Abstract
The Charcot-Marie-Tooth (CMT) disorders comprise a group of clinically and genetically heterogeneous hereditary motor and sensory neuropathies, which are mainly characterized by muscle weakness and wasting, foot deformities, and electrophysiological, as well as histological, changes. A subtype, CMT2, is defined by a slight or absent reduction of nerve-conduction velocities together with the loss of large myelinated fibers and axonal degeneration. CMT2 phenotypes are also characterized by a large genetic heterogeneity, although only two genes—NF-L and KIF1Bβ—have been identified to date. Homozygosity mapping in inbred Algerian families with autosomal recessive CMT2 (AR-CMT2) provided evidence of linkage to chromosome 1q21.2-q21.3 in two families (Zmax=4.14). All patients shared a common homozygous ancestral haplotype that was suggestive of a founder mutation as the cause of the phenotype. A unique homozygous mutation in LMNA (which encodes lamin A/C, a component of the nuclear envelope) was identified in all affected members and in additional patients with CMT2 from a third, unrelated family. Ultrastructural exploration of sciatic nerves of LMNA null (i.e., −/−) mice was performed and revealed a strong reduction of axon density, axonal enlargement, and the presence of nonmyelinated axons, all of which were highly similar to the phenotypes of human peripheral axonopathies. The finding of site-specific amino acid substitutions in limb-girdle muscular dystrophy type 1B, autosomal dominant Emery-Dreifuss muscular dystrophy, dilated cardiomyopathy type 1A, autosomal dominant partial lipodystrophy, and, now, AR-CMT2 suggests the existence of distinct functional domains in lamin A/C that are essential for the maintenance and integrity of different cell lineages. To our knowledge, this report constitutes the first evidence of the recessive inheritance of a mutation that causes CMT2; additionally, we suggest that mutations in LMNA may also be the cause of the genetically overlapping disorder CMT2B1.
Introduction
The Charcot-Marie-Tooth (CMT) disorders, also known as “hereditary motor and sensory neuropathies” (HMSNs), comprise the most common group of inherited neuropathies, affecting 10–40/100,000 individuals. A clinical, neuropathologic, electrophysiological, and genetic heterogeneity characterizes the CMT disorders (Skre 1974). Schematically, on the basis of nerve-conduction velocities (NCVs) at the median nerve, the CMT disorders can be divided into two main subtypes—demyelinating (i.e., type 1, or CMT1) and axonal (i.e., type 2, or CMT2) neuropathies, showing NCVs <38 m/s and >38 m/s, respectively (Garcia 1999).
Genetically, CMT2 is a heterogeneous group of peripheral neuropathies, and, in contrast to CMT1, for which seven genes and ⩽30 loci have been reported thus far (for review, see Reilly 2000), very little is known concerning their genetic bases. To date, only two specific genes responsible for autosomal dominant CMT2 (AD-CMT2) have been identified. Mutations in the neurofilament-light (NF-L) gene, which encodes a member of the intermediate filaments (IF) family of proteins, cause CMT2E (MIM 162280) (Mersiyanova et al. 2000), and mutations in KIF1Bβ, which encodes a microtubule motor protein that is a member of the kinesin superfamily of proteins, have recently been demonstrated to cause CMT2A (MIM 118210) (Zhao et al. 2001). Besides these mutations, gene defects in MPZ, which encodes myelin protein zero, which were initially reported to cause CMT1B (MIM 118200), Dejerine-Sottas disease (MIM 145900), and congenital hypomyelination (MIM 605253), also cause axonal phenotypes (Marrosu et al. 1998; Chapon et al. 1999; Misu et al. 2000).
Conversely, no genes responsible for autosomal recessive CMT2 (AR-CMT2) have yet been identified, although three loci have been localized on chromosomes 1q21.2-q21.3 (for CMT2B1 [MIM 605588]) (Bouhouche et al. 1999), 19q13.3 (for CMT2B2 [MIM 605589]) (Leal et al. 2001), and 8q21.3 (Barhoumi et al. 2001). AR-CMT2 is a rare and severe condition (De Jonghe et al. 1998; Gemignani et al. 2001).
Clinically, the main symptoms in 90% of cases are early onset, symmetrical muscle weakness and wasting (predominantly in the distal lower limbs), foot deformities, and walking difficulties associated with reduced or absent tendon reflexes (De Jonghe et al. 1998; Gemignani et al. 2001). Confirmation of diagnosis relies essentially on electrophysiology, which shows NCVs >38 m/s at the median nerve, and on histopathology after nerve biopsy, which evidences a loss of myelinated fibers with (for AD-CMT2) or without (for AR-CMT2) regenerative attempts (De Jonghe et al. 1998; Gemignani et al. 2001).
In the present study, we report a homozygous 892C→T mutation in exon 5 of LMNA, which encodes lamin A/C nuclear-envelope proteins. This mutation, causing an R298C amino acid substitution, segregates as a disease-causing founder mutation in individuals with AR-CMT2 from three Algerian families in which the disease was linked to chromosome 1q21.2-q21.3. Furthermore, the phenotype of Lmna knockout mice (Sullivan et al. 1999), as determined by analysis of peripheral nerves, was examined. Ultrastructural analysis of sciatic nerves of Lmna null (i.e., −/−) mice revealed neuropathic features—a strong reduction of axon density that was accompanied by an increase in axon diameter and the presence of nonmyelinated axons—that were highly similar to those found in human peripheral axonopathies.
