Abstract
In this unit, we describe a set of improvements we have made to the standard Illumina protocols to make the sequencing process more reliable in a high-throughput environment, reduce amplification bias, narrow the distribution of insert sizes, and reliably obtain high yields of data.
Keywords: Illumina, Next-Generation, Sequencer, Protocols
INTRODUCTION
Knowledge of the DNA sequence of an organism is the key to understanding how that organism functions. With it, we can define characteristics of genomes, and delineate differences between them, which, in turn, help us to understand genotype/phenotype relationships (Bentley et al., 2008; Mardis, 2008).
In the mid 1970s, several methods of sequencing DNA appeared around the same time (e.g., (Maxam and Gilbert, 1977; Sanger and Coulson, 1975), but it was dideoxy DNA sequencing (Sanger et al., 1977) that proved to be the most versatile and practical approach. Over the following decades, dideoxy sequencing continued to be developed, and thirty years later, it is still used widely as the standard sequencing technology in many laboratories. The drawback of the method is that its throughput is limited, as sequencing is performed on single isolated templates, which means that large-scale sequencing projects are expensive and laborious, requiring ligation of target DNAs into cloning vectors, and amplification in Escherichia coli. Consequently, the human genome sequence (International Genome Sequencing Consortium, 2004), which was generated entirely by capillary sequencing using dideoxy chemistry, took hundreds of sequencing machines several years and the final sequencing phase cost ~300 million US dollars.
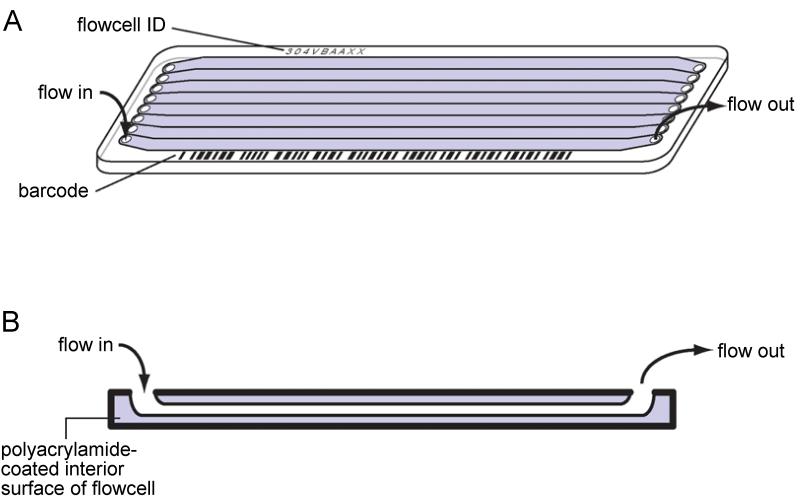
Currently, the Illumina SBS approach is the dominant sequencing technology. Here, molecules of DNA are hybridized to oligonucleotides that are attached to the polymer-coated glass surface of a flowcell (Figure 18.2.1A). Templates are amplified by flowing enzymes and reagents through the channels of the flowcell (Figure 18.2.1B). Once amplified, these molecules form clusters of amplicons, each of which is derived from a single template molecule. Clusters are then used as templates for sequencing-by-synthesis using fluorescent reversible-terminator deoxyribonucleotides (Bentley et al., 2008). This sequencing system has become dominant due to the high yield of high quality (Q30) total sequence obtained (several hundreds of gigabases per 8 lane flowcell on a HiSeq 2000), and the relatively low average cost per base ($41 per gigabase for large data sets (Quail et al., 2012c)).
Figure 18.2.1. Illumina flowcell.
(A) Flowcells are hollow glass slides, with 8 separate lanes, through which reagents and template DNA flow. Lanes have been shaded gray for clarity. (B) Cross-section of a single lane, showing the direction of reagent flow and polyacrylamide coating on the interior surface of the flowcell.
This dominance is, however, quite a recent thing. In the last 5 years, we have seen the rise and fall of alternative sequencing technologies, some of which are still quite frequently used today. For instance in 2005, the first of the next-generation DNA sequencers, 454’s GS20 (now Roche 454), became commercially available (Margulies et al., 2005). This method revolutionized the paradigm of DNA sequencing. Instead of a single sequencing reaction generating a single sequence, the 454 introduced massively parallel sequencing. The Roche 454 technology uses emulsion PCR to generate beads coated in amplicons that are derived from single template molecules. About a million of these beads are then sequenced in parallel by pyrosequencing (Ronaghi et al., 1998). Images of the beads are analyzed to generate high-quality sequences. In this way, throughput is increased, cost reduced and cloning avoided. This gave rise to megabases of sequence data per run, compared to <100 kb for a 96-well capillary machine. With evolutions of this technology, the maximum read length obtained from this platform increased to near 1000 bases, which is very similar to the maximum read length obtained from capillary sequencers.
Although long sequencing reads produced by the Roche 454 lend themselves particularly well to de novo assembly of smaller (e.g. bacterial) genomes, there are sequencing applications for which read length is of lesser importance than total output and cost per gigabase. For example, within-species genetic variation can be identified by mapping sequence reads to a reference sequence and identifying positions that differ. The availability of high-quality reference sequences for many organisms allows such resequencing experiments to be performed with relatively short-read (75, 150 or 250 base pair) sequences.
Furthermore with the maturation of the Illumina sequencing technology, a number of optimizations have been made that have increased the utility of this approach and helped establish its dominance e.g., introduction of longer (250 bp) read lengths, increasing yield, reliable and accurate size generation and selection methods for larger DNA fragments (e.g. Pippin Prep [www.sagescience.com]), new assemblers that use known insert size of paired-end libraries, and larger mate-pair libraries.
With increased yield the cost of sequencing per gigabase has dropped substantially and today most larger genomes have been sequenced in high quality. For some applications, however, e.g., small genomes and amplicon sequencing, high yield per run is of lesser importance. Instead smaller scale (personal, or bench-top) sequencers have emerged and have gained popularity. These bench-top sequencers combine relatively low start-up costs with fast turnaround time, while still yielding sufficient sequence data. The leading bench top sequencers currently available are Life Technologies Ion Personal Genome Machine (PGM) and Ion Proton; and Illumina’s MiSeq sequencers, each of which is capable of producing one to a few gigabases of sequence per run.
Life Technologies semiconductor sequencing, as with the Roche 454, exploits emulsion PCR to clonally amplify single template molecules onto beads. The beads are then deposited into millions of wells of a semiconductor chip. These beads are then used as templates for sequencing-by-synthesis in which separate nucleotides get flowed over the chip’s surface. This induces detectable pH changes and millions of beads are sequenced in parallel.
Illumina’s MiSeq is similar to the larger Illumina sequencer systems (e.g. Genome Analyzer and HiSeq), except that its flow cells have only one lane and therefore can run only one (potentially multiplexed) library at a time. Fortunately, these libraries are fully compatible to run on all Illumina sequencers, which makes the MiSeq suitable for large genome centers to quality check difficult templates, or quantify, independent libraries for accurate multiplexing before running on HiSeq.
Next-Generation DNA sequencing technology has enabled us to design genome-wide and ultra-deep sequencing projects that, because of their enormity, would not otherwise be possible. We are approaching a situation where whole-genome sequencing of complex organisms will be routine, allowing us to gain a deeper understanding of the full spectrum of genetic variation and to define its role in phenotypic variation and the pathogenesis of complex traits.
In this unit, we describe in detail the molecular biology underpinning sequencing on the Illumina platform. We also describe a set of improvements we have made to the standard Illumina protocols to make the library preparation more reliable in a high-throughput environment, to reduce bias, tighten the distribution of fragment sizes, and to obtain high yields of data more reliably.
In order to achieve successful cluster amplification, sample DNA must undergo a multi-step library preparation procedure. We found several of the standard Illumina sequencing protocols could be improved upon, and by making modifications to their standard laboratory pipeline, we have made our sequencing output more robust and reproducible, and have also increased the output of our sequencing runs (Quail et al., 2008). The preparation steps we now take are described below, and references to the updated protocols describing the optimized preparation steps are given.
Fragmentation
Cluster amplification is a relatively inefficient process, and there is an upper limit to the size of fragments that will amplify on the flowcell surface. Standard Illumina sequencing libraries currently tend to have a fragment size of 200 to 500 bp, excluding adapters (see Basic Protocol 1). Larger fragments up to 1000 bp will cluster, but with increasingly lower efficiency and lower yield.
End-repair
Random fragmentation produces double-stranded DNA with a mixture of blunt ends, recessed 3′ ends and recessed 5′ ends, with and without a 5′ phosphate moiety. These must be made uniform before adapters can be ligated, and so a mixture of enzymes is used to generate blunt-ended fragments with phosphorylated 5′ termini (Basic Protocol 3).
A-tailing
Addition of a single A nucleotide to the 3′ ends of fragments before adapter ligation deters concatemerization of templates and because the adapters are t-tailed, it increases the efficiency of adapter ligation (Basic Protocol 3).
Adapter ligation
Template strands must receive a different adapter sequence at either end to participate successfully in the cluster amplification and sequencing reactions. Adapter ligation to templates must be as efficient as possible, but at the same time, ligation of adapters to one another must be suppressed: adapter dimers will also generate clusters that can be sequenced, and will reduce the total proportion of desired sequence obtained from a run (see Basic Protocol 3).
Clean up
Clean up can be done using columns (e.g. QIAquick PCR Purification columns [Qiagen] or DNA Clean & Concentrator Columns [Zymo Research]) or using solid-phase reversible immobilization (SPRI) beads (DeAngelis et al., 1995), which use a carboxyl-coated magnetic particle that can reversibly bind DNA in the presence of polyethylene glycol (PEG) and salt. For high throughput library preparation, SPRI bead clean up up (using AMPure XP SPRI beads (Beckman Coulter, Inc.)), is preferred as it is more scalable than column-based clean up. An extra benefit of using AMPure beads is that when used after ligation, they allow the majority of adapter dimers to be removed from the library (see Alternate Protocol 1).
Size selection
Binding to solid-phase reversible immobilization (SPRI) beads (DeAngelis et al., 1995) can be controlled by altering the PEG/NaCl concentration (Borgstrom et al., 2011; Lundin et al., 2010). A single AMPure XP treatment can be used to remove DNA fragments below a certain size (typically 150-200bp). A double (upper and lower) size selection can be performed by two consecutive AMPure XP steps. First a low concentration of AMPure XP beads is added to the sample to bind larger DNA fragments. In this step the beads containing the larger fragments are discarded. More beads are then added to the supernatant, increasing the amount of PEG and NaCl, so smaller fragment sizes will be bound. Next the supernatant is discarded and the beads are washed and intermediate fragments are eluted. Depending on the concentrations of PEG and NaCl in the first and final SPRI step distinct size ranges can be generated (Borgstrom et al., 2011; Lundin et al., 2010). In Figure 18.2.2 examples are given. With increased SPRI bead to DNA ratios (1.5× to 2.5× Figure 18.2.2A) increasingly more smaller fragments from the original sheared sample are captured. Decreasing the ratio to 1.0× or lower decreases the amount of small fragments captured (see Figure 18.2.2B; Alternate Protocol 2).
Figure 18.2.2. Effect of AMPure XP ratios on fragment size selection.
1 μg of DNA was sheared to giver fragments from 20 to 400bp (average 160bp). Next the DNA was incubated with different AMPure ratios. The size distribution of fragments after AMPure clean up were analysed by electrophoresis using an Agilent Bioanalyzer High Sensitivity DNA chip. (A) AMPure bead to DNA ratios varying from 2.5× beads to DNA up to 1.0× beads to DNA (B) AMPure bead to DNA ratios varying from 1.5× beads to DNA up to 0.6× beads to DNA.
Sizing can also be modulated by changing conditions used for shearing, which is an effective method to increase the amount of fragments in the desired size range. In combination with SPRI/AMPure bead size selection, this method is both effective and amenable to automation but still produces quite a broad range of fragment sizes suitable for most applications. In cases, however where a very precise/narrow size distribution is necessary, we recommend using Sage Science’s Pippin Prep (Quail et al., 2012a; Figure 18.2.3; Alternate Protocol 3).
Figure 18.2.3.
Post-sequencing mapped insert-size distribution graphs for Illumina sequencing libraries prepared from genomic DNA with size-selection using; (A) agarose gel electrophoresis, (B) AMPure XP, (C) Caliper Labchip XT and (D) Sage Science Pippin Prep.
PCR
Following size selection and clean up, libraries are amplified by PCR (1) to enrich for properly ligated template strands—those that have an adapter at both ends, (2) to increase the amount of library available for sequencing (3) to generate enough DNA for accurate quantification, and (4) to add oligonucleotide sequences to the template strands that allow hybridization to the flowcell surface, since these sequences are not contained in the ligated adapter (see Basic Protocol 4). For direct sequencing of short amplicons, see Alternate Protocol 4. For production of non-PCR libraries see Alternate Protocol 6.
