization of the Human UGRP1 Gene
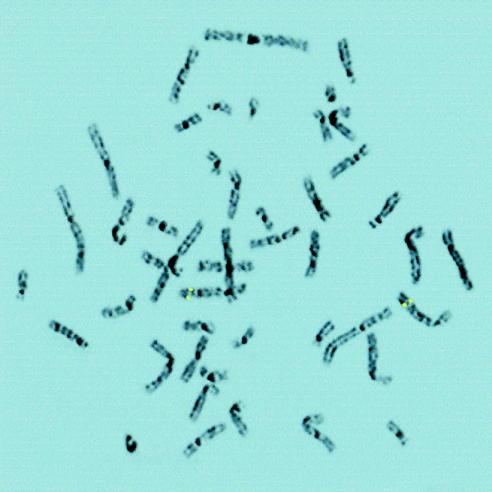
The human UGRP1 gene was mapped by FISH, on chromosomes prepared from normal human peripheral leukocytes (fig. 3). A symmetrical fluorescent signal on sister chromatids was observed only on chromosome 5 at identical sites in 38 of 40 metaphases recorded from two separate experiments. In 20 metaphases analyzed by imaging of 4′,6-diamidino-2-phenylindole (DAPI) G-like banding, the FISH signal was localized on chromosome 5 at region q31-q32, the region syntenic with mouse chromosome 18C-D (Searle et al. 1989; DeBry and Seldin 1996), to which the mouse Ugrp1 gene has been assigned (Niimi et al. 2001). To confirm the identity of chromosomes with specific signals, metaphases were rehybridized with a human chromosome-painting probe for chromosome 5. All metaphases with a specific signal were painted with this probe (data not shown).
Figure 3.
FISH chromosomal localization of the human UGRP1 gene. Chromosome 5 exhibits symmetrical fluorescent signals of the UGRP1 gene at region 5q31-q32, as depicted by enhanced DAPI-induced chromosome banding.
Polymorphism in the Human UGRP1 Gene Promoter
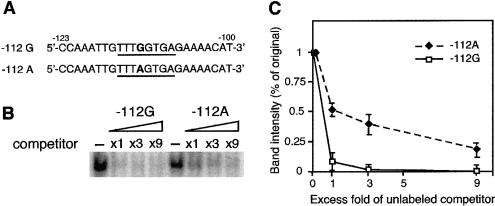
On the basis of our hypothesis that human UGRP1 may assume a role in asthma, an effort was focused on detecting possible sequence variations in the human UGRP1 gene that alter the concentration or activity of the coded protein, ultimately contributing to the development of clinical asthma. Initially, DNAs from 51 randomly selected individuals with or without asthma/allergy (rhinitis) were screened for possible sequence variation(s) in the coding region and in all the exon-intron boundaries. No variations were found. A 585-bp upstream region (GenBank accession number AF411473) of the gene was then screened using the same DNA samples. The G→A substitution was found at the −112-bp position, relative to the transcription start site of the gene (fig. 4A). Of the 51 samples, 2 were homozygous and 9 were heterozygous for the A allele.
Figure 4.
Human UGRP1 gene promoter analysis and the −112G/A polymorphism. A, Sequence of the promoter region used for transfection analysis. Numbers indicate nucleotide positions relative to the major transcription start site, marked by a bent arrow (+1). The nucleotide −112 is a polymorphic site, marked by an asterisk (*); the polymorphic G/A nucleotide is shown in boldface. The TATA box is boxed, and the ATG initiation codon is shown in boldface. B, Reporter-gene assays of human UGRP1 gene promoter constructs. Relative luciferase activities of constructs harboring the human UGRP1 gene sequence from −209 to +85 bp, with either G (−112G) or A (−112A) at −112 bp, were compared in transient transfection studies using NCI-H441 cells. Luciferase activities are shown; values are based on the activity obtained with the pGL3-Basic vector (GL3) as 1. The constructs were tested in duplicate in four independent experiments. Values are mean ± SD.
