Abstract
Lapatinib, a dual inhibitor of epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2 (HER2) tyrosine kinases, has shown promising results as a growth inhibitor of HER2-positive cancer cells in vitro. However, similar to other EGFR-targeting drugs, acquired resistance to lapatinib by HER2-positive cancer cells remains a major clinical challenge. To elucidate resistance mechanisms to EGFR/HER2-targeting agents, we performed a systematic quantitative comparison of the phosphoproteome of lapatinib-resistant (LR) human gastric cancer cells (SNU216-LR) versus parental cells (SNU216) using a titanium dioxide (TiO2) phosphopeptide enrichment method and analysis with a Q-Exactive hybrid quadrupole-Orbitrap mass spectrometer. Biological network analysis of differentially expressed phosphoproteins revealed apparent constitutive activation of the MET-axis phosphatidylinositide 3-kinase (PI3K)/α-serine/threonine-protein kinase (AKT) and mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) signaling pathways in SNU216-LR. Inhibition of the PI3K/AKT and MAPK/ERK signaling pathways in SNU216-LR also leads to cell cycle arrest, confirming the biological network analysis. Lapatinib sensitivity was restored when cells were treated with several molecular targeting agents in combination with lapatinib. Thus, by integrating phosphoproteomic data, protein networks and effects of signaling pathway modulation on cell proliferation, we found that SNU216-LR maintains constitutive activation of the PI3K/AKT and MAPK/ERK pathways in a MET-dependent manner. These findings suggest that pathway activation is a key compensatory intracellular phospho-signaling event that may govern gastric cancer cell resistance to drug treatment.
Keywords: drug resistance, HER2-positive gastric cancer, lapatinib, phosphoproteins, Q-Exactive, therapeutic targets
Introduction
Human epidermal growth factor receptor 2 (HER2) is one of the receptor tyrosine kinases activated through dimerization with members of the epidermal growth factor receptor (EGFR) family.1 Overexpression of HER2 leads to tumor progression via intracellular signal transduction cascades2 and is observed in ∼20−30% of high-risk breast cancer cases,3, 4 as well as in 6−35% of invasive gastric cancers.5 Lapatinib inhibits the proliferation of cancer cells overexpressing EGFR and/or HER2 both in vitro and in vivo, and it was approved by the United States Food and Drug Administration as a treatment for advanced HER2-positive breast cancer in 2007.6, 7, 8 The antitumor activity of lapatinib has also been examined in gastric cancer cells.9, 10 A phase III clinical trial comparing the effect of chemotherapy alone versus in combination with lapatinib in HER2-positive gastric cancer patients is currently on-going.11 However, acquired resistance to lapatinib has been and remains a limitation to its therapeutic use in the clinic because of a poor understanding of the underlying mechanisms.
One possible mechanism driving therapeutic resistance is the compensatory activation of an alternate receptor tyrosine kinase that would restore the downstream signaling pathways.12, 13 Many functional studies of cancers coexpressing hepatocyte growth factor receptor (MET) and EGFR have suggested that compensatory activation of MET in the presence of EGFR blockade may be responsible for EGFR tyrosine kinase inhibitor (TKI)-acquired resistance. For example, Engelman et al.14 detected MET amplification in patients with non-small-cell lung cancer who developed clinical resistance to the EGFR inhibitors gefitinib or erlotinib.14 Yano et al.15 also found that hepatocyte growth factor-mediated MET activation is a novel mechanism of gefitinib resistance in lung adenocarcinomas containing EGFR-activating mutations. These studies imply that the functional cross-talk between MET and the EGFR family of receptor tyrosine kinases plays an important role in the adaptive cellular response that may contribute to drug resistance. More recently, the activation of MET has been shown to confer resistance to lapatinib inhibition in HER2-positive gastric cancer cells.16
To identify the affected global cellular processes compensating for the selective pressure of lapatinib on EGFR/HER2, we performed a large-scale quantitative phosphoproteome analysis of lapatinib-resistant (LR) human gastric cancer cells generated from HER2-amplified gastric cancer cells during prolonged lapatinib treatment. By integrating differentially expressed phosphoproteins into the biological network analysis tools, we found that constitutive activation of the phosphatidylinositide 3-kinase (PI3K)/α-serine/threonine-protein kinase (AKT) and mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) signaling pathways induced by the MET axis may be a critical pathway conferring acquired lapatinib resistance. The phosphoproteomic results were confirmed by cell growth inhibition studies with a single or several molecular targeting agents in combination with lapatinib. These findings provide novel insights into adaptive changes in phospho-signaling networks that occur during the development of lapatinib resistance in human HER2-positive gastric cancer cells. Our data also demonstrate that network-based global phosphoproteomic analysis is a reliable approach for predicting cellular signaling events, which in turn can lead to the identification of alternative targeting strategies for LR gastric cancer.