Quantification
The number of clusters per lane of a flowcell is governed by the concentration of library that is added. Accurate quantification is vital, because too low a cluster density reduces the yield of data, and therefore increases the per-base cost of sequencing, whereas too high a cluster density results in a reduced yield of data due to cluster overlap (see Basic Protocol 5).
Denaturation
Libraries are rendered single stranded by incubation with sodium hydroxide, to allow efficient hybridization of the template strands to primers attached to the flowcell surface (see Basic Protocol 6).
After the denaturation, the samples are loaded on the cBot (for GAII or HiSeq 8 lane flowcells) or directly loaded onto the sequencer (MiSeq and HiSeq 2500 [fast flowcells only]). The subsequent steps are processsed by the sequencer or on the cBot. Except for the amplification quality control step (Support Protocol 2) all steps are covered by the manufacturer’s manual. Subsequent paragraphs are however crucial to understanding Illumina’s sequencing process.
Hybridization and extension
As a result of the PCR reaction, template strands possess a sequence at one end that matches exactly one of the flowcell primers, and at their opposite end they possess a sequence that is in the reverse complementary orientation to the other flowcell primer. Thus, only the complementary end of each template strand hybridizes to a flowcell primer. Flowcell primers are extended, by Phusion DNA polymerase (Thermo Scientific), generating a reverse complementary copy of the original template strand that is tethered to the flowcell surface. The original template strand is not tethered, and is removed by flushing with sodium hydroxide (Figure 18.2.4A).
Figure 18.2.4.
(A) Template hybridization, extension, and denaturation on the flowcell surface. Templates are prepared so as to possess tails that are complementary to primers on the flowcell surface. This allows one end of a template strand to hybridize to a flowcell primer. Flowcell primers are extended by Phusion DNA polymerase (Thermo Scientific), resulting in a reverse complementary copy of the original template strand, which is covalently attached to the flowcell surface. The original template strand is then removed by flushing 0.1 M NaOH though the flowcell. (B) Cluster amplification. The free end of the tethered reverse complementary copy of the original template strand can anneal to the other type of flowcell primer, forming a bridge. The flowcell primer is extended by Bst polymerase, in an isothermal reaction, which generates a double-stranded product. Formamide is used to denature these strands, which can then anneal to other primers on the flowcell surface, which extend in the next cycle. In this way, repeated cycles of extension and denaturation result in a cluster of strands, all of which are derived from a single template strand.
Cluster amplification
The free end of each newly generated tethered strand is complementary to the other primer on the flowcell surface, and can hybridize to it. This primer is extended, using Bst polymerase, generating a double-stranded product. The strands of this product are denatured by formamide, producing two single strands, the free end of each can anneal to another primer on the flowcell surface (Figure 18.2.4B). Repeated cycles of formamide denaturation, annealing, and extension result in a cluster of ~1000 strands, each of which derived from the same original template strand, and hence are clonal.
Linearization, blocking, and hybridization
As a consequence of the structure of the adapters used for the ligation step, ligation junctions have around 12 identical nucleotides at opposite ends of the fragments. Therefore, to ensure that each cluster is sequenced only in one direction during each read, and to allow more efficient hybridization of sequencing primers to strands within a cluster, one of the flowcell primers is cleaved, removing one strand selectively, and resulting in single-stranded clusters. To reduce background noise in the sequencing reaction, 3′ ends of both extended and unextended flowcell primers are blocked by incorporation of dideoxyribonucleotides, and sequencing primers are hybridized.
Sequencing-by-synthesis
Hybridized flowcells undergo repeated cycles of nucleotide incorporation, imaging, and cleavage (simultaneous deblocking of the 3′ end and removal of the fluorophore).
BASIC PROTOCOL 1
FRAGMENTATION
The first stage in a standard genomic DNA library preparation for Illumina libraries is fragmentation of DNA. There are various methods in use by which this can be achieved:
Nebulization
The initial method recommended by Illumina was nebulization with compressed nitrogen. During nebulization, approximately half of the original DNA sample is lost through vaporization, and only 50% of the original DNA will be available for subsequent library generation. An additional drawback of nebulization is that it is not possible to shift the peak of fragment sizes much further towards the smaller end, even if more extreme conditions are used. This means very little of the nebulized sample is within the desired size range.
Enzymatic shearing
An alternative approach is enzymatic shearing. Various enzymatic shearing kits are commercially available. Whilst these can be useful, the activity of these enzymes is dependent on a number of factors which make it impossible to have a defined set of reaction conditions that will work effectively on a range of samples from different sources. The degree of fragmentation is dependent on the DNA to enzyme ratio; therefore, varied amounts of input DNA can have a considerable effect on shearing size. Shearing size is also affected by GC content of DNA, and finally these enzymes can be inhibited by residual carryover (e.g., from DNA isolation steps), therefore cleanliness of the DNA is important.
Sonication
A third alternative is sonication. With sonication, it is possible to produce fragments with a smaller average size. Nevertheless, standard sonication still produces a relatively wide range of fragment sizes, so a large proportion of the fragmented DNA is wasted. In addition, it exposes the sample to a large amount of energy, a lot of which is converted into heat, which can lead to strand dissociation, DNA damage and bias.
Acoustic shearing
Acoustic shearing devices specifically designed for next generation sequencing have recently come to market (e.g. Epigentec’s EpiSonic Multi-Functional Bioprocessor or Diagenode’s Bioruptor). These give more consistent results than conventional sonicators and can shear DNA to tighter insert ranges. They do, however, still produce some heat, which, if not controlled by chilling, may cause DNA damage and bias.
We now routinely fragment all of our DNA samples using Covaris’ Adaptive Focused Acoustics technology (AFA), which focuses acoustic energy controllably into the aqueous DNA sample. The peak of fragment sizes can be tuned to below 400 bp, so a greater proportion of the DNA sample will contribute to the final library. Moreover, a lower proportion of the sample is lost as the sample is confined within a sealed microtube.
Materials
DNA sample
Covaris S2 or equivalent with chiller unit
Life Technologies Qubit Fluorometer (or other fluorometric based method, e.g. SYBR green)
6-mm × 16-mm AFA fiber vials (Covaris, cat. no. 520045)
Crimp caps (Covaris, cat no. 520028)
Allow the Covaris chiller to reach 4°C, and degas the water for at least 30 min.
- During this time, prepare the DNA sample:
- Obtain concentration using the Qubit Fluorometer
-
Dilute at least 500ng of DNA to 100 μl with EB buffer and transfer the DNA sample to a Covaris vial. Attach crimp cap by crimping.The aluminum lids perform better than the supplied plastic ones, as the tighter fit ensures that less acoustic energy is lost due to vibration.
-
Insert the sample vial into the holder, and for mean fragment sizes of 250 bp, run the Covaris with the settings:
Duty cycle 20% Intensity 55 Cycles per burst 200 Time 60 sec Proceed to DNA clean up (Basic Protocol 2).
BASIC PROTOCOL 2
DNA CLEAN UP USING COLUMNS (LOW THROUGHPUT)
After the DNA is sheared, a clean up step is performed to concentrate the sample and to remove the buffer in which the genomic DNA was originally suspended. Clean up can be done using columns (Basic Protocol 2) or SPRI beads (Alternate Protocol 1). When doing manual library preparation it is sometimes easier to use columns (e.g. DNA Clean & Concentrator Columns [Zymo Research] or QIAquick PCR purification columns [Qiagen]). In addition to general clean up, it is also possible to perform a rough size selection of the sheared DNA using AMPure XP beads. Use of a 1× AMPure cleanup after shearing, end-repair and A-tailing will remove all inserts below 150-200 bp approximately, such that if the DNA sample is sheared to an average size of 200bp the insert sizes in the final library will be mostly between 200 and 300 bp (see Alternate Protocol 1). If DNA is sheared to larger fragment sizes, the range of fragment sizes broadens and some fragments will be generated that will be too large for efficient clustering. In such a situation it can be useful to do a double SPRI bead selection (Alternate Protocol 2). For more accurate size selection, do gel size selection (Alternate Protocol 3) or preferably use Sage Science’s Pippin Prep (Figure 18.2.3).
Materials
- Qiagen QIAquick PCR purification kit (cat. no. 28104) containing:
- PB buffer
- PE buffer
- EB buffer
- Columns
-
Add 500-600 μl of PB buffer to the sheared DNA. The volume of added PB buffer should be at least 5× the volume of the initial sample.
In this case 500 μl of PB buffer should be enough, but higher amounts of PB do not seem to decrease yield. Since the column can only accommodate 750 μl, try not to exceed a total volume of 750 μl, else step 2 will have to be repeated until all sample is loaded.
Transfer the PB-DNA mix to a Qiagen QIAquick PCR cleanup column and spin down at 13,000 g for 1 minute. Discard flow through.
Pipette 750 μl PE buffer (containing ethanol) to the column and spin down at 13,000 g for 1 minute. Discard flow through.
Spin down the empty column for an additional minute at 13,000 g to dry the membrane.
Transfer the column to a new 1.5 ml eppendorf tube and add 30 μl of EB to the center of the column without touching the membrane and let it incubate at room temperature for at least 1 minute, but no more than 5 minutes.
Spin down the column at 13,000 g for 1 minute. Collect flow through. This contains the cleaned up DNA ready for further processing
Proceed to template preparation (Basic Protocol 3).
ALTERNATE PROTOCOL 1
DNA CLEAN UP USING AMPURE XP BEADS (AUTOMATION FRIENDLY)
For library preparation using a pipetting robot, the use of AMPure XP beads is preferred to columns. In addition to being automation-friendly, these beads make it possible to perform a rough size selection of the sheared DNA. For instance, the use of a 1× AMPure cleanup after shearing, end-repair and A-tailing will remove all inserts below 150-200 bp, such that if the DNA sample is sheared to an average size of 200bp the insert sizes in the final library will be mostly between 200 and 300 bp. If the DNA is sheared to larger fragment sizes, some fragments will be generated that will be too large for efficient clustering. In this case, it can be useful to do a double SPRI bead selection to remove these larger fragments (see Alternate Protocol 2).
Materials
Agencourt AMPure XP (Beckman Coulter Genomics: A63881)
80 % Ethanol
Magnetic stand, e.g., DynaMag Spin Magnet (Life Technologies cat no: 12320D)
Qiagen EB buffer
Let the AMPure XP beads come to room temperature before the experiment
Mix the beads well so that they appear homogeneous and consistent in color.
Add an equal volume of AMPure beads (e.g. 100 μl of AMPure XP to 100 μl of sheared DNA) to a new 1.5 ml Eppendorf tube.
Pipette DNA sample into this tube (e.g. 100 μl of sheared DNA). Mix well on a vortex mixer. Briefly centrifuge for 2 seconds to spin down residues on the side of the tube.
Incubate the DNA with the beads for 5 minutes.
Put the tube in the magnetic stand and wait for the solution to clear (~3 minutes).
-
Remove the supernatant from the tube and add 500 μl of 80% ethanol without disturbing the pellet.
The original AMPure protocol recommended use of a 70% ethanol wash solution. This needed to be freshly made, as over time it becomes more dilute since it absorbs atmospheric water and ethanol evaporates. When you rinse with more dilute ethanol more DNA gets washed away. We have found that this can be avoided by using an 80% ethanol solution instead. This washes just as well but does not need to be freshly prepared each time.
Repeat step 7.
Remove all ethanol, including residual droplets and dry the samples for 5 minutes in the magnetic stand on the bench with the tube lids open. Do not dry longer than 5 minutes. The beads get too dry and yield decreases.
Add 30 μl EB buffer, mix well on a vortex mixer, and incubate for 5 minutes at room temperature.
Put the tube in the magnetic stand and leave for 2 to 3 minutes.
Remove 30 μl of the supernatant to a fresh 1.5-ml tube. (Take care not to pipette any beads)
Proceed to template preparation (Basic Protocol 3).
ALTERNATE PROTOCOL 2
AMPURE BEAD DOUBLE DNA SIZE SELECTION
If fragments produced in Basic Protocol 1 are of larger sizes (400 bp and above), the library generally also contains a broader range of larger fragments. Since larger fragments do not cluster very well and therefore interfere with accurate cluster density prediction, it may be useful to perform a double size selection, after the fragmentation (to narrow insert-size distribution). In this way, larger fragments will be removed. For the majority of sequencing applications, however, with size ranges around 250 bp, this additional step is unnecessary, and we perform a single-size selection step (see Alternate Protocol 1).
The following method is designed to yield a library with the majority of inserts in the 400-600 bp size range with an average size of 500 bp. Other size ranges can be obtained by adjusting AMPure bead ratios.
Materials
Agencourt AMPure XP (Beckman Coulter Genomics: A63881)
80 % Ethanol
Magnetic stand e.g. DynaMag Spin Magnet (Life Technologies cat no: 12320D)
Qiagen EB buffer
Let the AMPure XP beads come to room temperature before the experiment.
Mix the beads well, so that they appear homogeneous and consistent in color.