Since LMNA mutations have been associated with limb-girdle muscular dystrophy type 1B (LGMD1B [MIM 159001]) (Muchir et al. 2000), autosomal dominant Emery-Dreifuss muscular dystrophy (AD-EDMD [MIM 181350]) (Bonne et al. 1999), dilated cardiomyopathy type 1A (CMD1A [MIM 115200]) (Fatkin et al. 1999), and autosomal dominant partial lipodystrophy (PLD [MIM 151660]) (Shackleton 2000), the specific association between the R298C amino acid substitution and AR-CMT2 suggests the existence of distinct functional domains in lamin A/C that are essential for the maintenance and integrity of different cell lineages. To our knowledge, the present article constitutes the first evidence of a mutation that causes CMT2 to be inherited as a recessive trait and, additionally, suggests that LMNA mutations may be the cause of the genetically overlapping disorder CMT2B1 (also called “AR-CMT2A”) (Bouhouche et al. 1999).
Subjects and Methods
Families and Phenotype Analysis
Twenty-three consanguineous families originating from Algeria were examined in the Services de Neurologie, Centre Hospitalier Universitaire Ben-Aknoun and Centre Hospitalier Universitaire Mustapha, Algiers, Algeria. Detailed clinical and electrophysiological examinations were performed, and inclusion criteria were those of axonal neuropathy, as described elsewhere (De Jonghe et al. 1998; Gemignani et al. 2001). Informed consent, for both blood collection and nerve biopsy, was obtained from all individuals participating in this study. This study complies with the ethical guidelines of the institutions involved.
Histology
Nerve biopsies were performed on the external branch of the musculocutaneous nerve in the foot of human subjects, and the samples were prepared for electron microscopy, as described elsewhere (Hahn et al. 2001). Sciatic nerves from mice were fixed in 2.5% glutaraldehyde, were dehydrated, were embedded in paraffin, and were sectioned before being stained in hematoxylin and eosin.
Genotyping
Genomic DNA was extracted from peripheral-blood lymphocytes by standard procedures. Genotyping experiments were performed using polymorphic microsatellite DNA markers that were retrieved from the Généthon genetic map. The following microsatellite markers were used to test linkage with chromosome 1q21.2-q21.3: D1S498 (AFM336xb1), D1S305 (AFM220xf8), D1S2715 (AFMa357ze5), D1S303 (AFM081zc5), D1S2777 (AFMb337zh1), D1S2721 (AFMb009zb9), D1S2624 (AFMa123wg5), and D1S2635 (AFMa133ye5). Marker informativity varied from 0.63 to 0.87. PCRs were set in a 30-ml total reaction volume that contained 0.05–0.1 mg genomic DNA, 0.2 mg each primer (one of which was 5′ labeled with IRD-800), 200 mM each dNTP, 3 ml 10× PCR buffer, and 1 ml Taq polymerase (Promega). After an initial 3-min cycle at 95°C, 30–38 cycles of amplification were performed for 30–60 s at 95°C, 55–60°C, or 72°C, depending on the primer pair; a 7-min elongation step (72°C) completed the reactions. PCR products were visualized under UV light after electrophoresis on a 1.5% agarose gel before genotyping, which was performed on a Li-Cor 4200 automated sequencing analyzer as described by Villard et al. (2001).
Linkage Analysis
Statistical analysis were performed assuming autosomal recessive mode of inheritance with full penetrance and with disease-allele frequency at 0.001, and two-point LOD scores were calculated using MLINK in the LINKAGE computer package (Lathrop et al. 1984). For linkage disequilibrium, allele frequencies in mutant chromosomes were compared to those observed in 100 control chromosomes from 50 unrelated individuals who originated from Algeria, and significance was established using the standard χ2 test.
Mutation Analysis
The public annotation database at the University of California–Santa Cruz Human Genome Project Working Draft (“Golden Path”) was used to integrate genetic, physical, and transcriptional data on the LMNA region from D1S2715 to D1S2624. PCR-amplified products corresponding to the coding sequences of A1U, FLJ12287, and LMNA (GenBank accession numbers AF188240, AK022349, and XM_044163, respectively), including exon-specific fragments and splice sites (all primers used in this study are listed in table 1), were purified with the QIAquick 50 PCR purification kit (Qiagen). M13 universal (GGTGTAAAACGACGGCCAGT) and reverse (GGCAGGAAACAGCTATGACC) plasmid sequences were added to the 5′ end of each forward primer and reverse primer, respectively. All the PCR products were further sequenced on both strands by use of either a dye terminator procedure (PE Biosystems), when sequencing reactions were loaded on ABI310 automated sequencing analyzer (Applied Biosystems), or a dye primer procedure, when sequencing reactions were loaded on Li-Cor 4200 automated sequencing analyzer (Li-Cor). All sequencing reactions were performed under conditions recommended by the manufacturers. The LMNA 892C→T mutation was specifically amplified with the following primers flanking exon 5: LMNA5-F (5′-CTCCCAGTCACCACAGTCCT-3′) and LMNA5-R (5′-GTGGTTGTGGGGACACTTTT-3′). PCR products were digested by the restriction endonuclease AciI and were loaded on 3% agarose electrophoresis gels. Sequence analysis was performed by comparing wild-type and patients' sequences by use of the Sequencher software (Genecodes).
Table 1.