The −112G→A Polymorphism Modulates the Transcription of the UGRP1 Gene
To examine whether the −112G→A polymorphism influences promoter activity of the UGRP1 gene, the promoter sequence, connected to a luciferase reporter gene, was transfected into human lung adenocarcinoma NCI-H441 cells. The reporter activities were compared between two constructs containing either G or A at −112 bp in the UGRP1 gene promoter region (fig. 4B). Significantly lower luciferase reporter activity was detected for the −112A construct, as compared with the −112G construct (24% decrease; P<.01). These results suggest that the −112A allele may be associated with the decreased transcriptional activity of the UGRP1 gene in lungs.
Computer analysis was performed using TF (transcription factor) Search (Heinemeyer et al. 1998), in an attempt to identify a nuclear protein that binds to the sequence around −112 bp and contributes to the transcriptional activation of the human UGRP1 gene. The C/EBP consensus sequence (T[G/T]TGG[A/T]NA) was identified as a candidate transcription factor. The C/EBPα/β consensus binding sequence (TTGCGCAAT) (Ryden and Beemon 1989), when used as a competitor, however, did not compete with the −112G oligonucleotide for a specific DNA-protein complex formation (data not shown), suggesting that the nuclear protein binding to the sequence around −112 bp may not be a member of the C/EBP family. It appears that an unknown nuclear protein binds to a sequence similar to the C/EBP binding site located around −112 bp in the human UGRP1 gene promoter and activates transcription of the gene.
Allele-Specific Binding of a Nuclear Protein to the −112G→A Polymorphic Site
Electrophoretic mobility shift analysis was then performed, to examine whether the G→A nucleotide substitution at −112 bp affects interaction of a nuclear protein(s) with the sequence around −112 bp, through use of NCI-H441 cell nuclear extracts and 24-bp double-stranded oligonucleotide probes that had a sequence from −123 to −100 bp of the UGRP1 gene promoter, with either G or A at −112 bp (fig. 5A). A single band due to a specific DNA-protein interaction was obtained at the same mobility for both oligonucleotide probes (data not shown). To determine whether this particular nuclear protein binds preferentially to one of the oligonucleotides, a series of competition assays was performed, where a radiolabeled −112G probe competed against an unlabeled −112G or −112A oligonucleotide (fig. 5B and C). The unlabeled −112G oligonucleotide was a more efficient competitor, having an approximately twofold-higher affinity for a specific DNA-protein complex formation, as compared with that of the −112A oligonucleotide. These results indicate that the A residue at −112 bp lowers the binding affinity, albeit not completely, of a particular nuclear protein to the site around −112 bp. Thus, the G→A point mutation at −112 bp in the human UGRP1 gene promoter decreases the affinity of a particular nuclear protein to the binding site around −112 bp, resulting in reduced transcriptional activity and ultimately leading to a lower expression of the UGRP1 protein.
Figure 5.
Electrophoretic mobility shift analysis of the −112G/A polymorphic site. A, Oligonucleotide sequences containing G (−112G) or A (−112A) at −112 bp (boldface), which were used as a probe or a competitor, are shown. The consensus sequence for C/EBP obtained by Transcription Factor Search (Heinemeyer et al. 1998) is underlined. B, A specific DNA-protein complex, formed between a nuclear protein in NCI-H441 cells and a 32P-labeled −112G fragment, was subjected to competition analysis in the presence of increasing concentrations (one to ninefold) of unlabeled −112G or −112A oligonucleotide, as a competitor. A representative result is shown. C, Graphic demonstration of the results obtained in B. -----⧫----- = −112A oligonucleotide; ——□—— = −112G oligonucleotide. Band intensity was quantitated using a PhosphoImager and ImageQuant program (Molecular Dynamics). Each value represents the mean of three independent experiments ± SD.