Add 0.65× (e.g. 65 μl of beads for 100 μl of sample) of AMPure XP beads to a new 1.5 ml Eppendorf tube.
Pipette DNA sample into this tube (e.g. all 100 μl of sheared DNA). Mix well on a vortex mixer. Briefly centrifuge for 2 seconds to spin down residues on the side of the tube.
Incubate the DNA with the beads for 5 minutes.
Put the tube in the magnetic stand and wait for the solution to clear (~3 minutes)
Transfer the supernatant into a fresh tube and discard the tube containing the beads.
Add 0.12× (e.g. 12 μl of beads for an original 100 μl of DNA sample) of AMPure XP beads to the supernatant. Mix well on a vortex mixer. Briefly centrifuge for 2 seconds to spin down residues on the side of the tube.
Incubate for 5 minutes.
Put the tube in the magnetic stand and wait for the solution to clear (~3 minutes)
Remove the supernatant from the tube and add 500 μl of 80% ethanol without disturbing the pellet.
Repeat step 11.
Remove all ethanol, including residual droplets and dry the samples for 5 minutes in the magnetic stand on the bench with the tube lids open. Do not dry longer than 5 minutes. The beads get too dry and yield decreases.
Add 30 μl EB buffer, mix well on a vortex mixer, and incubate for 5 minutes at room temperature.
Put the tube in the magnetic stand and leave for 2 to 3 minutes.
Remove 30 μl of the supernatant to a fresh 1.5-ml tube. (Take care not to pipette any beads)
Proceed to template preparation (Basic Protocol 3).
ALTERNATE PROTOCOL 3
GEL SIZE SELECTION
For certain sequencing applications, such as surveying for rare translocation events, the frequency of chimeric templates must be kept to a minimum. In such cases, we perform a double size selection to make the insert size distinct, after fragmentation and after PCR. Please note that because of this stringent size selection step, much of the sheared DNA will be lost and more starting DNA must be used to achieve reasonable yields.
We generally use the Pippin Prep electrophoresis platform for this application. It is however possible to perform this manually using an agarose gel (see below). Using double size selection any chimeric templates that do form during the ligation step will be far larger than the desired fragments and will be removed by the second size selection step. For the majority of sequencing applications, however, a low frequency of chimeras is tolerable, and, in these cases, we perform a single-size selection step (see Alternate Protocol 1), after adapter ligation to get rid of adapter dimers.
Materials
2% Ultra-pure agarose (Life Technologies, cat. no. 16500100)
5× TBE buffer (Severn Biotech, cat. no. 20-6005-10)
10 mg/ml ethidium bromide solution (Sigma, cat. no. E1510)
5× loading dye (e.g. Qiagen, cat. no. 239901)
Low-molecular-weight size standard ladder (e.g. New England Biolabs, cat. no. N3233S)
- Qiagen QIAquick gel extraction kit (cat. no. 28706) containing:
- Chaotropic buffer (QG buffer)
- PE buffer
- EB buffer
- Spin columns
Dark reader (Clare Chemical Research, cat. no. DR46)
Scalpel or razor blade
Perform size selection
Prepare a 2% Ultra-pure agarose gel in 1× TBE buffer, containing 0.4 μg/ml ethidium bromide (we typically use gels that are 12- to 15-cm long).
-
Following shearing, the DNA volume is usually 100 μl. Mix 100 μl DNA with 25 μl 5× loading dye and run in the gel alongside a 100-bp-size standard ladder, in 1× TBE containing 0.4 μg/ml ethidium bromide at 6 V/cm, for ~2 hours (this time may need to be adjusted depending upon the length of the gel tank), until the yellow dye is close to the bottom of the gel.
If loading samples into a gel that is submerged in TBE buffer, run one sample per gel to remove the risk of cross-contamination.
Alternatively, it is possible to fill the gel tank so that the buffer level is just below the upper surface of the gel, and to load multiple samples. Always exercise due care when loading samples to avoid spillover. After sample loading, top up all wells with buffer (including empty wells), being careful not to overfill, and run the gel for 10 min, so that the samples enter the gel. Then remove the lid of the gel tank, add sufficient 1× TBE buffer (also containing 0.4 μg/ml ethidium bromide) to submerge the gel completely, and run as normal.
-
Visualize the gel on a dark reader.
Ultraviolet light should not be used, as this can damage the DNA. It is helpful for visualization to make the room as dark as possible.
-
Using a clean scalpel or razor blade, cut a 2-mm high gel slice, corresponding to the desired range of fragment sizes. Take care to cut horizontally so as not to increase the size ranges of fragments.
When using this protocol for a size selection after PCR, increase size by an additional 90 bp to allow for the length of the ligated adapters.
Using a Qiagen Gel Extraction kit, dissolve gel slices in QG buffer at room temperature, rather than by heating. This typically takes 10 to 20 min with frequent mixing.
Transfer the QG-DNA mix to a Qiagen column and spin down at 13,000 g for 1 minute. Discard flow through.
Pipette 750 μl PE buffer (containing ethanol) onto the column and spin down at 13,000 g for 1 minute. Discard flow through.
Spin down the empty column for an additional 1 minute at 13,000 g to dry the membrane.
Transfer the column to a new 1.5 ml eppendorf tube and add 30 μl of EB to the center of the column without touching the membrane and let it incubate at room temperature for at least 1 minute.
Spin down the column at 13,000 g for 1 minute. Collect flow through. This contains the cleaned up DNA ready for further processing.
Proceed to preparation of the adapter-ligated DNA library (Basic Protocol 3).
BASIC PROTOCOL 3
PREPARATION OF THE ADAPTER-LIGATED DNA LIBRARY
Fragmentation generates templates with a mixture of blunt ends and 5′ and 3′ overhangs. So that adapters can be ligated, end repair and A-tailing reactions must be performed, following the manufacturer’s protocols. For certain sequencing applications, such as surveying for rare translocation events, the frequency of chimeric templates must be kept to a minimum. In such cases, we perform a double size selection, both after the fragmentation and after PCR using Pippin Prep or gel size selection (see Alternate Protocol 3). In this way, any chimeric templates that do form in the ligation step will be far larger than the desired fragments and will be removed by the second size selection step after PCR. For the majority of sequencing applications, however a low frequency of chimeras is tolerable.
Materials
- Purified fragmented DNA sample (see Basic Protocol 2 and Alternate Protocols 1-3)
- NEBnext Illumina Library prep kit; (New England Biolabs, cat. E6040S)
- 10× T4 DNA ligase buffer (+10 mM ATP)
- 10 mM dNTP mix
- T4 DNA polymerase
- Klenow DNA polymerase
- T4 polynucleotide kinase
- 10× Klenow buffer
- dATP
- Klenow DNA polymerase (3′→;5′ exo−)
- 2× Quick ligation reaction buffer
- Quick T4 DNA ligase
- Paired-end adapters:
-
PE_top_adapter primer:5′ ACACTCTTTCCCTACACGACGCTCTTCCGATC*T (*indicates phosphorothioate; (Bentley et al., 2008)
-
PE_bottom_adapter primer:5′ P-GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG (P-indicates phosphate; (Bentley et al., 2008)
-
- Agencourt AMPure XP (Beckman Coulter Genomics: A63881) or Qiagen QIAquick PCR purification kit (cat. no. 28104) containing:
- PB buffer
- PE buffer
- EB buffer
- Column
Perform end repair
-
Prepare the following reaction mix:
Purified fragmented DNA sample
(Basic Protocol 2 and Alternate Protocols 1-3)30 μl Water 45 μl 10× T4 DNA ligase buffer (+ 10 mM ATP) 10 μl 10 mM dNTP mix 4 μl T4 DNA polymerase 5 μl Klenow DNA polymerase 1 μl T4 polynucleotide kinase 5 μl Total volume = 100 μl Mix well and spin down. Incubate 30 min at room temperature.
-
3a.
Purify with a Qiagen QIAquick PCR purification kit, (see Basic Protocol 2) and elute in 32 μl of EB buffer.
-
or 3b.
Purify with 1× (100 μl) AMPure XP beads (see Alternate Protocol 1) and elute in 32 μl of EB buffer.
Perform A-tailing
-
4.Prepare the following reaction mix:
End-repaired DNA sample (from step 3) 32 μl 10× Klenow buffer 5 μl dATP 10 μl Klenow exonuclease (3′→5′ exo−) 3 μl Total volume = 50 μl Mix well and spin down. -
5.
Incubate 30 min at 37°C.
-
6a.
Purify using a QIAquick MinElute column, (see Basic Protocol 2, but use MinElute columns instead) and elute in 19 μl of EB buffer.
-
or 6b.
Purify with 1× (50 μl) AMPure XP beads (see Alternate Protocol 1) and elute in 19 μl of EB buffer.
-
7.
Save 1 μl of sample to run 1/10 diluted on an Agilent High Sensitivity DNA chip (see Support Protocol 1).
This is useful because this product can be compared to adapter ligated DNA to see whether the adapter ligation was successful.
Perform adapter ligation
-
8.Prepare the following reaction mix:
End-repaired, A-tailed DNA sample
(from step 6a or 6b)18 μl 2× Quick ligation reaction buffer 25 μl Paired-end adapters (at 10 μM dilution) 2 μl Please note, if starting with <100 ng of DNA use 2 μl of 0.5
μM paired-end adapters insteadLigase 5 μl Total volume = 50 μl Mix well and spin down. -
9.
Incubate 30 min at room temperature.
-
10a.
Purify on a QIAquick column, (see Basic Protocol 2), and elute in 30 μl of EB buffer. Proceed to PCR amplification (Basic Protocol 4).
-
or 10b.
Purify with 1× (50 μl) AMPure XP beads (see Alternate Protocol 1) and elute in 50 μl of EB buffer.
This step is only a buffer exchange. Because the Quick ligation reaction buffer contains PEG it interferes with the AMPure XP clean up protocol, and more fragments will be collected than normal.
The following size selection step is necessary to eliminate adapter dimers.
-
11.
Purify with 0.8× (40 μl) AMPure XP beads (see Alternate Protocol 1) and elute in 30 μl of EB buffer. Proceed to PCR amplification (Basic Protocol 4).
SUPPORT PROTOCOL 1
VERIFICATION OF ADAPTER LIGATION OF THE LIBRARY
Since adapter ligation is quite a crucial step, we compare pre-adapter ligated DNA with post-adapter ligated DNA by running the samples in tandem on an Agilent Bioanalyzer DNA High Sensitivity DNA chip. Successful adapter-ligated DNA is approximately 130 base pairs larger than unligated DNA.
Materials
- Agilent High Sensitivity DNA kit (cat. no. 5067-46264626) containing:
- DNA dye concentrate (blue-capped vial)
- Gel matrix (red-capped vial)
- DNA marker (green-capped vial)
- DNA ladder (yellow-capped vial)
- Spin filters
- DNA chips
- Syringe
Adapter ligated DNA library (see Basic Protocol 3)
Vortexer (supplied with the Bioanalyzer)
Agilent Bioanalyzer 2100
Verify adapter-ligated DNA
To prepare the gel/dye mix, first allow dye concentrate (blue-capped vial) and gel matrix (red-capped vial) to reach room temperature (takes 30 min). After they reach room temperature vortex and spin down.
Pipette 15 μl of the blue dye concentrate into the gel matrix vial, cap the tube, vortex for 5 sec, spin down, and transfer the gel/dye to the supplied spin filter (supplied with the Agilent High Sensitivity DNA kit). Centrifuge the spin filter 10 min at 2240 × g ± 20% at room temperature.
Store at 4°C, away from direct light when not in use for more than 2 hr.
Position a new chip onto the priming station, and pipette 9 μl of room-temperature gel/dye mix into the well marked with white G on black background. Close the priming station; making sure that the syringe clip is in the lowermost position.
Press the plunger of the syringe until it is held by the clip and wait 60 sec.
Release the clip. If the plunger does not rise ~0.6 ml within 5 sec, this suggests that the seal has failed. If so, replace the seal and repeat step 5.
Slowly pull the plunger back to the 1.0 ml position and open the priming station.
Pipette 9 μl gel/dye mix into all three wells marked G.
Load 5 μl of marker (green-capped lid) into the well marked with the ladder symbol and each well that you are using to run a sample, and pipette 6 μl of marker into unused wells.
-
Load 1 μl of ladder into the ladder well and 1 μl of sample into each sample well. 11. Vortex on the supplied vortexer for 1 min and run the chip on the Agilent Bioanalyzer 2100.
To prevent excessive evaporation from the chip, the run should be started within 5 min after the vortexing. To quantify broad peaks, it is usually necessary to perform manual integration of the trace (right click on trace), to add a new peak and to extend the width of the new peak to include the area of interest.