Materials and methods
Cell lines and reagents
Human gastric carcinoma SNU216 cells were obtained from the Seoul National University Cell Bank (Seoul, Korea). Cells were cultured in RPMI-1640 medium containing 10% (v/v) fetal bovine serum (Gibco, Carlsbad, CA, USA) at 37 °C under 5% CO2. Lapatinib was a gift from GlaxoSmithKline (Research Triangle Park, NC, USA). NVP-BEZ235, selumetinib, saracatinib and crizotinib were purchased from Selleck Chemicals (Houston, TX, USA). Stock solutions were prepared in dimethyl sulfoxide and stored at −20 °C. Ammonium bicarbonate, dithiothreitol (DTT), formic acid (FA) and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich (St Louis, MO, USA). The HPLC-grade acetonitrile (ACN) and water were from JT Baker (Phillipsburg, NJ, USA). Lyophilized trypsins were obtained from Promega (Madison, WI, USA).
Generation of LR clones from SNU216 cells in vitro
SNU216 cells were exposed to increasing concentrations of lapatinib over a period of 8 months, reaching a final concentration of 10 μM at the end of this period. Single-cell cloning was done by serial dilution or isolation with cloning cylinders, and isolated resistant cells were expanded in culture medium containing 1 μM lapatinib.
Cell growth inhibition assay
The viability of cells was assessed using MTT assays (Sigma-Aldrich). A total of 3 × 103 cells were seeded in 96-well plates, incubated at 37 °C for 24 h for recovery and treated for 72 h with specific drugs as indicated. After treatment, MTT solution was added to each well followed by incubation for 4 h at 37 °C. The medium was removed, dimethyl sulfoxide was added and the samples were mixed thoroughly for 30 min at room temperature. Cell viability was determined by measuring absorbance at 540 nm using a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Immunoblot analysis
Antibodies against p-EGFR (pY1068), p-HER2 (pY1221/1222), p-MET (pY1234/1235), p-AKT (pS473), p-MAPK (pY202/pY204), p-SRC (pY416), p-RAF (pS338), EGFR, HER2, MET, AKT, MAPK, SRC (proto-oncogene tyrosine-protein kinase Src) and RAF were purchased from Cell Signaling Technology (Beverley, MA, USA). Antibodies directed against β-catenin, cyclin D and p27 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Subconfluent cells (70−80%) were used for protein analyses. For the analysis of signaling after drug treatments, cells were treated with different concentrations of the indicated drugs for 48 h. Cell lysates were prepared with RIPA buffer, consisting of 50 mM Tris-HCl (pH 7.5), 1% (v/v) NP-40, 0.1% (w/v) sodium deoxycholate, 150 mM NaCl, 50 mM NaF, 1 mM sodium pyrophosphate, 1 mM sodium vanadate, 1 mM nitrophenylphosphate, 1 mM benzamidine, 0.1 mM PMSF, 0.1 mM aprotinin, 0.1 mM leupeptin and 0.1 mM pepstatin A, on ice for 10 min, after which the lysates were centrifuged at 14 000 × g for 20 min. Samples containing equal amounts of protein were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer of the proteins onto nitrocellulose membranes. The membranes were blocked for 1 h at room temperature with 5% (w/v) skim milk and incubated overnight at 4 °C with primary antibody. After probing with secondary antibody for 1 h at room temperature, detection was performed using an enhanced chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Cell cycle analysis
Cells were washed twice in phosphate-buffered saline, fixed in 70% (v/v) ethanol and stored at −20 °C until analysis was performed. Before analysis, thawed cell suspensions were washed with phosphate-buffered saline, digested with RNase A (50 μg ml−1) for 15 min at 37 °C and stained with propidium iodide (50 μg ml−1). The DNA content (10 000 cells per experimental group) was determined using a FACSCalibur flow cytometer (Becton Dickinson Biosciences, San Jose, CA, USA) equipped with a ModFit LT program (Verity Software House, Topsham, ME, USA), as previously described.10
One-dimensional SDS-PAGE fractionation and in-gel digestion
Cells were lysed by sonication in lysis buffer, consisting of 1% (w/v) sodium deoxycholate, 6 M urea, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM sodium orthovanadate, 0.5 mM PMSF and phosphatase and protease inhibitor (Roche, Mannheim, Germany). Protein concentration was determined using the Micro BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA). Protein samples (3 mg each) were dissolved in NuPAGE LDS sample buffer (Invitrogen, Carlsbad, CA, USA) containing 50 mM DTT and heated at 70 °C for 10 min. The cell lysates were separated on a 4−12% Bis-Tris NuPAGE gel (Invitrogen) and stained with Coomassie Brilliant Blue (Sigma-Aldrich). The entire gel lane was cut into six pieces and subjected to in-gel tryptic digestion.17 The excised gels were destained with 50% (v/v) ACN in 25 mM NH4HCO3 and in 100 mM NH4HCO3 for 15 min. After destaining, the gels were dried and reduced with 20 mM DTT at 60 °C for 1 h. They were alkylated with 55 mM iodoacetamide at room temperature for 45 min in the dark. The gel pieces were dehydrated for 15 min in 50% ACN in 25 mM NH4HCO3, and the proteins in the gels were digested overnight at 37 °C with 13 ng μl−1 sequencing-grade modified trypsin (Promega) in a volume of 50 mM NH4HCO3 sufficient to cover the gel pieces. Peptides were extracted from the gel pieces with 50% (v/v) ACN in 5% (v/v) FA and dried under a Centrivap concentrator (Labconco, Kansas City, MO, USA).