The −112A Allele Is Associated with an Increased Risk of Asthma
A case-control study was then performed, to examine whether the presence of the −112A allele is associated with an increased risk of asthma. This analysis included a total of 169 Japanese (98 males and 71 females); 84 patients with clinical diagnoses of asthma (case subjects) and 85 without asthma (healthy control subjects). The mean age of the 169 subjects was 45.6 years (range 18–81 years) and was similar between case subjects and healthy control subjects (46.3 vs. 44.8 years; P=.70). The mean IgE level was significantly higher among case subjects than among control subjects (679.4 IU/ml vs. 140.4 IU/ml; P=.0001). The IgE level, however, did not differ by sex (443.4 IU/ml in men and 356.0 IU/ml in women; P=.69). The prevalence of smoking was lower in case subjects as compared with control subjects (21% vs. 43%; P=.04) but was similar between the sexes (32% in men and 31% in women; P=.90). The allele frequency of the −112A variant in the UGRP1 gene among case subjects was 22.0%, compared with 10% among healthy control subjects (P=.003) (table 1). The prevalence of the A variant (G/A and A/A genotype combined) was 40.5% (34 of 84) among case subjects, compared with 17.6% (15 of 85) among control subjects (P=.001). With adjustment for IgE level, smoking status, age, and sex, a person with G/A or A/A genotype was 4.1 times more likely to be asthmatic, compared with those with G/G genotype. In addition, every 100-IU/ml increase in IgE level increased the risk of asthma 1.9-fold, on average (table 2). The correlation between serum IgE level and the clinical expression of allergy and asthma has been established (Burrows et al. 1989; Sears et al. 1991; Postma et al. 1995; Ruffilli and Bonini 1997).
Table 1.
Frequency of −112A Allele in the UGRP1 Gene among 169 Japanese Subjects with and without Asthma
|
No. of Individuals with Genotype |
||||
| Sample | −112G/G | −112G/A | −112A/A | Frequency of the A Allele(%) |
| Patients with asthma (n=84) | 50 | 31 | 3 | 22.0 |
| Healthy control individuals (n=85) | 70 | 13 | 2 | 10.0 |
Table 2.
The Association of Asthma with UGRP1 Variant and IgE Level[Note]
| Variable | OR (95% CI)a |
| G/A or A/A genotype | 4.11 (1.51–11.17) |
| IgE | 1.90 (1.41–2.58) |
Note.— Logistic-regression models include both variables shown and are adjusted for age, sex, and smoking status. Age and IgE level were continuous variables. Smoking status was either current or ex-/nonsmoker. The reference group for the UGRP1 genotype is the wild type (G/G).
The OR for the G/A or A/A genotype indicates that persons who have either of these genotypes are four times more likely to have asthma; the OR for IgE indicates that every increase of 100 IU/ml in IgE level elevates the risk of asthma 1.9-fold.
Discussion
We investigated the hypothesis that human UGRP1 plays a role in asthma, by screening patients with asthma and healthy control subjects for a possible sequence variation(s)—in exons, exon-intron boundaries, and promoter sequences of the UGRP1 gene—that alters the concentration or activity of the coded protein. The G→A substitution was found at position −112 bp relative to the transcription start site of the gene. Transfection and EMSA analyses demonstrated that the G→A point mutation at −112 bp in the human UGRP1 gene promoter decreases affinity of a particular nuclear protein to the binding site around −112 bp, resulting in reduced transcriptional activity and ultimately leading to a lower expression of UGRP1 protein. In a case-control study, subjects with asthma had a significantly higher A allele frequency, compared with healthy control subjects, suggesting the importance of the A allele in development of asthma.
UGRP1 is similar to CCSP/CC16, in its amino acid sequence (25% identity in mouse) and site of tissue-specific expression (Niimi et al. 2001). Both are secretory proteins and exhibit decreased expression in inflamed mouse lungs (Arsalane et al. 2000; Hayashida et al. 2000; Niimi et al. 2001). CCSP/CC16 is believed to function as an anti-inflammatory agent, on the basis of numerous in vivo and in vitro studies (Broeckaert and Bernard 2000; Mukherjee and Chilton 2000). Thus, decreased CCSP/CC16 expression has been found in humans with asthma (Shijubo et al. 1999) and after induction of airway inflammation in those with asthma (Lensmar et al. 2000). The CCSP/CC16 knockout mouse exhibited an increased level of inflammation, inflammatory cytokine production, and neutrophil infiltration in the lungs after infection with a virus or bacteria (Harrod et al. 1998; Hayashida et al. 2000). Although the exact function of UGRP1 is not known, it is possible that it may play a role as an anti-inflammatory agent, like CCSP/CC16, in the modulation of pulmonary inflammation. In this regard, it is interesting to note that the association of the −112A polymorphism with asthma appears to be independent of the increase in IgE level. Furthermore, the UGRP1 gene was mapped in the area where many proinflammatory cytokine genes are located, including IL-3, -4, -5, -9, and -13, as well as genes encoding granulocyte macrophage CSF–2 and the receptor for CSF–1 (Postma et al. 1995; The Collaborative Study on the Genetics of Asthma 1997; Ruffilli and Bonini 1997; Bleecker 1998; Cookson and Moffatt 2000; Ober and Moffatt 2000).
