Abstract
The hereditary spastic ataxias (HSA) are a group of clinically heterogeneous neurodegenerative disorders characterized by lower-limb spasticity and generalized ataxia. HSA was diagnosed in three unrelated autosomal dominant families from Newfoundland, who presented mainly with severe leg spasticity, dysarthria, dysphagia, and ocular-movement abnormalities. A genomewide scan was performed on one family, and linkage to a novel locus for HSA on chromosome 12p13, which contains the as-yet-unidentified gene locus SAX1, was identified. Fine mapping confirmed linkage in the two large families, and the third, smaller family showed LOD scores suggestive of linkage. Haplotype construction by use of 13 polymorphic markers revealed that all three families share a disease haplotype, which key recombinants and overlapping haplotypes refine to ∼5 cM, flanked by markers D12S93 and GATA151H05. SAX1 is the first locus mapped for autosomal dominant HSA.
Hereditary spastic ataxia (HSA) comprises a heterogeneous group of progressive neurodegenerative disorders characterized by lower-limb spasticity and generalized ataxia with dysarthria, with impaired ocular movements, and with gait disturbance (Mahloudji 1963; Bouchard et al. 2000). HSA presents as dominant (MIM 108600) and recessive (MIM 270500) types, the latter of which includes autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS [MIM 270550]) (De Braekeleer et al. 1993). The disease gene causing ARSACS has been identified on chromosome 13q11 (MIM 604490) and encodes sacsin, a protein that is thought to be involved in chaperone-mediated protein folding (Engert et al. 2000).
Three large families (13, 27, and 71) with autosomal dominant HSA were identified in Newfoundland (Grewal et al. 2001) and independently assessed, mostly by a single neurologist (M.S.), by use of a standard protocol. The majority of affected individuals initially present with progressive leg spasticity of variable degrees followed by ataxia in the form of involuntary head jerk, dysarthria, dysphagia, and ocular-movement abnormalities with no signs of amyotrophy. The lower limbs show hyperreflexia and hypertonicity. The ocular-movement abnormalities include slow saccades, impaired vertical gaze, and, in some cases, lid retraction. A few patients have additional features, such as dystonia, pes cavus, mild ptosis, and decreased vibration sense in lower limbs. This phenotype resembles that observed in ARSACS; however, these three families are from a different population and show a clear pattern of dominant inheritance with a later age at onset than is seen in ARSACS (Bouchard et al. 2000).
The severity of the phenotype in the three families varies greatly within and among the families, and the age at onset is from early childhood to early 20s, although the majority of patients start presenting with symptoms at age 10–20 years. The available neuropathological data indicate degeneration of the corticospinal tracts and posterior columns. The life span and cognition of patients is not affected, and there is no obvious kinship between these three families (Grewal et al. 2001; K.K.G., unpublished data).
Twenty-nine individuals have been collected from family 71, of whom 14 are clinically affected and 15 are unaffected (fig. 1). The largest family is family 13, from which 50 individuals have been collected: 21 affected and 29 unaffected (fig. 2). Finally, family 27 is a small collection of seven individuals, of whom three are affected (fig. 3). Blood was collected from consenting family members, age ⩾19 years, and DNA was extracted using a salting-out method (Miller et al. 1988).
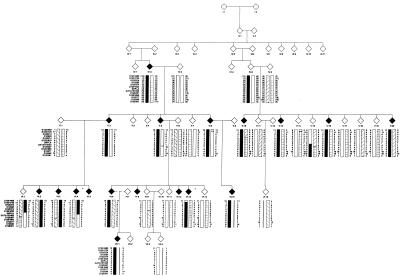
Figure 1.
Informative section of family 71, with the chromosome 12p13 haplotype. The disease haplotype for HSA is indicated by the blackened bar. Affected individuals are indicated by blackened symbols, unaffected individuals are indicated by unblackened symbols, and key recombinants (VI:4 and VI:13) are indicated by an asterisk (*). These recombinants determined the critical interval to be 10 cM, flanked by markers D12S1685 and GATA151H05.
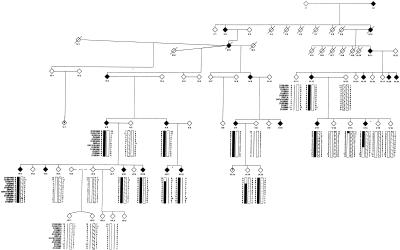
Figure 2.