PCR Primers for Sequence Analysis of LMNA, A1U, and FLJ12287
|
Primer(5′→3′) |
||
| Gene and Exon(s) | Forward | Reverse |
| LMNA: | ||
| Exon 1 | ccgagcagtctctgtccttc | ccctctcactcccttcctg |
| Exon 2 | gcactgtctaggcacacagact | gggagggcctaggtagaaga |
| Exon 3 | tgtgaccccttttcctcatc | cactagggcaagggactcag |
| Exons 4 and 5 | ggcctcccaggaactaattc | gtggggacacttttcatccc |
| Exon 5 (R298C) | ctcccagtcaccacagtcct | gtggttgtggggacactttt |
| Exons 6 and 7 | cttccccatacttagggccc | aaggatgttcctctctccac |
| Exons 8 and 9 | gcaagatacacccaagagcc | gctccgatgttggccatcag |
| Exon 10 | gtagacatgctgtacaaccc | ggccagcgagtaaagttcca |
| Exon 11 | ttgggcctgagtggtcagtc | gacccgcctgcaggatttgg |
| Exon 12 | atcgaggggtaggacgaggt | taaggcagatgtggagtttcc |
| A1U: | ||
| Exon 1 | agttctgggggttcctgttc | cccatctcttcgcagacc |
| Exons 2 and 3 | ccacagtgacctctgcctct | ccccaaacaagattgtgctc |
| Exon 4 | tcttgtggcagactgtgacc | agtgtcctccagctctccag |
| Exon 5 | gttggtgctcccctggtt | cactaccctggcccttgat |
| Exon 6 | aacccaaaacctttccctcct | gaccccactaagctccttcc |
| Exon 7 | tctggaccctttctctcctg | ctctcctcatgctggtttca |
| Exons 8–10 | aggtctccccctgctacct | cccccactgcctcatgta |
| Exon 11 | ctgtgcccacttcacaagg | gctgcagagggtaaggattg |
| FJL12287: | ||
| Exon 1 | cctgccaccaatacacacgc | ccacctccagctctctattg |
| Exon 2 | ccacttttatctcaggagtc | tattcatgcacaggcagcca |
| Exon 3 | cagctagaactgaggacct | tccagggatggtctagggat |
| Exons 4 and 5 | gggtccagcaatttgagtca | ccatatttcacagcgtgactg |
| Exon 6 | ccccgagactgatatgatg | ggacacattgtaccttccgc |
| Exons 7 and 8 | gttttcactcaggcaaaccc | tcatgtcaattcctgcctcc |
| Exon 9 | cttatcaacacagtgagggg | gctataccaggtgtaacgtct |
| Exon 10 | aaccaccaccctgaaatgag | gggcatcctctgctctagt |
| Exons 11 and 12 | ggaagggatctgtggatgag | cagtagcagaagctggctga |
| Exon 13 | cctccttcctcttcccactt | gtcttccagagcctttgcacc |
| Exon 14 | gctggggtccaaagataggt | tcctagtcagggctgtgctt |
Lamin A/C functional-domain predictions were obtained using the PROSITE database and the COILS (version 2) program. Amino acid comparisons were performed using the BLASTP program in the National Center for Biotechnology Information database (BLAST).
Results
Clinical and Electrophysiological Findings
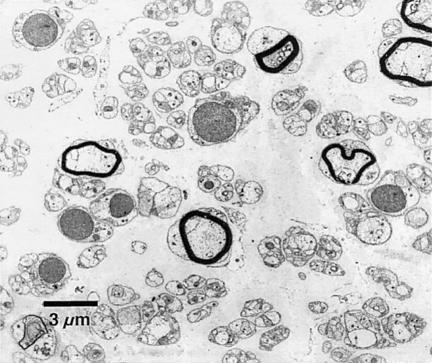
We examined 23 Algerian families in which the mode of inheritance was autosomal recessive and whose affected members presented with severe and early-onset CMT2. They showed abolished deep-tendon reflexes, as well as distal amyotrophy, mainly affecting the lower limbs. Motor deficits were distal in the upper limbs and both proximal and distal in the lower limbs. A patient's gait was thus characterized by marked lifting of the knees and waddling. Mean ± SD motor NCVs measured at the median nerve were 59±10 m/s, whereas sensory potentials were undetectable at sural, cubital, and median nerves. Ultrastructural analysis of peripheral nerves revealed a significant loss of large myelinated fibers (984/mm2 vs. 5,640/mm2 in normal controls), as well as the presence of abnormally myelinated axons (De Jonghe et al. 1998). Neither proliferations of Schwann cells (“onion bulbs”) nor regenerating “clusters” were observed (fig. 1). Further clinical, histological, and electrophysiological features associated with families with AR-CMT2 will be presented in a future article.
Figure 1.
Ultrastructural micrograph of peripheral nerve from an individual from family ALG.16-301 who is affected with CMT2. A severe rarefaction of myelinated and nonmyelinated fibers can be observed. Large myelinated fibers are almost totally lacking. Neither onion bulbs (proliferations of Schwann cells) nor regenerating clusters are present. Nerve biopsies were performed, and the samples were prepared for electron microscopy as described elsewhere (Hahn et al. 2001).