The G→A polymorphism was recently reported at the 38-bp position of the CCSP/CC16 gene, located within the exon 1 noncoding region that exhibits association with an increased risk of asthma (Laing et al. 1998, 2000). The CCSP/CC16 gene is one of the asthma-susceptibility gene candidates located on chromosome 11q13 (Ruffilli and Bonini 1997; Ober and Moffatt 2000). In a case-control study, the frequency of the 38A allele was significantly higher in case subjects (43.3%), compared with nonasthmatic control subjects (26.1%), suggesting an association between this allele and the risk of asthma. In a previous study, subjects with genotype 38A/A and G/A have been reported to have 6.9- and 4.2-fold increases, respectively, in risk of asthma (Laing et al. 1998). In this case, however, no direct relationship was reported between the G/A polymorphism and what it may cause, such as decreased activity of transcription and/or translation. Alternatively, this mutation may be in linkage disequilibrium with a locus that contributes to an inherited predisposition for asthma.
In conclusion, our studies demonstrate that UGRP1 is likely to be a gene candidate in asthma susceptibility and that the −112A polymorphism may cause a predisposition to asthmatic inflammation by reducing the levels of UGRP1 in the airway. However, the possibility that the −112A allele is in linkage disequilibrium with an allele in a neighboring gene that contributes to inflammation and asthma cannot be excluded. It is likely that other genes, in addition to UGRP1, contribute to asthma susceptibility. Furthermore, the possibility cannot be ruled out that the association of the −112A allele with asthma may be ethnicity dependent. Additional experiments will be required to demonstrate that UGRP1 expression is reduced in asthma and to confirm the association of this polymorphism with asthma.
Acknowledgments
We thank Drs. Frank Gonzalez and Robert Biggar for helpful discussions and critical review of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for a 585-bp upstream sequence of the human UGRP1 gene [submitted under accession number AF411473])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for UGRP1 [MIM 606531])
References
- Arsalane K, Broeckaert F, Knoops B, Wiedig M, Toubeau G, Bernard A (2000) Clara cell specific protein (CC16) expression after acute lung inflammation induced by intratracheal lipopolysaccharide administration. Am J Respir Crit Care Med 161:1624–1630 [DOI] [PubMed] [Google Scholar]
- Bleecker ER (1998) Mapping susceptibility genes for asthma and allergy. Clin Exp Allergy 28 Suppl 5:6–12; discussion 26–28 [DOI] [PubMed] [Google Scholar]
- Breslow N, Day N (1980) The analysis of case-control studies. In: International Agency for Research on Cancer (ed) Statistical methods in cancer research, vol 1. International Agency for Research on Cancer, Lyon, pp 192–242 [Google Scholar]
- Broeckaert F, Bernard A (2000) Clara cell secretory protein (CC16): characteristics and perspectives as lung peripheral biomarker. Clin Exp Allergy 30:469–475 [DOI] [PubMed] [Google Scholar]
- Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG (1989) Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med 320:271–277 [DOI] [PubMed] [Google Scholar]
- Cookson WO, Moffatt MF (2000) Genetics of asthma and allergic disease. Hum Mol Genet 9:2359–2364 [DOI] [PubMed] [Google Scholar]
- Collaborative Study on the Genetics of Asthma, The (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15:389–392 [DOI] [PubMed] [Google Scholar]