Informative section of family 13, with the chromosome 12p13 haplotype. The disease haplotype for HSA is indicated by the blackened bar. Affected individuals are indicated by blackened symbols, unaffected individuals are indicated by unblackened symbols, key recombinants (V:14 and VI:11) are indicated by an asterisk (*), and possibly affected individuals are indicated by a question mark (?). These recombinants determined the critical region to be 8 cM, flanked by markers D12S1725 and D12S397.
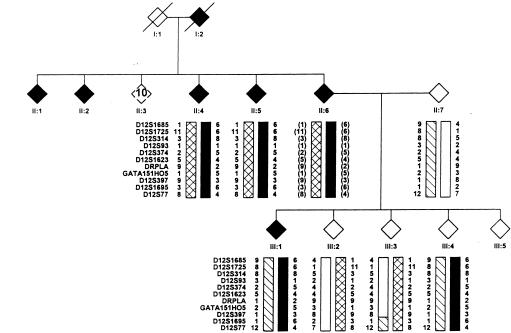
Figure 3.
Informative section of family 27, with the chromosome 12p13 haplotype. The disease haplotype for HSA is indicated by the blackened bar. Affected individuals are indicated by blackened symbols, and unaffected individuals are indicated by unblackened symbols. Individual III:4 shares the complete haplotype and is unaffected.
To map the gene causing HSA in these three dominant kindreds, a genomewide scan of the 22 autosomes was performed on family 71. Although this family is not the largest pedigree, the individuals in it are more closely related, and fewer individuals were required for significant linkage results. Approximately 400 evenly spaced polymorphic microsatellite markers throughout the genome were used. Two-point parametric linkage analysis was performed for each marker by use of the FASTLINK (version 4.1) software package. The parameters in the linkage analysis were (a) an estimated disease gene frequency of 1/10,000, (b) a penetrance of 90%, (c) equal male and female recombination frequencies, and (d) Centre d'Étude du Polymorphisme Humain (CEPH) marker-allele frequencies. Additional polymorphic markers were genotyped to further investigate the positive linkage results. These markers and their order were obtained from the Marshfield genetic map (Center for Medical Genetics, Marshfield Medical Research Foundation), and the primer sequences were obtained from the Genome Database and the Cooperative Human Linkage Center database. These polymorphic markers were amplified by PCR, which incorporated a radiolabeled nucleotide (S35-dATP ) in the product. The PCR products were separated on 6% denaturing polyacrylamide gels and detected by exposure to autoradiographic film. The alleles were assigned on the basis of their size, with comparison to a M13mp18 sequence ladder. The MLINK program of the FASTLINK software package was then used to perform two-point parametric linkage analysis on the genotype data (Cottingham et al. 1993). Haplotypes of individuals with inferred genotypes were constructed under the assumption of minimal recombination.
Linkage analysis of the genomewide-scan data identified linkage of family 71 to marker D12S374 on chromosome 12, with a LOD score slightly greater than 3 at θ=0; no other LOD scores greater than 2 were found. Following the genome scan, seven additional markers over an 11-cM interval were genotyped, and the results confirmed linkage to a novel locus for HSA on chromosome 12p13 (table 1). The highest LOD score was obtained for markers D12S374 (4.8 at θ=0) and D12S93 (5.1 at θ=0). The locus for dentatorubral pallidoluysian atrophy (DRPLA) (MIM 125370) is in the vicinity of marker D12S374, but two individuals from each of the three families tested negative for expansions in the DRPLA gene (G.A.R., unpublished data).
Table 1.
LOD Scores for Chromosome 12 in Family 71
|
LOD Score at θ = |
||||||||
| Marker | Position(cM) | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D12S1725 | 9.52 | .909 | 1.007 | 1.190 | 1.223 | 1.031 | .667 | .241 |
| D12S314 | 11.37 | 1.439 | 1.450 | 1.455 | 1.401 | 1.174 | .846 | .420 |
| D12S93 | 12.60 | 5.092 | 5.026 | 4.729 | 4.294 | 3.265 | 2.057 | .766 |
| D12S374 | 14.23 | 4.884 | 4.819 | 4.521 | 4.085 | 3.057 | 1.852 | .575 |
| D12S1623 | 15.69 | −1.248 | −1.056 | −.676 | −.450 | −.229 | −.118 | −.051 |
| D12S397 | 17.72 | −4.083 | .903 | 1.834 | 2.070 | 1.820 | 1.178 | .396 |
| D12S1695 | 19.68 | −1.447 | 1.704 | 2.429 | 2.535 | 2.119 | 1.322 | .373 |
| D12S77 | 20.27 | −1.773 | 2.314 | 2.920 | 2.907 | 2.316 | 1.397 | .371 |
Once linkage of family 71 was confirmed, all markers were also tested in families 13 and 27. Family 13 showed highly significant linkage to the new locus, with a highest LOD score, 7.2, at θ=0 for markers D12S93 and D12S374 both, which are 1.6 cM apart (table 2). The highest LOD score for family 27 was 0.45 at θ=0 for marker D12S1623. The SLINK program of the LINKAGE software program was used to simulate linkage in family 27, and the highest LOD score attainable was predicted to be 1.18 at θ=0.01. Interestingly, none of the markers showed a negative LOD score, and, given the small number of samples, these results indicate suggestive linkage of this family to the new locus (table 3).