Linkage Study and Linkage Disequilibrium
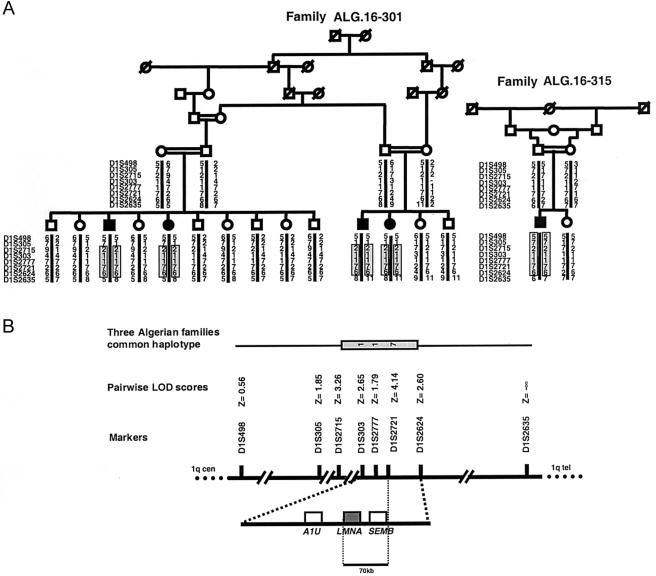
Genotyping analysis showed significant linkage to chromosome 1q21.2-q21.3, from D1S305 to D1S2635 (Zmax=4.14 at θ=0 at D1S2721), in two families (ALG.16-301 and ALG.16-315). All affected individuals in both families were born of consanguineous parents and were homozygous for each marker within this region. Additionally, each affected individual shared a 3.2-cM common haplotype (2-1-1-7-6 at D1S2715, D1S303, D1S2777, D1S2721, and D1S2624, respectively) in both of these families (fig. 2). In a third, unrelated family, which was not included in the linkage analysis because DNA from only two affected patients was available, we subsequently observed homozygosity for D1S303, D1S2777, D1S2721, and D1S2624, possibly indicating linkage to the same interval. All patients in these three families shared a smaller homozygous haplotype (1-1-7 at D1S303, D1S2777, and D1S2721, respectively) and were homozygous for allele 7 at D1S2721, for which homozygosity is rare (<2%; P<.001) in Algerian populations. These observations (1) suggested that an ancestral haplotype, flanked by D1S2715 and D1S2624, segregated with AR-CMT2 that was linked to chromosome 1q21.2-q21.3 and (2) were consistent with a founder effect in families of Algerian descent. Moreover, we hypothesized that, owing to linkage-disequilibrium mapping, the disease-causing gene mapped near or at D1S2721. In the remaining 20 families, linkage to chromosome 1q was clearly excluded, thus confirming the genetic heterogeneity that is associated with CMT2 disorders.
Figure 2.
Pedigree of largest Algerian family with AR-CMT2, mutant haplotypes, and integrated map at the LMNA locus. A, Pedigrees and genotypes of Algerian families ALG.16-301 and ALG.16-315, both with AR-CMT2. Blackened symbols represent subjects with clinical, electrophysiological, and histological diagnosis of CMT2. The homozygosity interval in each affected individual is boxed and shaded. B, Genetic, physical, and partial transcriptional map of 1q21.2-q21.3 region. The common haplotype for D1S303, D1S2777, and D1S2721 that is shared by all affected individuals from the three Algerian families is shown (see “Results” section). Position of genetic markers is indicated with two-point LOD-score values. The positions of A1U, SEMB, and LMNA are indicated.
Sequencing and Mutation Analysis
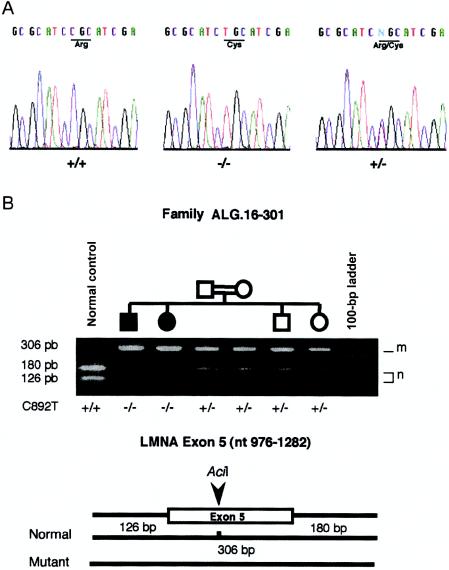
Flanked by D1S2715 and D1S2624 and encompassing the linkage-disequilibrium region, an ∼1.5-Mb physical and transcriptional BAC/PAC contig of sequenced genomic DNA was thus electronically screened. Among the numerous potentially transcribed units, including several characterized human genes, we initially focused our efforts on the examination of three genes that represented good candidates owing to their function and localization—near D1S2721 within the linkage-disequilibrium region. A1U, which encodes ataxin 1–interacting protein, a protein interacting with ataxin (Davidson et al. 2000), and FLJ12287, a noncharacterized transcribed unit homologous to the mouse Semaphorin B (SemB) gene (GenBank accession number X85991) (Puschel et al. 1995), were initially analyzed, but sequencing did not reveal any sequence variation among the patients. LMNA, which encodes the nuclear-envelope proteins lamin A/C (Moir et al. 2000) and is located 70 kb proximal to D1S2721 (fig. 2), was then screened for mutations. We sequenced exons and intron-exon junctions that correspond to the four transcripts (lamin A, AD10, C, and C2) that were differentially spliced from LMNA (Furukawa et al. 1994; Machiels et al. 1996), and we identified a single homozygous C→T mutation in exon 5, at position 892 of the coding sequence (892C→T). This mutation resulted in an R→C amino acid substitution at position 298 (R298C), which was localized in the LMNA rod domain and thus affected all four transcripts produced by LMNA (fig. 3). The homozygous 892C→T mutation was present in all affected individuals from the three families and cosegregated with the disease in the two linked families (Z=5.97 at θ=0 at LMNA, when we included its mutation as a polymorphic marker in the pairwise linkage study), whereas the heterozygous carriers remained clinically asymptomatic regardless of their age at clinical examination.
Figure 3.