BASIC PROTOCOL 4
PCR AMPLIFICATION OF THE LIBRARY
After the libraries have been shown to be adapter ligated we typically amplify the DNA with 8 to 18 cycles of PCR to increase the quantity of the library, enrich for fully ligated fragments, and tail fragments with the nucleotide sequences necessary for cluster amplification. Excessive PCR cycling should be avoided as it can lead to bias in the library and can result in unusual size profiles when libraries are analyzed, for example, on an Agilent Bioanalyzer chip. Over-amplification depletes the primer pool, resulting in an accumulation of non-extended single stranded template, which after denaturation anneals at the common adapter sequences forming bubble structures that migrate slower during electrophoresis. We would therefore recommend optimization of cycle number if starting a new project. Here we use Kapa HiFi polymerase as we have shown this to amplify complex fragment libraries with the least amount of bias (Quail et al., 2012b); Figure 18.2.5). Since not all samples will require a full flowcell lane’s worth of sequence, it is possible to barcode libraries with an index sequence during this PCR step. Whilst there are multiple strategies available for this we routinely use a set of 96 alternative PCR_R 5′ primers (see Appendix 1).
Figure 18.2.5. Genome browser screenshots of selected regions in four genomes demonstrating superior coverage when using kapa HiFi polymerase.
Genome browser screenshots of selected regions in the genomes of: (A) B. pertussis (GC-rich region); (B) S. pullorum; (C) S. aureus and (D) P. falciparum (AT-rich var gene region of chromosome 11). Libraries were prepared without PCR (green line), with 14 cycles of PCR using Phusion polymerase (blue line) and with 14 cycles of PCR using Kapa HiFi polymerase (purple line). In each window the top graph shows the percentage GC content at each position, with the numbers on the right denoting the minimum and maximum values. The middle graph in each window (purple, green and blue traces) is a coverage plot showing depth of reads (unnormalised) mapped at each position and below that are the coordinates of the selected region in the given genome.
Materials
Adapter-ligated DNA library (see Basic Protocol 3)
KAPA HiFi HotStart ReadyMix (KAPA Biosystems, cat. no KK2601)
- Paired-end PCR primers at 100 μM:
- PCR_F
- 5′ AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T 3′ (*indicates phosphorothioate; (Bentley et al., 2008)
- PCR_R
5′ CAAGCAGAAGACGGCATACGAGATXXXXXXXXCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T 3′ (*indicates phosphorothioate (Bentley et al., 2008); XXXXXXXX can be used to insert index tags)
Thermal cycler
1.5-ml microcentrifuge tubes
Agencourt AMPure XP (Beckman Coulter Genomics, cat. no A63881)
Magnetic separator (e.g. DynaMag-Spin Life Technologies cat. no. 12320D)
80% ethanol
Perform PCR
-
1a.
For libraries that were made from 500ng input DNA or more use 8 cycles of PCR in a 50-μl PCR reaction.
-
or 1b.
For libraries that were made from 200ng to 500ng input DNA use 12 cycles of PCR in a 50-μl PCR reaction..
-
or 1c.
For libraries that were made from 50ng to 200ng input DNA use 14 cycles of PCR in a 50-μl PCR reaction.
-
or 1d.
For libraries that were made from less than 50ng input DNA use 18 cycles of PCR in a 50-μl PCR reaction.
These quantities give the optimal compromise between clean libraries and a low frequency of duplicate sequences, but are only given as guideline amounts. Some optimization may be necessary.
-
2.
In a 50-μl PCR reaction, mix 25 μl KAPA HiFi HotStart ReadyMix, with 1 μM primers and template DNA. Typically we use 25 μl KAPA HiFi HotStart ReadyMix, 1 μl of each primer at 10 μM, 13 μl of water and 10 μl of adapter-ligated DNA.
-
3.
Mix well and spin down.
-
4.
Carry out the amplification (according to guidelines given in 1a to 1d), using the following cycling conditions in a thermal cycler:
| 1 cycle | 2 min | 95°C | (initial denaturation) |
| 8 to 18 cycles | 15 sec | 98°C | (denaturation) |
| 30 sec | 62°C | (annealing) | |
| 30 sec | 72°C | (extension) | |
| 1 cycle | 10 min | 72°C | (final extension) |
| indefinitely | 4°C | (hold) |
Clean up PCR
Please note, when doing standard PCR clean up, follow the protocol below. When doing a dual size selection as suggested in Alternate Protocol 3 or 4, please follow steps from Alternate Protocol 3 or 4 again.
-
5.
Let the AMPure XP beads come to room temperature before the experiment
-
6.
Mix the beads well so that they appear homogeneous and consistent in color.
-
7.
Add 30 μl of AMPure XP beads to a new 1.5 ml Eppendorf tube.
-
8.
Pipette into this tube all 50 μl of PCR reaction. Mix well on a vortex mixer. Briefly centrifuge for 2 seconds to spin down residues on the side of the tube.
-
9.
Incubate the DNA with the beads for 5 minutes.
-
10.
Put the tube in the magnetic stand and wait for the solution to clear (~3 minutes)
-
11.
Remove the supernatant from the tube and add 500 μl of 80% ethanol without disturbing the pellet.
-
12.
Repeat step 11 twice.
-
13.
Remove all ethanol, including residual droplets and dry the samples for 5 minutes in the magnetic stand on the bench with the tube lids open. Do not dry longer than 5 minutes. The beads get too dry and yield decreases.
-
14.
Add 30 μl EB buffer, mix well on a vortex mixer, and incubate for 5 minutes at room temperature.
-
15.
Put the tube in the magnetic stand and leave for 2 to 3 minutes.
-
16.
Remove 30 μl of the supernatant to a fresh 1.5-ml tube. (Take care not to pipette any beads) This now contains the DNA. Discard the beads.
-
17.
Proceed to quantification (Basic Protocol 5).
ALTERNATE PROTOCOL 4
DIRECT SEQUENCING OF SHORT AMPLICONS
Amplicons can be prepared for sequencing in multiple ways. Amplicons that are of a longer length than the combined sequencing read length can be fragmented and libraries made as described above. Shorter amplicons can either i) be ligated to adapters using the standard library protocol described above except that fragmentation is not necessary or ii) Illumina adapter sequences can be added during PCR by utilizing tailed PCR primers.
Here locus specific primers are used that possess tails that are analogous to the adapter sequences added by ligation in Basic Protocol 3. These can then be extended in a secondary PCR reaction as described in Basic Protocol 4 wherein the full sequences required for flowcell annealing and sequence priming are added along with unique multiplexing barcodes if required.
Note: Unnecessary PCR amplification steps will exacerbate amplification biases, and so they should be avoided wherever possible.
Materials
Genomic DNA template
Thermal cycler
Gene specific primers (please see below)
Amplicon generation and amplification
-
Design tailed PCR primers:
5′ ACACTCTTTCCCTACACGACGCTCTTCCGATCT-[gene-specific forward] 3′
5′ TCGGCATTCCTGCTGAACCGCTCTTCCGATCT-[gene-specific reverse] 3′
This will enable the resulting amplicons to be amplified using the standard Illumina library PCR amplification primers (Basic Protocol 4) wherein they can be barcoded to enable amplicons from multiple samples to be pooled. Note cluster registration does not work well if all the fragments within a library have the same sequence. Therefore experiments should be designed such that mixtures of amplicons from different regions are sequenced together.
Amplify the amplicons using the gene specific primers with enough cycles to give an end concentration of 10 ng/μl.
-
Amplify 5-10 μl of this PCR product with 6 additional PCR cycles according to Basic Protocol.
This second PCR is to add index tags and the sequences that allow the library to bind to the Illumina flowcell. These quantities and PCR cycles are given as a rough guide. Some further optimization might be necessary.
ALTERNATE PROTOCOL 5
DIRECT SEQUENCING OF LOW AMOUNTS OF DNA USING ILLUMINA’S NEXTERA KIT
An alternative to the standard library preparation kit is the Illumina’s Nextera kit. This kit utilizes a modified Tn5 transposon that simultaneously shears DNA and introduces short adapters that are pre-complexed to the transposon. This has several benefits:
-
-
It is very quick, taking just five minutes to shear and introduce the Illumina adapter sequences, generating fragments ready for PCR amplification.
-
-
No specific equipment is required for shearing
-
-
As it is a relatively efficient process, less starting material is required allowing sequencing to be performed with low input DNA amounts (50ng or less)
Currently the cost per library is higher than when performing the standard library protocols described above. Because Nextera avoids the need to the purchase expensive equipment, such as a Covaris focused-ultrasonicator (or expensive enzymatic shearing reagents), the protocol might be cheaper for those labs that want to sequence only a few samples from low amounts of DNA. Whilst this approach works well for most genomes, poor results have been obtained with some genomes, particularly those that are very AT rich (Quail et al., 2012c).
We therefore think this protocol has its uses and we feel it deserves to be mentioned in this protocol as a viable alternative to the standard library approach. For more details, please consult the protocols available on the Illumina website (http://www.illumina.com/products/nextera_dna_sample_prep_kit.ilmn).
ALTERNATE PROTOCOL 6
SEQUENCING WITHOUT PCR
PCR amplification during library prep allows lower amounts of starting material to be used, but introduces bias (Quail et al., 2012b). To eliminate this, we developed a PCR-free library method (Kozarewa et al., 2009). Here, full-length Illumina Y adapters are ligated directly to A-tailed fragments. Since these adapters contain the full complement of sequencing primer hybridization sites for all types of reads, the finished product is ready for sequencing. This works well but requires larger amounts of starting material (generally at least >1μg [or 5μg if size selection is required], these quantities are given as guidelines, and some optimization might be necessary). Since these adapters are longer than the standard adapter, primer dimers are consequently longer, and harder to remove; so we recommend two 0.7× AMPure cleanups, or Pippin Prep size selection, following adapter ligation.
We have designed a set of 96 PCR free adapters all with unique barcodes enabling multiple libraries to be multiplexed in a single Illumina lane. These adapters can be ordered pre-duplexed and PAGE purified (e.g., from IDT). (See Appendix 2) If used at a working concentration of 4 μM add 2 μl of adapter instead of the 2 μl 10μM paired end adapter used in Basic Protocol 3.
Commercial kits for PCR-free library preparation are also available from Illumina and Bioo Scientific. Bioo Scientific also sell their Next Flex PCR-free adapters separately.
BASIC PROTOCOL 5
QUANTIFICATION USING SYBR GREEN
To obtain an optimal yield from a sequencing run, it is critical to quantify the library accurately before amplification on the flowcell surface. This is best achieved by quantitative PCR. The qPCR uses forward and reverse primers that anneal to the flowcell-specific primer regions of library DNA, and can therefore be used to quantify any Illumina library. The concentration of the template DNA is measured by comparison with a standard of known concentration. For optimal dilution, the molar concentration of the library can be determined by running 1 μl of the library on an Agilent High Sensitivity chip (see Support Protocol 1) or Caliper GX. For normal libraries, qPCR and Agilent traces will strongly agree, however for more difficult samples (e.g., no-PCR libraries) these can be discrepant. In this case we rely on qPCR results to accurately quantify the libraries. We recommend using this protocol initially to get the best possible sequencing yield. Only if experience shows certain types of libraries (e.g., standard murine genomes) consistently agree between qPCR and Agilent traces, we feel this step can be omitted to reduce library generation costs.
Materials
Library quantification kit (e.g., - Illumina/Universal Library Quantification Kit [KAPA Biosystems, cat. no. K4824])
- This kit contains a range of libraries of standard concentrations and the primers required (see below) to amplify Illumina libraries. If an alternative kit is used then the user may have to supply the following PCR primers at 10μM:
- Syb_FP5 (desalted) ATGATACGGCGACCACCGAG
- Syb_RP7 (desalted) CAAGCAGAAGACGGCATACGAG
Template DNAs of unknown concentration (from Basic Protocol 4)
96-well qPCR plates (Life Technologies, cat. no. 4346906)
Adhesive plate sealers (Life Technologies, cat. no. 4311971)
Life Technologies StepOne Quantitative PCR machine (or equivalent)
qPCR reaction set-up
-
1a.
On first use add the PCR primer premix to the SYBR green mastermix to create an amplification mastermix.
-
or 1b.
Alternatively, add 4 μl of each primer above per 100 μl SYBR green mastermix.
-
2.
Prepare 1/1000 and/or 1/10,000-fold dilutions of the template DNA that is to be quantified.
-
3.Make sure there is enough amplification master mix for all reactions, to ensure consistency. For example, if you have two templates of unknown concentration, each at 1000- and 10,000-fold dilution, thus four samples, and six concentration standards, you have a total of ten samples. Each will be assayed in triplicate, so you will have 30 qPCR reactions. Therefore, make sure there is enough amplification master mix for 33 reactions (an additional 10%). Below we give the volume for 1 sample in triplicate (3 reactions).
Amplification Master Mix (as prepared in step 1) 39 μl Water 15 μl Final volume 54 μl Mix thoroughly. -
4.
For each sample, prepare a triplicate qPCR reaction mix by adding 52 μl of the mix prepared in the preceding step to 13 μl of 1/1000 or 1/10000 diluted sample (or qPCR standard) and mix thoroughly. This will give you 65 μl of reaction mix (three reactions plus an additional 10%, to dispense in the following step).