Table 2.
LOD Scores for Chromosome 12 in Family 13
|
LOD Score at θ = |
||||||||
| Marker | Position(cM) | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D12S1725 | 9.52 | 2.904 | 6.058 | 6.180 | 5.691 | 4.241 | 2.527 | .882 |
| D12S314 | 11.37 | −2.461 | 5.086 | 5.997 | 5.9142 | 4.948 | 3.500 | 1.776 |
| D12S93 | 12.60 | 7.293 | 7.142 | 6.526 | 5.736 | 4.104 | 2.447 | .905 |
| D12S374 | 14.23 | 7.223 | 7.324 | 7.325 | 6.9685 | 5.749 | 4.106 | 2.135 |
| D12S1623 | 15.69 | 1.107 | 1.265 | 1.489 | 1.545 | 1.374 | .981 | .458 |
| D12S397 | 17.72 | −4.342 | 1.562 | 2.398 | 2.384 | 2.261 | 1.553 | .713 |
| D12S1695 | 19.68 | 2.487 | 6.808 | 7.204 | 6.932 | 5.719 | 4.028 | 2.017 |
| D12S77 | 20.27 | .679 | 5.291 | 5.948 | 5.918 | 5.071 | 3.678 | 1.917 |
Table 3.
LOD Scores for Chromosome 12 in Family 27
|
LOD Score at θ = |
||||||||
| Marker | Position(cM) | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D12S1725 | 9.52 | .226 | .241 | .273 | .276 | .217 | .123 | .037 |
| D12S314 | 11.37 | .355 | .367 | .391 | .380 | .285 | .154 | .042 |
| D12S93 | 12.60 | .280 | .271 | .238 | .197 | .122 | .059 | .016 |
| D12S374 | 14.23 | .354 | .367 | .389 | .378 | .281 | .148 | .039 |
| D12S1623 | 15.69 | .448 | .437 | .391 | .333 | .217 | .110 | .030 |
| D12S397 | 17.72 | .259 | .273 | .303 | .301 | .230 | .126 | .036 |
| D12S1695 | 19.68 | .333 | .346 | .370 | .359 | .266 | .137 | .035 |
| D12S77 | 20.27 | .349 | .362 | .386 | .374 | .279 | .148 | .039 |
A disease haplotype was established for each family. In family 71, key recombinants (VI:4 and VI:13) determined the smallest critical interval to be 10 cM, flanked by markers D12S1685 and GATA151H05. The haplotype was constructed by following the order of markers available from the Human Genome Project Working Draft database. Haplotype analysis for the same critical markers in families 13 and 27 identified an identical disease haplotype over an 11-marker interval for both families. It is highly unlikely that two families share the same alleles over an 11-marker interval by chance. For family 13, the key recombinants V:14 and VI:11 determined the critical interval to be 8 cM, flanked by markers D12S1725 and D12S397.
When the disease haplotypes of the three families are overlapped, the smallest common disease haplotype is 5.1 cM, flanked by markers D12S93 and GATA151H05 (table 4). This interval contains the as-yet-unidentified disease gene, which has been assigned the symbol SAX1 by the HUGO Gene Nomenclature Committee. All affected individuals in each family inherited this identical segment of DNA. The shared haplotype in the three Newfoundland families suggests that they originate from a common ancestor in Newfoundland. It therefore is likely that a common mutation will be identified for these three families, which would facilitate a genetic-testing program in Newfoundland. However, there is currently no genealogical evidence that these families are related, and available family records will be further studied in search of a common ancestor. Furthermore, there are a few individuals (individuals VI:14 and VI:15 in family 13 and individual III:4 in family 27) who are presently unaffected but share part of or the entire 5.1-cM region. These individuals may be presymptomatic or asymptomatic, which can be explained by age-dependent or reduced penetrance of HSA.
Table 4.