Lamin A/C R298C mutation. A, Sequence analysis and identification of R298C mutation in LMNA. DNA sequences from normal control (+/+), unaffected heterozygote (+/−), and homozygote (−/−) are presented. The R→C amino acid substitution at position 298 of human lamin A/C peptidic sequence is indicated. B, Segregation of the nucleotidic 892C→T mutation in a branch of family ALG.16-301 assayed by digestion with AciI. The 306-bp PCR product encompassing exon 5 includes one invariant AciI restriction site in wild-type alleles, which disappears when transition C→T is present, thereby leading to a 306-bp digestion fragment compared with the 126-bp and 180-bp bands observed for the wild-type allele. Normal (n) and mutant (m) fragments, as well as the mutation status (+/+, +/−, and −/−), are indicated for each individual. Undigested and digested fragment sizes, respectively, are 40 and 20 bp larger than described, owing to the M13 universal and reverse primers' sequencing tags (see “Subjects and Methods” section).
Since the 892C→T mutation removed an AciI restriction site, the genomic status of each individual was confirmed after AciI digestion of a specific PCR product encompassing the complete exon 5 (fig. 3). This test, applicable as a preliminary 892C→T mutation screening, was performed in >300 control chromosomes, including 150 from the Algerian population, without the identification of any mutation. To obtain further evidence that the 892C→T mutation was not a polymorphism segregating on a particular Algerian haplotype, we genotyped 100 Algerian subjects (200 chromosomes). We found that six subjects shared the 1-1-7 haplotype with patients with AR-CMT2 for D1S303, D1S2777, and D1S2721 on at least one chromosome without carrying the 892C→T mutation.
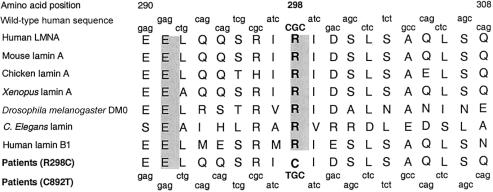
Arginine Conservation at Position 298
The lamin A/C amino acid substitution (R298C) that is associated with the CMT2 phenotype that we here report lies in the rod domain that is essential for protein-protein interactions. This domain is highly conserved through evolution from Caenorhabditis elegans to Homo sapiens, and the arginine at position 298 is also conserved in lamin B1 (fig. 4). We predict that the R298C mutation impairs proteins' interactions that are essential for the maintenance of cellular function; calculation of the consensus value at the PROSITE database by the COILS (version 2) program revealed a consensus value for the mutated domain (0.41) that was lower than that for the wild-type equivalent (1.00).
Figure 4.
Alignment of amino acid sequence of human lamin A/C with its orthologs from various species and human lamin B1, showing conservation of arginine at position 298. Fully conserved amino acids are shown atop a shaded background. The conservation of arginine at position 298 and its substitution in patients with AR-CMT2 are shown in boldface. The DNA sequences encoding the normal and mutant proteins are shown above and below the amino acid sequences, respectively.
Axonopathy in Lmna Knockout Mice
Homozygous Lmna knockout mice (Sullivan et al. 1999) display an abnormal gait with a stiff walking posture, which is characterized by splayed hind legs and the inability to hang on to structures with their forepaws. During development, they start to exhibit distinct scoliosis/kyphosis and become progressively more hunched. These aspects are not observed in heterozygous littermates and are extremely evocative of a severe peripheral neuropathy, since they are also reported in animal models of peripheral neuropathies (Huxley et al. 1996; Sereda et al. 1996; Norreel et al. 2001).
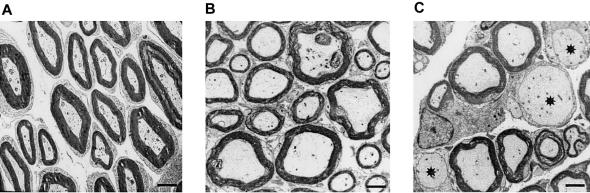
The responsibility of LMNA homozygous defects for the impairment of peripheral axons was further analyzed at the ultrastructural level by the examination of the sciatic nerves of Lmna knockout mice as compared to age-matched controls (fig. 5). Whereas heterozygous (i.e., +/−) knockout mice showed a preserved peripheral-nerve histology, null mice—having lost the full-length forms of Lmna transcripts (Sullivan et al. 1999) and showing a strong reduction of axon density, an increase in axon diameter, and the presence of nonmyelinated axons—were histologically similar to human patients with CMT2.
Figure 5.
Electron micrographs from transverse sections of sciatic nerves of Lmna knockout and control mice at age 7 wks. A, Sciatic nerve of Lmna +/+ control. B, Sciatic nerve of Lmna +/− mouse. No abnormality was detected, although a slight accumulation of neurofilaments was observed. Axon density, size, and myelination were preserved. C, Sciatic nerve of Lmna −/− mouse. Axon density was reduced, and several pathological axons are indicated by asterisks (*). The axons' diameters are enlarged, and no myelin sheath is visible; additionally, they present focal accumulations of neurofilaments, as observed in nerves of Lmna +/− mouse (bars correspond to 1 μm).
Discussion
We report linkage to chromosome 1q21.2-q21.3 and segregation of a specific LMNA 892C→T homozygous founder mutation in three Algerian families that are affected with AR-CMT2. The R298C amino acid substitution induced by the mutation was determined to have caused AR-CMT2 in 3 (13%) of 23 inbred Algerian families and thus is, apparently, a frequent cause of axonal autosomal recessive neuropathies. Additionally, we report the observation that Lmna null mice present an axonal pathological phenotype that is highly similar to that presented by patients with AR-CMT2.