- DeBry RW, Seldin MF (1996) Human/mouse homology relationships. Genomics 33:337–351 [DOI] [PubMed] [Google Scholar]
- Harrod KS, Mounday AD, Stripp BR, Whitsett JA (1998) Clara cell secretory protein decreases lung inflammation after acute virus infection. Am J Physiol 275:L924–L930 [DOI] [PubMed] [Google Scholar]
- Hayashida S, Harrod KS, Whitsett JA (2000) Regulation and function of CCSP during pulmonary Pseudomonas aeruginosa infection in vivo. Am J Physiol Lung Cell Mol Physiol 279:L452–L459 [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing IA, Goldblatt J, Eber E, Hayden CM, Rye PJ, Gibson NA, Palmer LJ, Burton PR, Le Souef PN (1998) A polymorphism of the CC16 gene is associated with an increased risk of asthma. J Med Genet 35:463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing IA, Hermans C, Bernard A, Burton PR, Goldblatt J, Le Souef PN (2000) Association between plasma CC16 levels, the A38G polymorphism, and asthma. Am J Respir Crit Care Med 161:124–127 [DOI] [PubMed] [Google Scholar]
- Lensmar C, Nord M, Gudmundsson GH, Roquet A, Andersson O, Jornvall H, Eklund A, Grunewald J, Agerberth B (2000) Decreased pulmonary levels of the anti-inflammatory Clara cell 16 kDa protein after induction of airway inflammation in asthmatics. Cell Mol Life Sci 57:976–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden ER Jr, Gilbert IA (1992) Asthma. N Engl J Med 327:1928–1937 [DOI] [PubMed] [Google Scholar]
- Mukherjee AB, Chilton BS (eds) (2000) The uteroglobin/Clara cell protein family. Proceedings of the Uteroglobin/Clara Cell 10kDa Family of Proteins conference, Bethesda, April 14–16. Ann NY Acad Sci 923:1–35611193749 [Google Scholar]
- Niimi T, Keck-Waggoner CL, Popescu NC, Zhou Y, Levitt RC, Kimura S (2001) UGRP1, a uteroglobin/Clara cell secretory protein-related protein, is a novel lung-enriched downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor. Mol Endocrinol 15:2021–2036 [DOI] [PubMed] [Google Scholar]
- Ober C, Moffatt MF (2000) Contributing factors to the pathobiology. The genetics of asthma. Clin Chest Med 21:245–261 [DOI] [PubMed] [Google Scholar]
- Postma DS, Bleecker ER, Amelung PJ, Holroyd KJ, Xu J, Panhuysen CI, Meyers DA, Levitt RC (1995) Genetic susceptibility to asthma—bronchial hyperresponsiveness coinherited with a major gene for atopy. N Engl J Med 333:894–900 [DOI] [PubMed] [Google Scholar]
- Ruffilli A, Bonini S (1997) Susceptibility genes for allergy and asthma. Allergy 52:256–273 [DOI] [PubMed] [Google Scholar]
- Ryden TA, Beemon K (1989) Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein. Mol Cell Biol 9:1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle AG, Peters J, Lyon MF, Hall JG, Evans EP, Edwards JH, Buckle VJ (1989) Chromosome maps of man and mouse. IV. Ann Hum Genet 53:89–140 [DOI] [PubMed] [Google Scholar]
- Sears MR, Burrows B, Flannery EM, Herbison GP, Hewitt CJ, Holdaway MD (1991) Relation between airway responsiveness and serum IgE in children with asthma and in apparently normal children. N Engl J Med 325:1067–1071 [DOI] [PubMed] [Google Scholar]
- Shijubo N, Itoh Y, Yamaguchi T, Imada A, Hirasawa M, Yamada T, Kawai T, Abe S (1999) Clara cell protein-positive epithelial cells are reduced in small airways of asthmatics. Am J Respir Crit Care Med 160:930–933 [DOI] [PubMed] [Google Scholar]
- Zimonjic DB, Rezanka L, DiPaolo JA, Popescu NC (1995) Refined localization of the erbB-3 proto-oncogene by direct visualization of FISH signals on LUT-inverted and contrast-enhanced digital images of DAPI-banded chromosomes. Cancer Genet Cytogenet 80:100–102 [DOI] [PubMed] [Google Scholar]