SAX1 Locus on Chromosome 12p13 Determined by Key Recombinants and Overlapping of Haplotypes
|
Familya |
||||
| Marker | 71 | 13 | 27 | MinimalCommonlyInheritedSegment [Size]b |
| D12S1685 | 13 |
6 | 6 | |
| D12S1725 | 1 | 6 |
6 | |
| D12S314 | 5 | 8 | 8 | |
| D12S93 | 7 | 1 | 1 | |
| D12S374c | 5 | 5 | 5 | 5 [209 bp] |
| D12S1623 | 4 | 4 | 4 | 4 [127 bp] |
| DRPLA | 2 |
2 | 2 | 2 [180 bp] |
| GATA151H05 | 5 | 5 |
5 | |
| D12S397 | 3 | 3 | 3 | |
| D12S1695 | 6 | 6 | 6 | |
| D12S77 | 4 | 4 | 4 | |
| D12S89c | 8 | 8 | … | |
| D12S391 | 5 | 4 | … | |
The boxed regions in these columns indicate the minimal disease interval determined by key recombinants. Note that there were no key recombinants identified for family 27 in this interval.
The boxed region in this column represents the minimal commonly inherited disease haplotype of 5.1 cM, which was obtained by overlapping the critical disease intervals of each family. To standardize the results, the size of the marker alleles is provided.
All three families share part of their haplotypes from D12S374 to D21S89.
According to the Human Genome Project Working Draft sequence, the smallest disease haplotype, 5.1 cM, corresponds to ∼3.7 Mb on the physical map and contains three large unsequenced gaps. There are 40 known genes and 52 predicted genes in this interval. Additional polymorphic dinucleotide repeats can be identified using the Human Genome Project Working Draft database. Furthermore, single-nucleotide polymorphisms in the known genes can also be used to further refine the disease haplotype. Finally, the collection of affected family members—for family 27 in particular and possibly for new families—will continue, in an effort to identify more recombinations to further narrow the region. Meanwhile, interesting candidate genes will be screened, with priority placed on expression profiles and possible molecular function in disease pathogenesis.
HSA is clinically related to the hereditary spastic paraplegias (HSPs) and the spinocerebellar ataxias (SCAs) (Bouchard et al. 2000; Tallaksen et al. 2001). Lower-limb spasticity is marked in HSA but is also the hallmark of HSP. The main pathological feature is axonal degeneration of the terminal ends of the corticospinal tracts and the dorsal-column–pathway tracts (McDermott et al. 2000). At present, the defective proteins implicated in HSP pathogenesis are the L1 cell-adhesion molecule (L1CAM), which is involved in the development of the nervous system; the proteolipid protein (PLP), which is involved in myelin-sheet compaction and maintenance; and two AAA (ATPase with various cellular activities) family members, paraplegin and spastin. Paraplegin is a mitochondrial metalloprotease, and spastin is thought to act as a chaperone (Casari and Rugarli 2001). Recently, HSP-causing mutations have also been identified in a GTPase gene, which encodes atlastin, a protein thought to be involved in vesicle trafficking (Zhao et al. 2001).
SCAs are another group of clinically heterogeneous neurodegenerative disorders. This group is characterized by degeneration of the cerebellum, brain stem, and spinal cord (Harding 1993). Evidence suggests that trinucleotide-repeat expansions are the main pathogenic mechanism for autosomal dominant cerebellar ataxias, including DRPLA and Machado-Joseph disease (Nakamura et al. 2001; Tan and Ashizawa 2001), although the underlying cause of the cell degeneration observed remains unknown (Klockgether et al. 2000).
Genes homologous to any of the known HSP, SCA, or ARSACS genes; AAA family members; L1CAM; PLP; GTPases; and sacsin present in the 3.7-Mb interval will be investigated. Interestingly, DRPLA cannot be excluded as a candidate gene and will be considered for point-mutation screening. Anticipation has not been observed in any of the three families, which suggests that a DNA-repeat expansion is less likely to be responsible for SAX1. Other interesting candidates in the region are vesicle-associated–membrane protein 1 (VAMP1), a synaptobrevin involved in vesicle transport to the synapse (Archer et al. 1990), and γ-enolase (ENO2), a neuron-specific enzyme with increased expression during cell death (Craig et al. 1990; Lafon-Cazal et al. 1992).
In an effort to find the molecular cause of a phenotypically distinct clinical disorder (HSA), we identified the first locus for dominant HSA on chromosome 12p13. This suggests that, in HSA, there is locus heterogeneity with two identified loci—SAX1 and ARSACS. Pinpointing the disease gene would facilitate clinical diagnosis, which is often difficult in HSA. In addition, a genetic-testing program and improved clinical diagnosis in Newfoundland would be feasible with the evidence of a common ancestor. The characterization of the disease-causing gene and defective protein will provide more insight into the disease pathogenesis of this and other related neurodegenerative diseases, such as HSP, SCA, and amyotrophic lateral sclerosis.