LMNA was regarded as a good candidate in AR-CMT2 linked to chromosome 1q21, because it had been mapped to the linkage-disequilibrium region, because of its expression pattern during neuronal development (Pierce et al. 1999) and its membership in the IF multigenic family that also includes NF-L, which is involved in CMT2E (Mersiyanova et al. 2000; De Jonghe et al. 2001), and because of the wide spectrum of phenotypes associated with its mutations (Bonne et al. 1999; Fatkin et al. 1999; Muchir et al. 2000; Shackleton 2000). Additionally, the clinical phenotype presented by Lmna null mice, including locomotor difficulties and kyphoscoliosis (Sullivan et al. 1999), was thought to be reminiscent of human motor and sensory neuropathies. These phenotypic features, which were specific to homozygous knockout mice, were confirmed by the neuropathologic analysis of sciatic nerves, which provided evidence of an axonopathy with a strong reduction of axon density, axonal enlargement, and the presence of nonmyelinated axons, all of which were highly similar to the phenotypes of human peripheral axonopathies. It is relevant that these features were not observed in heterozygous knockout mice, thereby indicating that loss of function of all lamin A/C proteins is necessary for the expression of the neuropathic disease, since it appears to happen in patients who carry the R298C amino acid substitution.
Although most of the pathogenic mutations previously reported in LMNA-associated disorders are expressed when carried by heterozygous individuals, two patients with EDMD have recently been reported as being either homozygous or compound heterozygous for LMNA mutations (Raffaele di Barletta et al. 2000). In one case, the mutation (H222Y) lies in linker 2 of the lamin A/C protein, connecting the coiled 1b and coiled 2a domains; it is interesting that it had been determined that the patient carried either an atypical EDMD or a congenital muscular dystrophy. At age 40 years, the patient had severe and diffuse muscle wasting and was confined to a wheelchair, and, moreover, he showed neither a dilated cardiomyopathy nor any other cardiac disease; his heterozygous relatives were unaffected (Raffaele di Barletta et al. 2000). In the second case, the patient carried two different mutations, one of which (E358K) lies in the coiled 2b domain and the other of which (R624H) lies in the tail domain (Brown et al. 2001); muscle weakness was localized proximally in the upper limbs and distally in the lower limbs, and the patient did not show any cardiac disease. In patients with AR-CMT2, the R→C amino acid substitution at position 298 (R298C) is localized in the lamin A/C rod domain and thus affects all four known isoforms (A, AΔ10, C, and C2) that are produced by LMNA. Ultrastructural analysis of the peripheral nerves of the patients whom we studied showed an axonal neuropathic process; additionally, at clinical examination, patients presented with a proximal involvement of lower-limb muscles, whereas no cardiac-conduction disturbances or dilatation among among patients aged 12–52 years were observed. These clinical observations are consistent with those reported for the consanguineous Moroccan family in which linkage to the 1q21.2-q21.3 region was first described (Bouhouche et al. 1999).
LMNA mutations have been reported as being associated with numerous genetic disorders, including LGMD1B (Muchir et al. 2000), AD-EDMD (Bonne et al. 1999), CMD1A (Fatkin et al. 1999), and autosomal dominant PLD (Shackleton et al. 2000). Since LMNA is ubiquitously expressed, the finding of site-specific amino acid substitutions in AR-CMT2, as well as in the disorders described elsewhere, indicates the existence of distinct functional domains in lamin A/C that are essential for the maintenance and integrity of different cell lineages. At the same time, much care is needed in the definition of clinical phenotypes that may share features with different disorders. For example, muscle wasting is observed as a common feature in CMT phenotypes and is usually thought to be secondary to nerve deterioration; that various mutations in a single gene may lead to neuronal (CMT2) and muscular (CMD1A, EDMD, and LGMD1B) phenotypes suggests that muscular phenotypes may be primarily associated with nerve fibers’ loss or degeneration and should not be considered only as a delayed consequence of nerve degeneration. On the basis of these considerations, a closer examination of the muscular phenotype of patients with AR-CMT2 and LMNA mutations will be performed.
These considerations may also be correlated with the recent studies performed on the myotubularin protein multigene family (Laporte et al. 2001). Mutations in MTM1 cause myotubular myopathy (Laporte et al. 1996), a severe muscular disorder, whereas mutations in MTMR2, one of its close homologs, are associated with CMT4B, a demyelinating motor and sensory neuropathy (Bolino et al. 2000).
The 892C→T mutation has never been reported as being associated with other phenotypes, and, most likely, it is a founder mutation that segregates in Algerian families that are affected with AR-CMT2 linked to the 1q21.2-q21.3 region. Our results add to the evidence of clinical heterogeneity associated with LMNA mutations that has been described elsewhere and confirm that different mutations in this gene may lead to tissue-specific phenotypes. LMNA thus seems to have a fundamental function in the regulation of axon development and/or survival. These data are supported by the observation that neuronal LMNA expression seems to vary in different neuronal subtypes (Cance et al. 1992) and to decrease with the progression of the differentiation state (Pierce et al. 1999).
The observation that LMNA mutations cause AR-CMT2 broadens the range of disorders for which members of the nuclear envelope may be considered as candidate genes and underscores the role that IFs play in nerve-fiber survival and regeneration. Besides NF-L and LMNA, members of the IF family may thus be considered as candidates for axonal HMSNs. The association between LMNA mutations and neuronal disorder not only is being reported for the first time but also significantly expands our previous views on lamin A/C–associated pathologies that are restricted to muscle and lipid tissues. Also, this association greatly advances the progress made toward comprehension of (1) how mutations in a nuclear structural protein can result in so many different diseases and (2) how nuclear structure relates to cellular functions and genetic diseases. Toward this end, a complete examination of Lmna null mice, which is specifically focused on peripheral-nerve function, is being performed. Additionally, a knockin-mouse line carrying the R298C substitution is being generated to determine whether this mutation has a specific effect on peripheral nerves only or affects other tissues. These mice will also serve as a model to determine how a single missense mutation impacts protein interactions, since the founder mutation reported here is predicted to destabilize the lamin interaction domain, and, consequently, nuclear structure and function. The determination of the function of lamin A/C during the normal and abnormal development of peripheral nervous system and skeletal muscle will shed light on nerve-muscle interactions, the understanding of which is essential for the establishment of a comprehensive approach to HMSNs.