-
5.
Pipette 20 μl of each reaction into a separate well of a 96-well PCR plate and seal with a plate seal.
-
6.
Centrifuge the plate 1 min at 1200 × g, room temperature, and transfer to the qPCR machine. Cycling conditions are:
1 cycle: 10 min 95°C 40 cycles: 30 sec 95°C 1 min 60°C
qPCR analysis
-
7.
Determine the concentration of each sample relative to the standard curve using the software on the real time PCR instrument. Then calculate the concentration of the original library using the following formula:
Concentration (nM) = qPCR value × (452/average fragment length of library) × dilution factor
Note: The average fragment length is that that was determined by running the library on an Agilent Bioanalyzer. The Kapa standards are a 452 base pair fragment. The dilution factor is what was used in the qPCR (e.g 1000 or 10,000).
-
8.
Proceed to denaturation (Basic Protocol 6).
BASIC PROTOCOL 6
DENATURATION OF TEMPLATES
We denature all double-stranded DNA libraries in 0.1 M NaOH, before transferring an aliquot into 1 ml of hybridization buffer. It is essential not to transfer >8 μl of the denatured library into the hybridization buffer, because the pH becomes too high for efficient hybridization of the template DNA to the oligonucleotides on the flowcell surface.
Materials
1 M NaOH
Hybridization buffer (Illumina)
UltraPure water or EB buffer (Qiagen)
Ice
DNA library (see Basic Protocol 3), concentration determined in Basic Protocol 5
EB buffer (supplied with Qiagen QIAquick PCR purification kit, cat. no. 28104)
200-μl tubes
1.5-ml microcentrifuge tubes
Vortexer
Template denaturation
-
1.
Make a 10-fold dilution of the supplied 1M NaOH solution by adding 10 μl 1M NaOH to 90 μl UltraPure water and mixing thoroughly.
-
2.
Add 1 ml hybridization buffer into a 1.5-ml microcentrifuge tube and put on ice.
-
3.
Dilute the DNA library to 2 nM using EB buffer.
-
4.
Add 10 μl of the resulting 2 nM DNA library to 10 μl of the 0.1 M NaOH solution (prepared in step 1).
This minimizes pipetting inconsistencies. For libraries that are weaker than 2 nM one can add the same amount (1μmol) of more concentrated NaOH (e.g. 5 μl of 0.2M to 15 μl 1nM DNA library). Be careful to not exceed the recommended amount of NaOH when loading this neat, as this will affect the clustering reaction.
-
5.
Vortex thoroughly and spin down.
-
6.
Leave for 5 min at room temperature.
-
7.
Transfer 8 μl to 1 ml of the hybridization buffer on ice (from step 2).
This generates a 8 pM dilution. This is generally a good starting loading concentration, but may need to be adjusted up or down to achieve Illumina recommended cluster density.
-
8a.
On a MiSeq load 800 μl into port 17 and follow the manufacturer’s protocol.
-
or 8b.
On a HiSeq or Genome Analyzer proceed to cluster amplification.
For cluster amplification, follow the manufacturer’s protocol. After completing cluster amplification, proceed with the sequencing.
SUPPORT PROTOCOL 2
AMPLIFICATION QUALITY CONTROL
Following cluster amplification, DNA on the flowcell is double stranded and can be stained by an intercalating dye and detected on a fluorescence microscope. This is a useful quality control (QC) step, which we use for all HiSeq and GA flowcells prior to linearization and blocking to confirm that the cluster density is appropriate. We generally do not sequence flowcells that have too high or too low a cluster density (Figure 18.2.6).
Figure 18.2.6. SYBRGreen QC.
Although the most accurate method to measure cluster density is to perform a first-base incorporation on the flowcell, it is more economical to stain flowcells with SYBRGreen I immediately after amplification, and to examine cluster density qualitatively, using a fluorescence microscope. When coupled with qPCR quantification, this method is usually sufficiently accurate.
Materials
0.1 M Tris·HCl pH 8.0 (APPENDIX 2D)
Sodium ascorbate (Sigma, cat. no. A4034)
10,000× SYBRGreen I (Life Technologies, cat. no. S-7567)
Amplified flowcell
PR2 buffer (Illumina, supplied with sequencing kits)
15-ml Falcon tubes
0.2-μm syringe filter
cBot (Illumina)
Fluorescence microscope, set up to detect SYBRGreen I
Prepare 5 ml of a solution of 0.1 M Tris·HCl, pH 8.0, and 0.1 mM sodium ascorbate, and filter into a 15-ml Falcon tube using a 0.2-μm syringe filter.
Transfer 1960 μl of this Tris-ascorbate solution to a clean 15-ml Falcon tube, and add 40 μl of 100× SYBRGreen I (dilute from 10,000× stock solution with water), to produce a working concentration of 2×.
-
Pipette 220 μl into each tube of a strip tube. Place the strip tube into the cBot proceed with cluster amplification only.
This is a custom recipe. It is possible to split the standard Illumina cluster amplification and linearization (blocking) and hybridization into two separate steps using custom recipes. Your Illumina field application specialist should be able to adivise.
Visualize clusters by fluorescence microscopy to check for an appropriate density.
-
Return the flowcell to the cBot and flush through with PR2 buffer (150 μl per channel over 10 min), before storage or linearization and blocking.
If the cluster density is out of range of the 3 tiles shown in figure 18.2.6, it may be uneconomical to proceed with the sequencing run.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2D; for suppliers, see SUPPLIERS APPENDIX.
COMMENTARY
Background Information
The sequencing reaction on Illumina platforms takes place on the interior surfaces of a hollow glass slide, named a flowcell. On a MiSeq the various types of flowcells have only 1 lane, whereas generic flowcells for a GA and HiSeq are divided physically into eight lanes (Figure 18.2.1A; GAII flowcell), allowing up to eight different sequencing libraries (or sets of libraries), to be sequenced in a single run. On the recently introduced HiSeq 2500 an additional flow cell is available, with only two lanes allowing up to two different sequencing libraries to be sequenced in a single run. Sequencing libraries consist of a collection of DNA fragments, with a specific range of sizes, which are ready to be sequenced. The interior surfaces of a flowcell are coated in polyacrylamide (Figure 18.2.1B), to which two oligonucleotides are attached, creating a random lawn of both oligos (Figure 18.2.1 and Figure 18.2.4). These act as forward and reverse primers for the exponential, isothermal cluster amplification reaction, which is performed by repeated cycles of extension, denaturation, and annealing. Because primers are covalently attached to the polyacrylamide, cluster amplicons are tethered to a fixed position on the flowcell surface. Amplified clusters consist of double-stranded DNA, and one strand is removed selectively before sequencing.
During the sequencing process clusters undergo a sequencing-by-synthesis reaction using reversible fluorescent terminator deoxyribonucleotides. Being terminator nucleotides, each DNA strand within a cluster can only incorporate a single nucleotide during each chemistry cycle, and being clonal, each strand within a cluster incorporates the same nucleotide. Clusters are imaged, blocking groups and fluorophores are removed by chemical cleavage, and the next round of nucleotide incorporation begins. Images are analyzed, generating a separate sequence for each cluster. Sequence length is identical for all clusters, as it is governed by the number of cycles of nucleotide incorporation, imaging, and cleavage.
Library preparation
The purpose of the library preparation reactions is to introduce adapter sequences onto template molecules that allow amplification onto the flowcell surface. Here we have described a number of modifications that allow for more efficient library preparation, and which enable a stable workflow in a production environment.
Fragmentation
We now routinely fragment all of our DNA samples using Covaris’ Adaptive Focused Acoustics technology (AFA). Here, acoustic energy is focused controllably into the aqueous DNA sample by a dish-shaped transducer, which creates cavitation events within the sample. The collapse of bubbles in the suspension creates multiple, intense, localized jets of water, which disrupt the DNA molecules in a reproducible and predictable way.
Following disruption, 200-bp fragments comprise 17% of the total fractionated DNA by mass, but in contrast to nebulization, very little DNA is lost during the fragmentation process, generating a 4- to 5-fold higher yield of the intended fragment size range than nebulization (Fig. 18.2.7). In addition, because the size distribution of DNA fragmented by AFA is narrow, we generally omit size selection from the library preparation, decreasing the workload and increasing yields further.
Figure 18.2.7. Comparison of sample fragmentation by nebulization with Covaris AFA.
4.5 μg human genomic DNA was fragmented by nebulization (red line) and AFA (blue line). Both were purified using a spin column and eluted in 30 μl EB buffer (Qiagen). 1 μl of each eluate was run on an Agilent Bioanalyzer DNA 2100 chip. Image adapted with permission from Macmillan Publishers Ltd. (Quail et al., 2008).
A-tailing, adapter ligation and size selection
Analysis of paired-end sequence data — in which each cluster is sequenced in both forward and reverse directions — reveals several artifacts that can be attributed to the library-preparation protocol:
Bias in the base composition of sequences: The mean GC content, e.g., of the sequences obtained differs from that of the organism from which the sequences were derived.
Chimeric sequences: These are sequences for which the two paired-end reads map to regions of the genome that are separated by far more than the intended insert size. Though this could indicate a genuine deletion or translocation in the sample, which can be confirmed by PCR, a high frequency of chimeric sequences is most likely to be a library preparation artifact.
Imperfect distribution of insert sizes: A perfect distribution should be Gaussian, with a peak at the expected position.
These artifacts can be reduced through the use of several protocol modifications.
A-tailing and adapter ligation
Prior to adapter ligation, templates are given an A-overhang on the 3′ end of each strand, which complements a 3′ T-overhang on the adapter. This makes ligation more efficient than if it were blunt ended. A-tailing also hinders blunt-ended self-ligation of templates, which would otherwise generate chimeric sequences. The ligation adapters themselves are modified on one strand using a phosphorothioate modification between the T-overhang and the penultimate base at the 3′ end (Bentley et al., 2008). This reduces removal of the T-overhang by any contaminating exonuclease activity in the ligase preparation, so preventing blunt-ended self-ligation of adapters. The other strand is phosphorylated at the 5′ end, allowing efficient ligation to templates.
Paired-end adapters are partially complementary, so the end that ligates to the template is double stranded, whereas the opposite end is not. Essentially, the adapters consist of the nucleotide sequences to which the sequencing primers hybridize during the sequencing-by-synthesis reaction. These are ligated onto the A-tailed fragments (Sambrook et al., 1989) via their T-overhang. Their structure ensures that each template strand receives different sequences at the opposite ends (Bentley et al., 2008; Smith, 2007), and works in a similar way to a vectorette (Riley et al., 1990). Paired-end libraries can be amplified and sequenced on both paired- and single-end flowcells.
Sizes selection
Despite all the preventative measures used above, adapter dimers do still form during the ligation step. This is possibly due to some remaining exonuclease activity, although the actual sequence data obtained appears to be more consistent with the T-overhangs annealing to one another. Adapter dimers should be removed, if possible, so as not to waste the sequencing capacity of a flowcell. Using 0.7× AMPure XP SPRI beads size selection is a convenient way of achieving this, and at the same time, allowing fragments of larger insert size to be selected.
Size selection for tighter insert sizes
If the fragments produced in Basic Protocol 1 are of large size (500 bp and up) the library generally contains a broader range of fragments. Since larger fragments do not cluster very well and therefore interfere with accurate cluster density prediction, it may be useful to restrict the range of fragment sizes within a library. One way to achieve this is to do a double AMPure bead selection step on both fragmented DNA, and again after PCR. During the first double-bead selection, the insert size of the fragments is restricted by binding and discarding the larger fragments in the first bead step, while washing away small fragments during the subsequent second bead step. Using this double size selection step, the range of fragment sizes for the subsequent library preparation is decreased.
In addition to narrowing the library size after shearing, this procedure may be useful after PCR too. Here it has the added benefit of reducing the shoulder of larger fragments, which is sometimes evident, which do not cluster well. In addition, this reduced size range leads to clusters with more uniform diameter, which improves sequencing. Even though this method narrows library sizes for most applications, in certain cases (e.g., to determine translocation events in cancer) this method is not stringent enough. In such cases we use the size selection method below.
Size selection to reduce chimeric molecules
Template molecules that have not been A-tailed at the 3′ ends of both strands possess one or two blunt ends, and so are substrates for blunt-ended ligation. This results in a chimeric template molecule. Because (in the general protocol) ligation is performed before any size selection step, the full range of fragment sizes will be present. If, for example, two blunt 100-bp fragments ligate together, and if the desired fragment size is 200 bp, during a single size selection step, the chimera will be isolated along with the fragments that were originally that size. For many sequencing applications, a low frequency of chimeric sequences can be tolerated, and can be removed using bioinformatics, as they will map to distant parts of the genome. For other applications, such as screening for translocations, these in vitro translocations can be falsely interpreted as genuine structural variants, and they will create a larger amount of subsequent confirmatory work.