Acknowledgments
We thank the families for their participation in the study and Dr. A. Toulouse for carefully reading the manuscript. We are also grateful for the assistance of Drs. T. Hudson, C. Brewer, and A. Verner, of the McGill University Genome Centre. This work was supported by the Canadian Institutes for Health Research. E.J.I. and K.K.G. were also supported by the Atlantic Canada Opportunities Agency and the Department of Health and Community Services of Newfoundland and Labrador.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Cooperative Human Linkage Center, The, http://lpg.nci.nih.gov/CHLC/
- Fondation Jean Dausset–CEPH, http://www.cephb.fr/
- Genome Database, The, http://www.gdb.org/
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for spastic ataxia [MIM 108600; MIM 270500], DRPLA [MIM 125370], and ARSACS [MIM 604490])
- UCSC Human Genome Project Working Draft (“Golden Path”), http://genome.ucsc.edu/
References
- Archer BTI, Ozcelik T, Jahn R, Francke U, Sudhof TC (1990) Structures and chromosomal localizations of two human genes encoding synaptobrevins 1 and 2. J Biol Chem 265:17267–17273 [PubMed] [Google Scholar]
- Bouchard JP, Richter A, Melancon SB, Mathieu J, Michaud J (2000) Autosomal recessive spastic ataxia (Charlevoix-Saguenay). In: Klockgether T (ed) Handbook of ataxia disorders. Marcel Dekker, New York, pp 312–323 [Google Scholar]
- Casari G, Rugarli E (2001) Molecular basis of inherited spastic paraplegias. Curr Opin Genet Dev 11:336–342 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Craig SP, Day INM, Thompson RJ, Craig IW (1990) Localisation of neurone-specific enolase (ENO2) to 12p13. Cytogenet Cell Genet 54:71–73 [DOI] [PubMed] [Google Scholar]
- De Braekeleer M, Giasson F, Mathieu J, Roy M, Bouchard JP, Morgan K (1993) Genetic epidemiology of autosomal recessive spastic ataxia of Charlevoix-Saguenay in northeastern Quebec. Genet Epidemiol 17–25 [DOI] [PubMed] [Google Scholar]
- Engert JC, Berube P, Mercier J, Dore C, Lepage P, Ge B, Bouchard J-P, Mathieu J, Melancon SB, Schalling M, Lander ES, Morgan K, Hudson TJ, Richter A (2000) ARSACS, a spastic ataxia common in northeastern Quebec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat Genet 24:120–125 [DOI] [PubMed] [Google Scholar]
- Grewal KK, Stefanelli MG, Meijer IA, Hand CK, Rouleau GA, Ives EJ (2001) Evidence for a common ancestor in two large families with phenotypically variable spastic ataxia. Paper presented at the annual meeting of The American Society of Human Genetics. San Diego, October 12–16 [Google Scholar]
- Harding AE (1993) Clinical features and classifications of inherited ataxias. Adv Neurol 61:1–14 [PubMed] [Google Scholar]
- Klockgether T, Wullner U, Spauschus A, Evert B (2000) The molecular biology of the autosomal dominant cerebellar ataxias. Mov Disord 15:604–612 [DOI] [PubMed] [Google Scholar]
- Lafon-Cazal M, Bougault I, Steinberg R, Pin J, Bockaert J (1992) Measurement of gamma-enolase release, a new method for selective quantification of neurotoxicity independently from glial lysis. Brain Res 593:63–68 [DOI] [PubMed] [Google Scholar]
- Mahloudji M (1963) Hereditary spastic ataxia simulating disseminated sclerosis. J Neurol Neurosurg Psychiatry 26:511–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott CJ, White K, Bushby K, Shaw PJ (2000) Hereditary spastic paraparesis: a review of new developments. J Neurol Neurosurg Psychiatry 69:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Jeong S, Uchihara T, Anno M, Nagashima K, Nagashima T, Ikeda S, Tsuji S, Kanazawa I (2001) SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet 10:1441–1448 [DOI] [PubMed] [Google Scholar]
- Tallaksen CM, Durr A, Brice A (2001) Recent advances in hereditary spastic paraplegia. Curr Opin Neurol 14:457–463 [DOI] [PubMed] [Google Scholar]
- Tan E, Ashizawa T (2001) Genetic testing in spinocerebellar ataxias. Arch Neurol 58:191–195 [DOI] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L, Bui M, Fink JK (2001) Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 29:326–331 [DOI] [PubMed] [Google Scholar]