Acknowledgments
We wish to thank the patients and their families, for their invaluable participation and cooperation, and the cell and DNA bank of the Hôpital d’Enfants de la Timone. We are indebted to Dominique Recan, for providing us with control DNA samples, and Michel Fontés and Pierre Cau, for helpful comments and discussions. We are grateful to Jerome Belougne for excellent technical assistance. During the revision process, the scientific expertise of Josue Feingold has been of invaluable help. This work was supported by fellowships from the Association Française contre les Myopathies and the Association CMT-France (to A.D.-G.) and by the Institut National de la Sante et de la Recherche Medicale (INSERM).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for BLASTP)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for A1U [accession number AF188240], FLJ12287 [accession number AK022349], LMNA [accession number XM_044163], mouse SemB [accession number X85991], lamin A/C mRNA [accession number XM_044163], lamin B1 mRNA [accession number AAC37575], mouse lamin A [accession number P48678], chicken lamin A [accession number P13648], X. laevis lamin A [accession number P11048], D. melanogaster DMO [accession number P08928], C. elegans lamin [accession number S42257], and genomic LMNA sequence contained in a BAC contig [accession number NT004858])
- Généthon, ftp://ftp.genethon.fr/pub/Gmap/Nature-1995/ (for genetic map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT2E [MIM 162280], CMT2A [MIM 118210], CMT1B [MIM 118200], Dejerine-Sottas disease [MIM 145900], congenital hypomyelination [MIM 605253], CMT2B1 [MIM 605588], CMT2B2 [MIM 605589], LGMD1B [MIM 159001], AD-EDMD [MIM 181350], CMD1A [MIM 115200], and PLD [MIM 151660])
- PROSITE: Database of Protein Families and Domains, http://www.expasy.ch/prosite/
- UCSC Human Genome Project Working Draft (“Golden Path”), http://genome.ucsc.edu/
References
- Barhoumi C, Amouri R, Ben Hamida C, Ben Hamida M, Machghoul S, Gueddiche M, Hentati F (2001) Linkage of a new locus for autosomal recessive axonal form of Charcot-Marie-Tooth disease to chromosome 8q21.3. Neuromuscul Disord 11:27–34 [DOI] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, Christodoulou K, Hausmanowa-Petrusewicz I, Mandich P, Schenone A, Gambardella A, Bono F, Quattrone A, Devoto M, Monaco AP (2000) Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet 25:17–19 [DOI] [PubMed] [Google Scholar]
- Bonne G, Raffaele di Barletta M, Varnous S, Bécane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 21:285–288 [DOI] [PubMed] [Google Scholar]
- Bouhouche A, Benomar A, Birouk N, Mularoni A, Meggouh F, Tassin J, Grid D, Vandenberghe A, Yahyaoui M, Chkili T, Brice A, LeGuern E (1999) A locus for an axonal form of autosomal recessive Charcot-Marie-Tooth disease maps to chromosome 1q21.2-q21.3. Am J Hum Genet 65:722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CA, Lanning RW, McKinney KQ, Salvino AR, Cherniske E, Crowe CA, Darras BT, Gominak S, Greenberg CR, Grosmann C, Heydemann P, Mendell JR, Pober BR, Sasaki T, Shapiro F, Simpson DA, Suchowersky O, Spence JE (2001) Novel and recurrent mutations in lamin A/C in patients with Emery-Dreifuss muscular dystrophy. Am J Med Genet 102:359–367 [DOI] [PubMed] [Google Scholar]
- Cance WG, Chaudhary N, Worman HJ, Blobel G, Cordon-Cardo C (1992) Expression of the nuclear lamins in normal and neoplastic human tissues. J Exp Clin Cancer Res 11:233 [Google Scholar]
- Chapon F, Latour P, Diraison P, Schaeffer S, Vandenberghe A (1999) Axonal phenotype of Charcot-Marie-Tooth disease associated with a mutation in the myelin protein zero gene. J Neurol Neurosurg Psychiatry 66:779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JD, Riley B, Burright EN, Duvick LA, Zoghbi HY, Orr HT (2000) Identification and characterization of an ataxin-1-interacting protein: A1Up, a ubiquitin-like nuclear protein. Hum Mol Genet 9:2305–2312 [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Mersivanova I, Nelis E, Del Favero J, Martin JJ, Van Broeckoven C, Evgrafov O, Timmermann V (2001) Further evidence that neurofilament light chain gene mutations can cause Charcot-Marie-Tooth disease type 2E. Ann Neurol 49:245–249 [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Timmerman V, Van Broeckhoven C (1998) 2nd Workshop of the European CMT Consortium: 53rd ENMC International Workshop on Classification and Diagnostic Guidelines for Charcot-Marie-Tooth Type 2 (CMT2–HMSN II) and Distal Hereditary Motor Neuropathy (Distal HMN–Spinal CMT), September 26–28, 1997, Naarden, The Netherlands. Neuromuscul Disord 8:426–431 [PubMed] [Google Scholar]
- Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ Jr, Spudich S, De Girolami U, Seidman JG, Seidman C, Muntoni F, Muehle G, Johnson W, McDonough B (1999) Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 341:1715–1724 [DOI] [PubMed] [Google Scholar]
- Furukawa K, Inagaki H, Hotta Y (1994) Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp Cell Res 212:426–430 [DOI] [PubMed] [Google Scholar]
- Garcia CA (1999) A clinical review of Charcot-Marie-Tooth. Ann NY Acad Sci 883:69–76 [PubMed] [Google Scholar]