Performing two size selection steps can reduce the frequency of such chimeric templates. The first size selection will be done immediately after fragmentation, concurrent with purification of the sample DNA, reducing library insert size varriation. A second size selection will be completed after PCR. Consequently, most of the chimeric templates that will be generated during the ligation step will fall out of the size range during the second size selection after PCR. Nowadays, we generally use Pippin Prep for this application (Quail et al., 2012a). Even though we prefer this method (Figure 18.2.3), it is possible to do this manually using an agarose gel (see Alternate Protocol 3). This additional size selection reduces the incidence of chimeras from ~5% to 0.02%.
PCR
To produce a good sequencing library, it is advantageous to use optimized quantities of template in the PCR. We routinely analyze our post-PCR sequencing libraries by performing microchip capillary electrophoresis, using an Agilent Bioanalyzer 2100. This allows us to quantify the library, but also to detect products that differ in size from the expected amplicons. In this way, we noticed that the quality of a post-PCR library decreases as the amount of template DNA used in the PCR increases. Too much template DNA often results in the generation of an apparently high molecular weight peak. This is typically twice the size of the expected product, as measured by the Bioanalyzer, and may represent a product that forms as primers become depleted. Conversely, if too little DNA is used in the PCR, the smaller the pool of original templates, and the greater the incidence of PCR duplicates in the resulting sequences. PCR duplicates are pairs of sequences for which the ends map to identical positions in the genome. Some duplicate sequences will inevitably arise by chance, when two molecules in the sample are sheared at the same position at both ends, but the frequency of this is very low and predictable, and depends upon the read length and depth of sequence coverage. A low frequency of duplicate sequences (~0.1%) also arises by the cluster detection software misinterpreting single clusters as pairs (optical duplicates). However, the vast majority of observed duplicates arise during the PCR. Thus, it is essential to choose the appropriate set of conditions for each PCR.
PCR yield
The standard Illumina PCR uses Phusion polymerase, but by using KAPA high-fidelity polymerase, we have been able to increase the yield of the enrichment PCR reaction 5- to 10-fold, which allows fewer cycles of amplification to be performed. In addition, we found this enzyme to have a decreased amplification bias towards genomes with higher and lower GC content ((Quail et al., 2012b; Quail et al., 2012c) and Figure 18.2.5).
PCR cleanup
Surplus PCR primers may interfere with quantification and will compete with amplicons for hybridization to the flowcell surface. However, a more significant problem is presented by adapter dimers that enter the PCR reaction. Here, dimers also receive the full-length nucleotide tails that allow hybridization and amplification on the flowcell surface, and so will form clusters that are sequenced alongside the desired templates. Consequently, it is advantageous to remove dimers, as well as unextended oligos after the PCR. We have found that solid-phase reversible immobilization (SPRI) technology (DeAngelis et al., 1995; Hawkins et al., 1994) removes a higher proportion of primers and adapter dimers than spin columns, without compromising on yield. SPRI beads also allow elution in a wider variety of buffers (Fig. 18.2.8). In addition, they give the possibility of doing a double size selection to remove larger fragments when necessary (Alternate Protocol 2).
Figure 18.2.8. PCR cleanup.
We prepared a paired-end PhiX library using conditions that would promote the formation of adapter and primer dimers and unextended PCR primers. After PCR, we divided the library in two: half was purified using a QIAquick spin column, as in the standard Illumina protocol (left), whereas the other half was purified using AMPure SPRI beads (right). Gels are shown after staining and excision of the gel slice corresponding to the desired size range of fragments. Image adapted with permission from Macmillan Publishers Ltd. (Quail et al., 2008).
Library quantification
Accurate quantification of DNA prior to cluster amplification is essential. For fragment sizes undergoing a given number of cycles of cluster amplification, there is a concentration range of DNA that will yield clusters in the optimal density range, enabling the maximum amount of data to be obtained (Fig. 18.2.9). For fragments with a mean insert size of 500 bp or lower, we aim for ~800,000 clusters per mm2 on the HiSeq and MiSeq, giving ~750,000 purity-filtered (PF) clusters mm2.
Figure 18.2.9. Cluster throughput as a function of total clusters.
The graph shows analyzable purity filtered (PF) clusters per mm2, versus total raw clusters per mm2. It can be seen that above the optimal cluster density of ~800,000 per mm2, the fraction of filters passing purity filtering is more variable and begins to reduce. With higher cluster densities, more clusters overlap. This can give rise to underestimation of real cluster density. Mixed clusters, however are discarded by the image analysis software causing a great drop in purity filtered clusters when flowcells are over-clustered. Therefore accurate library quantification is essential for the sequencing run to generate the maximum yield of data. Red line: Theoretical maximum of PF clusters per cluster density. Black line: trendline fitting all PF clusters per cluster density.
Electrophoresis QC (Agilent Bioanalyzer or Caliper GX)
Cluster density values achieved by spectrophotometry measurements tends to be inconsistent, but are typically 5- to 10-fold lower than expected for a given library concentration, presumably because spectrophotometry cannot distinguish between differently sized DNA species. Spectral methods measure not only the intended amplicon but also adapter dimers and unextended primers. Spectrophotometry also struggles to measure low DNA concentrations accurately.
Using an Agilent Bioanalyzer 2100 or Caliper GX for general library quantification, we can achieve a much more consistent cluster density, provided the library is diluted to between 1 and 15nM and concentration is measured by integrating library fragments up to 600bp, since larger fragments cluster much less efficiently. These instruments can determine the size of DNA species allowing the quality of the sample preparation to be determined. In spite of this, however, for a small proportion of libraries, we obtain suboptimal cluster densities, and consequently far less useful data, than the measured concentration value would predict. There can be many reasons for this. It could be a consequence of the presence of single-stranded DNA generated in the PCR. The Bioanalyzer or Caliper GX cannot quantify single-stranded DNA accurately, making libraries with high amounts of single stranded DNA unquantifiable. Although optimized PCR conditions can help us to avoid the generation of single-stranded DNA, we also prefer a quantification assay that can detect all amplifiable template molecules in a library, i.e., quantitative PCR (see below). Electrophoretic methods can only estimate the quantity and size of the DNA fragments present in the library they cannot differentiate between fragments that have correctly ligated adapters and those that do not. Following PCR, virtually all library fragments will have adapters at both ends. However this will not be the case for PCR-free libraries where the use of qPCR is recommended.
Quantitative PCR
Quantitative PCR should be capable of detecting and quantifying all amplifiable molecules—i.e., those with adapters at either end (for discussion see (Meyer et al., 2008)). We designed amplification primers to anneal to the Illumina paired-end adapter sequences. Because the amplification of Illumina libraries is rarely 100% efficient, we quantify unknown libraries against a series of known commercially available concentration standards. This allows us to predict cluster number much more accurately.
Denaturation
After quantification, double-stranded DNA libraries are denatured with 0.1 M NaOH before diluting and loading onto the flowcell. Because a high pH prevents efficient hybridization, Illumina recommends that the final denatured library, in1 ml hybridization buffer, should have no more than 1mM NaOH. More NaOH will induce pH changes that will interfere with effective clustering. Given that the optimal loading concentration generally is 8 pM, denaturation of libraries that are below 0.5 nM becomes problematic, as these require 15μl of library and 5 μl of 0.2 M NaOH to be added to the hybridization buffer. Denaturation by heating is an alternative, but has the potential both to damage the DNA and to introduce anti-GC bias (Mandel and Marmur, 1968). Consequently, we still prefer to denature dilute templates with NaOH, using Basic Protocol 6.
Critical Parameters
Fragmentation and post-fragmentation QC
Prior to working on real samples it is recommended that a series of fragmentation experiments be performed upon various concentrations of a test DNA sample (e.g., human genomic DNA; Promega, cat. no. G1471). After fragmentation, results can be visualized by running the sheared DNA on an agarose gel or Agilent Bioanalyzer High Sensitivity DNA chip allowing the identification of optimal shearing parameters that give a maximal proportion of DNA within the desired size range.
The starting quantity of DNA can have a critical effect on the success of library preparation. For standard paired-end libraries, we use at least 500 ng, though we recommend that 5 μg genomic DNA be used if performing double size selection. Quantification of genomic DNA tends to be unreliable and varies depending on the method used. This can lead to suboptimal amounts of DNA being available for library preparation. We do not encourage the use of spectrophotometric methods for quantification as these can be rendered inaccurate by small nucleic acids and contaminating chemicals; we have found fluorometric quantitation assays to be more trustworthy (e.g., Life Technologies’ Qubit 2.0 fluorometer—order code Q32866), though even here, concentrations can be overestimated in samples containing excessive amounts of contaminating RNA.
If sufficient genomic DNA (>2 μg) is available, the sample can be diluted 1/10 and analyzed after fragmentation on an Agilent Bioanalyzer High Sensitivity DNA chip to assess the size distribution that has been generated, and to confirm quantification. If necessary, samples that have not sheared successfully can be subjected to further fragmentation.
End-repair, A-tailing, and adapter ligation
These reactions are generally very robust, although it is essential to ensure that buffers have been thawed completely, and that any particulate matter has fully dissolved (especially T4 DNA ligase buffer, which seems to have crystals that dissolve slowly). To achieve this, leave buffers, oligos, and adapters at room temperature for 30 min prior to use. Check for any precipitated material by visual inspection, and if this is present, warm the buffer in your hand, vortex, and spin down. Avoid repeated freezing and thawing of these buffers. Store enzymes at −20°C and take out of the freezer, spin down, and place tubes on ice just before use. Return enzymes to −20°C immediately after use.
When setting up reactions, we recommend using a tick list to record when each component has been added. Enzymes should be added last, and all reactions should be mixed well by gently pipetting up and down three times.
A potentially very useful feature of Illumina’s TruSeq library prep kits is their spike-in controls. These are three types of fragments of DNA that contain: DNA that still needs to be end repaired; blunt DNA fragments; and DNA with overhanging A. After successful library generation all these fragments will be sequenced and give an indication of the efficacy of the independent library preparation steps.
Another possible quality control step, is to save some of the fragmented, end repaired and A-tailed sample and attempt it to self-ligate. When any of the library creation steps is inefficient, (especially the A-tailing), this will lead to ligation of these products to each other and library fragments that are larger compared to the original non-ligated sample.
Pre-PCR QC
The quantity of template used in the PCR can affect library quality. We quantify the adapter-ligated template prior to PCR using an Agilent Bioanalyzer High Sensitivity DNA chip.
If template concentration is too low to be measured, either insufficient genomic DNA was used to begin with, or too much loss has occurred during the column cleanup steps. In either event, it is necessary to repeat the library prep. To enhance yields from column cleanup steps, ensure that the PB buffer is mixed thoroughly with the sample before adding to the column, and that the ethanol has evaporated after the wash step (leaving at room temperature for 5 min is usually sufficient). For optimal results, wait for 5 min after adding EB buffer to the column before eluting.
When using AMPure XP beads, be careful to use specific ratios (e.g. 0.7×, 1.0× of beads as outlined in this protocol. Even though most of the DNA binds to and elutes from the beads within 1-3 minutes, we still suggest incubating and eluting the DNA for 5 minutes for optimal yield. The longer the fragments of DNA, the more difficult it is to elute them from the beads, especially after intense drying, so we recommend not drying at 37°C nor drying for longer than 5 minutes.
It is also possible to quantify the amount of adapter-ligated DNA at this stage by qPCR to determine what fraction of the library has adapters on both ends.
Post-PCR QC
A 1-μl aliquot of each library should be run on an Agilent Bioanalyzer High Sensitivity DNA chip to check concentration and gauge library quality. This reveals the amount of adapter dimer present in the library (observed as a sharp peak at ~130 bp), and the distribution of fragment sizes present in the library.
Quantification
Because the Agilent Bioanalyzer (or Caliper LabChipGX) is useful in determining general library concentrations, library production costs can be reduced by omitting the subsequent quantitative PCR step. In most cases, when standard libraries are generated from standard human or mouse DNA, the Agilent or Caliper traces are accurate enough to give a good estimate for pooling.
When more difficult libraries (e.g. PCR-free libraries or libraries with differing insert size ranges) are quantified, however, this method cannot be relied upon completely. The concentration measurement should be considered an initial estimate, which allows samples to be diluted to a point that is within the range of the standard curve for quantification by qPCR. We dilute libraries to 10 pM, based upon their Bioanalyzer concentration, and quantify them by qPCR using commercial KAPA Library Quantification DNA Standards.
A third method, which is still being validated in our lab, is to pool equal volumes of independent libraries, run the pool on an Agilent High Sensitivity chip for quantification and subsequently on a MiSeq using a 50bp kit. Cluster density and distribution of different multiplexes will help give an accurate pooling strategy for HiSeq runs. This second pool can then be run on an Agilent high sensitivity chip or quantified by qPCR to validate final molarity before running it on the HiSeq.
Cluster amplification QC
Too high a cluster density will result in a lower yield of purity-filtered data, because clusters will overlap to a greater extent. Conversely, too low a cluster density can result in a lower yield of data, and additional sequencing lanes being required. Figure 18.2.6 shows the range of cluster densities, determined by SYBR green that will yield a good quantity of purity filtered clusters, whereas outside of this range, decreased yields are inevitable.
Troubleshooting
Some problems that may be encountered in carrying out the protocols described in this unit, along with their possible causes and solutions, are described in Table 18.2.1.
Table 18.2.1. Troubleshooting Guide for Preparing DNA Libraries Used in the Illumina Sequencing System.
| Problem | Possible cause | Solution |
|---|---|---|
| Insufficient DNA after fragmentation |
Inaccurate quantification of genomic DNA |
Fragment additional genomic DNA |
| Poor recovery from cleanup column |
Mix sample and buffer thoroughly before adding to column; allow column to dry thoroughly after rinsing with wash buffer; incubate column with elution buffer for 5 min before eluting. |
|
| Poor recovery from Ampure beads |
Mix sample and Ampure beads thoroughly; allow to incubate for 5 minutes after adding; dry for 5 minutes, but do not dry further, since longer DNA will elute slower. |
|
| Unexpected size distribution after fragmentation |
Inappropriate settings used on Covaris. Incorrect vials used on Covaris. |
Check settings and vials |
| DNA is contaminated | Clean up DNA using columns or Ampure beads |
|
| Insufficient DNA after adapter ligation (<25 ng total) |
Poor recovery from cleanup column/gel |
Mix sample and buffer thoroughly before adding to column; allow column to dry thoroughly after rinsing with wash buffer; incubate column with elution buffer for 5 min before eluting. |
| Insufficient DNA after adapter ligation (<25 ng total) |
Poor recovery from cleanup AMPure XP beads |
Mix sample and AMPure beads thoroughly and incubate for 5 minutes, before putting on a magnet; Don’t over dry the beads; Incubate beads with elution buffer for 5 min before putting on the magnet. |
| Large peak at 130 bp after PCR |
Adapter dimer contamination—size range of selected fragments not optimal |
Cut gel slice corresponding to larger fragment size, or purify with AMPure beads using a bead:DNA ratio of 0.8:1 |
| Insufficient DNA after PCR (<2 ng/μl) |
PCR failure | Repeat PCR with fresh reagents |
| Fragment size range too broad |
Perform qPCR quantification on stock and 1/10 dilution |
|
| No clusters visible in lane 8, column 2 of the GAII |
Insufficient oil added to right- hand side of flowcell |
Add more oil to right-hand side of flowcell |
| Clusters blurred on the GAII | Poor focusing Oil on flowcell |
Repeat focusing steps Clean flowcell gently with lens tissue and ethanol |
| Low cluster intensity | Failure of primer hybridization | No/not enough/the wrong sequencing primer was added The primer melting temperature of the custom primer needs optimization |
| Low % of PF clusters | Cluster density too high | Repeat amplification with lower concentration |
| Too wide a range of fragment sizes |
Repeat size selection, taking narrower gel slice |
|
| Low number of raw clusters detected |
Cluster density too high | Repeat amplification with lower library concentration |
| Cluster density too low | Repeat amplification with higher library concentration |
|
| Degradation of concentration standard used in qPCR |
Repeat qPCR with fresh dilutions of concentration standards |
|
| Spiky IVC plots | Adapter contamination | If % adapter is too high (e.g., >10%) repeat size selection step of library prep |
| High A signal in basecall plots |
Low cluster density | Repeat amplification with higher library concentration |
| Low coverage of AT-rich regions |
Heating during gel step | Dissolve gel slice at room temperature |
| Too many PCR cycles | Repeat using a maximum of 10 cycles |
|
| High % duplicate sequences | Too little ligated DNA in PCR | Repeat PCR with higher quantity of ligated DNA |
| Inefficient end repair/A- tailing/adapter ligation |
Repeat library prep using fresh reagents |
Anticipated Results
The Illumina library preparation protocols and kits, when used in conjunction with the recommendations discussed here, result in a robust approach that enables the preparation of high-quality libraries of adapter-ligated fragments with the correct concentration and quality for sequencing. Libraries should have the desired range of insert sizes and be virtually free of adapter dimers, and should be at a concentration in the nanomolar range, that will allow for preparation of multiple flowcells for sequencing, with cluster densities of ~800,000 clusters per mm2 for HiSeq or MiSeq flowcells.
Time Considerations
The Illumina library preparation protocol can be completed manually within 1 day if processing 1 sample, and 2 days if processing multiple (2 to 8) samples. The protocol can be stopped after any column or AMPure cleanup step, and samples can be stores in low-bind tubes at −20°C until required. Larger sample numbers can be processed manually using multichannel pipettes in microtitre plates at a rate of about a plate per 3-4 days, or by automated liquid handlers in 4-5 hours, utilizing SPRI-bead cleanup and 96 well plate magnets.
Agilent Bioanalyzer QC and qPCR take 0.5 days.
Cluster amplification, linearization, blocking, primer hybridization, and setting up the sequencing run can be performed in a single day, but if desired, flowcells can be stored after amplification. Once the sequencing primer has been hybridized, the sequencing run should be started within 4 hr.
Acknowledgements
This work was supported by the Wellcome Trust [grant number 098051], and the European Union 7th Framework Programme, European Sequencing and Genotyping Infrastructure (ESGI),[grant number 262055].
We are grateful to all members of the Research and Development and Illumina Sequencing teams at the Wellcome Trust Sanger Institute.
Appendix 1: PCR indexing primers.
Index sequencing is performed as a separate read using the iPCRtagseq sequencing oligo (see below for sequence). Spike 4 μl of a 100μM dilution into the standard Illumina index sequencing primer mix.
iPCRtagseq: AAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTC
| Oligo name | sequence obtained | PCR primers |
|---|---|---|
| iPCRtagT1 | ATCACGTTAT | CAAGCAGAAGACGGCATACGAGATAACGTGATGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT2 | CGATGTTTAT | CAAGCAGAAGACGGCATACGAGATAAACATCGGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT3 | TTAGGCATAT | CAAGCAGAAGACGGCATACGAGATATGCCTAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT4 | TGACCACTAT | CAAGCAGAAGACGGCATACGAGATAGTGGTCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT5 | ACAGTGGTAT | CAAGCAGAAGACGGCATACGAGATACCACTGTGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT6 | GCCAATGTAT | CAAGCAGAAGACGGCATACGAGATACATTGGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT7 | CAGATCTGAT | CAAGCAGAAGACGGCATACGAGATCAGATCTGGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT8 | ACTTGATGAT | CAAGCAGAAGACGGCATACGAGATCATCAAGTGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT9 | GATCAGCGAT | CAAGCAGAAGACGGCATACGAGATCGCTGATCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT10 | TAGCTTGTAT | CAAGCAGAAGACGGCATACGAGATACAAGCTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT11 | GGCTACAGAT | CAAGCAGAAGACGGCATACGAGATCTGTAGCCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT12 | CTTGTACTAT | CAAGCAGAAGACGGCATACGAGATAGTACAAGGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT13 | TGGTTGTTAT | CAAGCAGAAGACGGCATACGAGATAACAACCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT14 | TCTCGGTTAT | CAAGCAGAAGACGGCATACGAGATAACCGAGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT15 | TAAGCGTTAT | CAAGCAGAAGACGGCATACGAGATAACGCTTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT16 | TCCGTCTTAT | CAAGCAGAAGACGGCATACGAGATAAGACGGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT17 | TGTACCTTAT | CAAGCAGAAGACGGCATACGAGATAAGGTACAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT18 | TTCTGTGTAT | CAAGCAGAAGACGGCATACGAGATACACAGAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT19 | TCTGCTGTAT | CAAGCAGAAGACGGCATACGAGATACAGCAGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT20 | TTGGAGGTAT | CAAGCAGAAGACGGCATACGAGATACCTCCAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT21 | TCGAGCGTAT | CAAGCAGAAGACGGCATACGAGATACGCTCGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT22 | TGATACGTAT | CAAGCAGAAGACGGCATACGAGATACGTATCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT23 | TGCATAGTAT | CAAGCAGAAGACGGCATACGAGATACTATGCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT24 | TTGACTCTAT | CAAGCAGAAGACGGCATACGAGATAGAGTCAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT25 | TGCGATCTAT | CAAGCAGAAGACGGCATACGAGATAGATCGCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT26 | TTCCTGCTAT | CAAGCAGAAGACGGCATACGAGATAGCAGGAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT27 | TAGTGACTAT | CAAGCAGAAGACGGCATACGAGATAGTCACTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT28 | TACAGGATAT | CAAGCAGAAGACGGCATACGAGATATCCTGTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT29 | TCCTCAATAT | CAAGCAGAAGACGGCATACGAGATATTGAGGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT30 | TGTGGTTGAT | CAAGCAGAAGACGGCATACGAGATCAACCACAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT31 | TAGTCTTGAT | CAAGCAGAAGACGGCATACGAGATCAAGACTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT32 | TTCCATTGAT | CAAGCAGAAGACGGCATACGAGATCAATGGAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT33 | TCGAAGTGAT | CAAGCAGAAGACGGCATACGAGATCACTTCGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT34 | TAACGCTGAT | CAAGCAGAAGACGGCATACGAGATCAGCGTTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT35 | TTGGTATGAT | CAAGCAGAAGACGGCATACGAGATCATACCAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT36 | TGAACTGGAT | CAAGCAGAAGACGGCATACGAGATCCAGTTCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT37 | TACTTCGGAT | CAAGCAGAAGACGGCATACGAGATCCGAAGTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT38 | TCTCACGGAT | CAAGCAGAAGACGGCATACGAGATCCGTGAGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT39 | TCAGGAGGAT | CAAGCAGAAGACGGCATACGAGATCCTCCTGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT40 | TAAGTTCGAT | CAAGCAGAAGACGGCATACGAGATCGAACTTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT41 | TCCAGTCGAT | CAAGCAGAAGACGGCATACGAGATCGACTGGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT42 | TGTATGCGAT | CAAGCAGAAGACGGCATACGAGATCGCATACAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT43 | TCATTGAGAT | CAAGCAGAAGACGGCATACGAGATCTCAATGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT44 | TGGCTCAGAT | CAAGCAGAAGACGGCATACGAGATCTGAGCCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT45 | TATGCCAGAT | CAAGCAGAAGACGGCATACGAGATCTGGCATAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT46 | TCAGATTCAT | CAAGCAGAAGACGGCATACGAGATGAATCTGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT47 | TACTAGTCAT | CAAGCAGAAGACGGCATACGAGATGACTAGTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT48 | TTCAGCTCAT | CAAGCAGAAGACGGCATACGAGATGAGCTGAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT49 | TGTCTATCAT | CAAGCAGAAGACGGCATACGAGATGATAGACAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT50 | TATGTGGCAT | CAAGCAGAAGACGGCATACGAGATGCCACATAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT51 | TTACTCGCAT | CAAGCAGAAGACGGCATACGAGATGCGAGTAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT52 | TCGTTAGCAT | CAAGCAGAAGACGGCATACGAGATGCTAACGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT53 | TACCGAGCAT | CAAGCAGAAGACGGCATACGAGATGCTCGGTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT54 | TGTTCTCCAT | CAAGCAGAAGACGGCATACGAGATGGAGAACAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT55 | TTCGCACCAT | CAAGCAGAAGACGGCATACGAGATGGTGCGAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT56 | TTGCGTACAT | CAAGCAGAAGACGGCATACGAGATGTACGCAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT57 | TCTACGACAT | CAAGCAGAAGACGGCATACGAGATGTCGTAGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT58 | TGACAGACAT | CAAGCAGAAGACGGCATACGAGATGTCTGTCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT59 | TAGAACACAT | CAAGCAGAAGACGGCATACGAGATGTGTTCTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT60 | TCATCCTAAT | CAAGCAGAAGACGGCATACGAGATTAGGATGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT61 | TGCTGATAAT | CAAGCAGAAGACGGCATACGAGATTATCAGCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT62 | TAGACGGAAT | CAAGCAGAAGACGGCATACGAGATTCCGTCTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT63 | TGTGAAGAAT | CAAGCAGAAGACGGCATACGAGATTCTTCACAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT64 | TCTCTTCAAT | CAAGCAGAAGACGGCATACGAGATTGAAGAGAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT65 | TTGTTCCAAT | CAAGCAGAAGACGGCATACGAGATTGGAACAAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT66 | TGAAGCCAAT | CAAGCAGAAGACGGCATACGAGATTGGCTTCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT67 | TACCACCAAT | CAAGCAGAAGACGGCATACGAGATTGGTGGTAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT68 | TGCGTGAAAT | CAAGCAGAAGACGGCATACGAGATTTCACGCAGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT69 | GGTGAGTTAT | CAAGCAGAAGACGGCATACGAGATAACTCACCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT70 | GATCTCTTAT | CAAGCAGAAGACGGCATACGAGATAAGAGATCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT71 | GTGTCCTTAT | CAAGCAGAAGACGGCATACGAGATAAGGACACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT72 | GACGGATTAT | CAAGCAGAAGACGGCATACGAGATAATCCGTCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT73 | GCAACATTAT | CAAGCAGAAGACGGCATACGAGATAATGTTGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT74 | GGTCGTGTAT | CAAGCAGAAGACGGCATACGAGATACACGACCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT75 | GAATCTGTAT | CAAGCAGAAGACGGCATACGAGATACAGATTCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT76 | GTACATCTAT | CAAGCAGAAGACGGCATACGAGATAGATGTACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT77 | GAGGTGCTAT | CAAGCAGAAGACGGCATACGAGATAGCACCTCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT78 | GCATGGCTAT | CAAGCAGAAGACGGCATACGAGATAGCCATGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT79 | GTTAGCCTAT | CAAGCAGAAGACGGCATACGAGATAGGCTAACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT80 | GTCGCTATAT | CAAGCAGAAGACGGCATACGAGATATAGCGACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT81 | GGAATGATAT | CAAGCAGAAGACGGCATACGAGATATCATTCCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT82 | GAGCCAATAT | CAAGCAGAAGACGGCATACGAGATATTGGCTCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT83 | GCTCCTTGAT | CAAGCAGAAGACGGCATACGAGATCAAGGAGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT84 | GTAAGGTGAT | CAAGCAGAAGACGGCATACGAGATCACCTTACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT85 | GAGGATGGAT | CAAGCAGAAGACGGCATACGAGATCCATCCTCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT86 | GTTGTCGGAT | CAAGCAGAAGACGGCATACGAGATCCGACAACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT87 | GGATTAGGAT | CAAGCAGAAGACGGCATACGAGATCCTAATCCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT88 | GATAGAGGAT | CAAGCAGAAGACGGCATACGAGATCCTCTATCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT89 | GTGTGTCGAT | CAAGCAGAAGACGGCATACGAGATCGACACACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT90 | GCAATCCGAT | CAAGCAGAAGACGGCATACGAGATCGGATTGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT91 | GACCTTAGAT | CAAGCAGAAGACGGCATACGAGATCTAAGGTCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT92 | GCCTGTTCAT | CAAGCAGAAGACGGCATACGAGATGAACAGGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT93 | GCACTGTCAT | CAAGCAGAAGACGGCATACGAGATGACAGTGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT94 | GCTAACTCAT | CAAGCAGAAGACGGCATACGAGATGAGTTAGCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT95 | GATTCATCAT | CAAGCAGAAGACGGCATACGAGATGATGAATCGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
| iPCRtagT96 | GTCTTGGCAT | CAAGCAGAAGACGGCATACGAGATGCCAAGACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC*T |
Appendix 2: PCR free adapters.