- Gemignani F, Marbini A (2001) Charcot-Marie-Tooth disease (CMT): distinctive phenotypic and genotypic features in CMT type 2. J Neurol Sci 184:1–9 [DOI] [PubMed] [Google Scholar]
- Hahn AF, Ainsworth PJ, Bolton CF, Bilbao JM, Vallat JM (2001) Pathological findings in the X-linked form of Charcot-Marie-Tooth disease: a morphometric and ultrastructural analysis. Acta Neuropathol 101:129–139 [DOI] [PubMed] [Google Scholar]
- Huxley C, Passage E, Manson A, Putzu G, Figarella-Branger D, Pellissier JF, Fontes M (1996) Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum Mol Genet 5:563–569 [DOI] [PubMed] [Google Scholar]
- Laporte J, Blondeau F, Buj-Bello A, Mandel JL (2001) The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet 17:221–228 [DOI] [PubMed] [Google Scholar]
- Laporte J, Hu LJ, Kretz C, Mandel JL, Kioschis P, Coy JF, Klauck SM, Poustka A, Dahl N (1996) A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet 13:175–182 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Leal A, Morera B, Del Valle G, Heuss D, Kayser C, Berghoff M, Villegas R, Hernandez E, Mendez M, Hennies HC, Neundorfer B, Barrantes R, Reis A, Rautenstrauss B (2001) A second locus for an axonal form of autosomal recessive Charcot-Marie-Tooth disease maps to chromosome 19q13.3. Am J Hum Genet 68:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels BM, Zorenc AH, Endert JM, Kuijpers HJ, van Eys GJ, Ramaekers FC, Broers JL (1996) An alternative splicing product of the lamin A/C gene lacks exon 10. J Biol Chem 271:9249–9253 [DOI] [PubMed] [Google Scholar]
- Marrosu MG, Vaccargiu S, Marrosu G, Vannelli A, Cianchetti C, Muntoni F (1998) Charcot-Marie-Tooth disease type 2 associated with mutation of the myelin protein zero gene. Neurology. 50:1397–1401 [DOI] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, Sitnikov VF, Dadali EL, Oparin RB, Petrin AN, Evgrafov OV (2000) A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am J Hum Genet 67:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misu K, Yoshihara T, Shikama Y, Awaki E, Yamamoto M, Hattori N, Hirayama M, Takegami T, Nakashima K, Sobue G (2000) An axonal form of Charcot-Marie-Tooth disease showing distinctive features in association with mutations in the peripheral myelin protein zero gene (Thr124Met or Asp75Val). J Neurol Neurosurg Psychiatry 69:806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD (2000) Review: the dynamics of the nuclear lamins during the cell cycle—relationship between structure and function. J Struct Biol 129:324–334 [DOI] [PubMed] [Google Scholar]
- Muchir A, Bonne G, van der Kooi AJ, van Meegen M, Baas F, Bolhuis PA, de Visser M, Schwartz K (2000) Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum Mol Genet 9:1453–1459 [DOI] [PubMed] [Google Scholar]
- Norreel JC, Jamon M, Riviere G, Passage E, Fontes M, Clarac F (2001) Behavioural profiling of a murine Charcot-Marie-Tooth disease type 1A model. Eur J Neurosci 13:1625–1634 [DOI] [PubMed] [Google Scholar]
- Pierce T, Worman HJ, Holy J (1999) Neuronal differentiation of NT2/D1 teratocarcinoma cells is accompanied by a loss of lamin A/C expression and an increase in lamin B1 expression. Exp Neurol 157:241–250 [DOI] [PubMed] [Google Scholar]
- Puschel AW, Adams RH, Betz H (1995) Murine semaphorin D/collapsin is a member of a diverse gene family and creates domains inhibitory for axonal extension. Neuron 14:941–948 [DOI] [PubMed] [Google Scholar]
- Raffaele di Barletta M, Ricci E, Galluzzi G, Tonali P, Mora M, Morandi L, Romorini A, Voit T, Orstavik KH, Merlini L, Trevisan C, Biancalana V, Housmanowa-Petrusewicz I, Bione S, Ricotti R, Schwartz K, Bonne G, Toniolo D (2000) Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery-Dreifuss muscular dystrophy. Am J Hum Genet 66:1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MM (2000) Classification of the hereditary motor and sensory neuropathies. Curr Opin Neurol 13:561–564 [DOI] [PubMed] [Google Scholar]
- Sereda M, Griffiths I, Puhlhofer A, Stewart H, Rossner MJ, Zimmerman F, Magyar JP, Schneider A, Hund E, Meinck HM, Suter U, Nave KA (1996) A transgenic rat model of Charcot-Marie-Tooth disease. Neuron 16:1049–1060 [DOI] [PubMed] [Google Scholar]
- Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, Gregory S, O'Rahilly S, Trembath RC (2000) LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet 24:153–156 [DOI] [PubMed] [Google Scholar]
- Skre H (1974) Genetic and clinical aspects of Charcot-Marie-Tooth's disease. Clin Genet 6:98–118 [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147:913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villard L, Levy N, Xiang F, Kpebe A, Labelle V, Chevillard C, Zhang Z, Schwartz CE, Tardieu M, Chelly J, Anvret M, Fontes M (2001) Segregation of a totally skewed pattern of X chromosome inactivation in four familial cases of Rett syndrome without MECP2 mutation: implications for the disease. J Med Genet 38:435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hayashi Y, Hirokawa N (2001) Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell 105:587–597 [DOI] [PubMed] [Google Scholar]