Order these PCR-free adapters PAGE-purified and preduplexed from IDT with bottom strand from table below and common top oligo (Top_no_PCR). To obtain the index sequence, sequence the index read with iPCRtag seq primer as described in Appendix 1. Working concentration is 4μM.
Top_no_PCR: A*ATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T
| Tag | sequence obtained | no-PCR bottom strand adapter sequences |
|---|---|---|
| tag1 | ATCACGTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCATCACGTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag2 | CGATGTTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCCGATGTTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag3 | TTAGGCATAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTAGGCATATCTCGTATGCCGTCTTCTGCTT*G |
| tag4 | TGACCACTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGACCACTATCTCGTATGCCGTCTTCTGCTT*G |
| tag5 | ACAGTGGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCACAGTGGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag6 | GCCAATGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCCAATGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag7 | CAGATCTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCCAGATCTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag8 | ACTTGATGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCACTTGATGATCTCGTATGCCGTCTTCTGCTT*G |
| tag9 | GATCAGCGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGATCAGCGATCTCGTATGCCGTCTTCTGCTT*G |
| tag10 | TAGCTTGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAGCTTGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag11 | GGCTACAGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGGCTACAGATCTCGTATGCCGTCTTCTGCTT*G |
| tag12 | CTTGTACTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCCTTGTACTATCTCGTATGCCGTCTTCTGCTT*G |
| tag13 | TGGTTGTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGGTTGTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag14 | TCTCGGTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCTCGGTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag15 | TAAGCGTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAAGCGTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag16 | TCCGTCTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCCGTCTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag17 | TGTACCTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGTACCTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag18 | TTCTGTGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTCTGTGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag19 | TCTGCTGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCTGCTGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag20 | TTGGAGGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTGGAGGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag21 | TCGAGCGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCGAGCGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag22 | TGATACGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGATACGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag23 | TGCATAGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGCATAGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag24 | TTGACTCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTGACTCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag25 | TGCGATCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGCGATCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag26 | TTCCTGCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTCCTGCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag27 | TAGTGACTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAGTGACTATCTCGTATGCCGTCTTCTGCTT*G |
| tag28 | TACAGGATAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTACAGGATATCTCGTATGCCGTCTTCTGCTT*G |
| tag29 | TCCTCAATAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCCTCAATATCTCGTATGCCGTCTTCTGCTT*G |
| tag30 | TGTGGTTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGTGGTTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag31 | TAGTCTTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAGTCTTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag32 | TTCCATTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTCCATTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag33 | TCGAAGTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCGAAGTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag34 | TAACGCTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAACGCTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag35 | TTGGTATGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTGGTATGATCTCGTATGCCGTCTTCTGCTT*G |
| tag36 | TGAACTGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGAACTGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag37 | TACTTCGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTACTTCGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag38 | TCTCACGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCTCACGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag39 | TCAGGAGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCAGGAGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag40 | TAAGTTCGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAAGTTCGATCTCGTATGCCGTCTTCTGCTT*G |
| tag41 | TCCAGTCGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCCAGTCGATCTCGTATGCCGTCTTCTGCTT*G |
| tag42 | TGTATGCGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGTATGCGATCTCGTATGCCGTCTTCTGCTT*G |
| tag43 | TCATTGAGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCATTGAGATCTCGTATGCCGTCTTCTGCTT*G |
| tag44 | TGGCTCAGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGGCTCAGATCTCGTATGCCGTCTTCTGCTT*G |
| tag45 | TATGCCAGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTATGCCAGATCTCGTATGCCGTCTTCTGCTT*G |
| tag46 | TCAGATTCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCAGATTCATCTCGTATGCCGTCTTCTGCTT*G |
| tag47 | TACTAGTCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTACTAGTCATCTCGTATGCCGTCTTCTGCTT*G |
| tag48 | TTCAGCTCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTCAGCTCATCTCGTATGCCGTCTTCTGCTT*G |
| tag49 | TGTCTATCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGTCTATCATCTCGTATGCCGTCTTCTGCTT*G |
| tag50 | TATGTGGCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTATGTGGCATCTCGTATGCCGTCTTCTGCTT*G |
| tag51 | TTACTCGCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTACTCGCATCTCGTATGCCGTCTTCTGCTT*G |
| tag52 | TCGTTAGCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCGTTAGCATCTCGTATGCCGTCTTCTGCTT*G |
| tag53 | TACCGAGCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTACCGAGCATCTCGTATGCCGTCTTCTGCTT*G |
| tag54 | TGTTCTCCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGTTCTCCATCTCGTATGCCGTCTTCTGCTT*G |
| tag55 | TTCGCACCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTCGCACCATCTCGTATGCCGTCTTCTGCTT*G |
| tag56 | TTGCGTACAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTGCGTACATCTCGTATGCCGTCTTCTGCTT*G |
| tag57 | TCTACGACAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCTACGACATCTCGTATGCCGTCTTCTGCTT*G |
| tag58 | TGACAGACAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGACAGACATCTCGTATGCCGTCTTCTGCTT*G |
| tag59 | TAGAACACAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAGAACACATCTCGTATGCCGTCTTCTGCTT*G |
| tag60 | TCATCCTAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCATCCTAATCTCGTATGCCGTCTTCTGCTT*G |
| tag61 | TGCTGATAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGCTGATAATCTCGTATGCCGTCTTCTGCTT*G |
| tag62 | TAGACGGAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTAGACGGAATCTCGTATGCCGTCTTCTGCTT*G |
| tag63 | TGTGAAGAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGTGAAGAATCTCGTATGCCGTCTTCTGCTT*G |
| tag64 | TCTCTTCAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTCTCTTCAATCTCGTATGCCGTCTTCTGCTT*G |
| tag65 | TTGTTCCAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTTGTTCCAATCTCGTATGCCGTCTTCTGCTT*G |
| tag66 | TGAAGCCAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGAAGCCAATCTCGTATGCCGTCTTCTGCTT*G |
| tag67 | TACCACCAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTACCACCAATCTCGTATGCCGTCTTCTGCTT*G |
| tag68 | TGCGTGAAAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCTGCGTGAAATCTCGTATGCCGTCTTCTGCTT*G |
| tag69 | GGTGAGTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGGTGAGTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag70 | GATCTCTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGATCTCTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag71 | GTGTCCTTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTGTCCTTATCTCGTATGCCGTCTTCTGCTT*G |
| tag72 | GACGGATTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGACGGATTATCTCGTATGCCGTCTTCTGCTT*G |
| tag73 | GCAACATTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCAACATTATCTCGTATGCCGTCTTCTGCTT*G |
| tag74 | GGTCGTGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGGTCGTGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag75 | GAATCTGTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGAATCTGTATCTCGTATGCCGTCTTCTGCTT*G |
| tag76 | GTACATCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTACATCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag77 | GAGGTGCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGAGGTGCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag78 | GCATGGCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCATGGCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag79 | GTTAGCCTAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTTAGCCTATCTCGTATGCCGTCTTCTGCTT*G |
| tag80 | GTCGCTATAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTCGCTATATCTCGTATGCCGTCTTCTGCTT*G |
| tag81 | GGAATGATAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGGAATGATATCTCGTATGCCGTCTTCTGCTT*G |
| tag82 | GAGCCAATAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGAGCCAATATCTCGTATGCCGTCTTCTGCTT*G |
| tag83 | GCTCCTTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCTCCTTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag84 | GTAAGGTGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTAAGGTGATCTCGTATGCCGTCTTCTGCTT*G |
| tag85 | GAGGATGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGAGGATGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag86 | GTTGTCGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTTGTCGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag87 | GGATTAGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGGATTAGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag88 | GATAGAGGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGATAGAGGATCTCGTATGCCGTCTTCTGCTT*G |
| tag89 | GTGTGTCGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTGTGTCGATCTCGTATGCCGTCTTCTGCTT*G |
| tag90 | GCAATCCGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCAATCCGATCTCGTATGCCGTCTTCTGCTT*G |
| tag91 | GACCTTAGAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGACCTTAGATCTCGTATGCCGTCTTCTGCTT*G |
| tag92 | GCCTGTTCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCCTGTTCATCTCGTATGCCGTCTTCTGCTT*G |
| tag93 | GCACTGTCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCACTGTCATCTCGTATGCCGTCTTCTGCTT*G |
| tag94 | GCTAACTCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGCTAACTCATCTCGTATGCCGTCTTCTGCTT*G |
| tag95 | GATTCATCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGATTCATCATCTCGTATGCCGTCTTCTGCTT*G |
| tag96 | GTCTTGGCAT | /5PHOS/G*ATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTCGTCTTGGCATCTCGTATGCCGTCTTCTGCTT*G |
Literature Cited
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgstrom E, Lundin S, Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6:e19119. doi: 10.1371/journal.pone.0019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- DeAngelis MM, Wang DG, Hawkins TL. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins TL, O’Connor-Morin T, Roy A, Santillan C. DNA purification and isolation using a solid-phase. Nucleic Acids Res. 1994;22:4543–4544. doi: 10.1093/nar/22.21.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin S, Stranneheim H, Pettersson E, Klevebring D, Lundeberg J. Increased throughput by parallelization of library preparation for massive sequencing. PLoS One. 2010;5:e10029. doi: 10.1371/journal.pone.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M, Marmur J. [109] Use of ultraviolet absorbance-temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol. 1968;12:195–206. [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Briggs AW, Maricic T, Hober B, Hoffner B, Krause J, Weihmann A, Paabo S, Hofreiter M. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 2008;36:e5. doi: 10.1093/nar/gkm1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Gu Y, Swerdlow H, Mayho M. Evaluation and optimisation of preparative semi-automated electrophoresis systems for Illumina library preparation. Electrophoresis. 2012a;33:3521–3528. doi: 10.1002/elps.201200128. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Otto TD, Gu Y, Harris SR, Skelly TF, McQuillan JA, Swerdlow HP, Oyola SO. Optimal enzymes for amplifying sequencing libraries. Nat Methods. 2012b;9:10–11. doi: 10.1038/nmeth.1814. [DOI] [PubMed] [Google Scholar]
- Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012c;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Malek J. Asymmetrical adapters and methods of use thereof. Publication number US20070172839 A1. USPTO Application no. US 11/338,620. United States patent application. 2007