Phosphopeptide enrichment by TiO2 affinity column chromatography
Phosphopeptides were enriched using the TitanspherePhospho-TiO Kit (GL Sciences, Tokyo, Japan) following the manufacturer's protocol. Briefly, the titanium dioxide (TiO2) affinity column was conditioned with 20 μl of Buffer A (0.4% w/v TFA in 80% v/v ACN) by centrifuging twice at 3000 × g for 2 min and equilibrated with Buffer B (25% w/v lactic acid and 0.3% w/v TFA in 60% v/v ACN) by again centrifuging twice as before. The extracted peptides, which had been resuspended in 150 μl Buffer B, were loaded onto the column. For sample adsorption, the column was first centrifuged at 1000 × g for 10 min, and the flow-through was loaded onto the column and centrifuged again. The column was washed three times with 20 μl Buffer B by centrifugation at 3000 × g for 2 min, followed by washing with 20 μl Buffer A by centrifugation under the same conditions. Phosphorylated peptides were eluted twice with 50 μl 5% (v/v) NH4OH and 50 μl 30% ACN by centrifugation at 1000 × g for 5 min. The eluate was evaporated in a Centrivap concentrator and stored at −20 °C until liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Mass spectrometric analysis
Peptides were reconstituted in 2% (v/v) ACN in 0.4% (v/v) FA and separated on an in-house packed micro-capillary Michrom Magic C18 column (75 μm inner diameter, 12 cm length) by elution with a linear gradient of 2−32% Solvent B (0.1% v/v FA in ACN) in Solvent A (0.1% v/v FA in water) for 65 min at a flow rate of 300 nl min−1. Samples were analyzed in triplicate on a Q-Exactive (Thermo Fisher Scientific, San Jose, CA, USA) hybrid quadrupole-Orbitrap mass spectrometry (MS), interfaced with a nano-ultra performance liquid chromatography system (Easy nLC, Thermo Fisher Scientific). The spray voltage was set to 2.0 kV, and the temperature of the heated capillary was set to 250 °C. The Q-Exactive was operated in data-dependent mode with one survey MS scan followed by 10 MS/MS scans and a dynamic exclusion time of 60 s. The full scans were acquired in the mass analyzer at 400−1600 m/z with the resolution of 70 000, and the MS/MS scans were obtained with a resolution of 17 500 by using a normalized collision energy of 27% for high-energy collisional dissociation fragmentation. The automatic gain control target was 5e,4 the maximum injection time was 120 ms and the isolation window was set to 3 m/z.
Database search and identification of the phosphoproteome
Collected raw files were converted into mzXML files through the Trans Proteomic Pipeline (version 4.5; Seattle Proteomic Center, Seattle, WA, USA). The peptides were assigned by the SEQUEST algorithm (Thermo Fisher Scientific, version 27, rev. 11) against the decoy IPI.HUMAN database (v.3.18, 120180 entries), and the false discovery rate (FDR) was estimated by Sorcerer (Sage-N Research, Milpitas, CA, USA). All searches were carried out based on trypsin specificity, allowing two missed cleavages. Carbamidomethylation of cysteine was set as fixed, and oxidation of methionine, phosphorylation of serine, threonine and tyrosine were set as variable modifications. The search considered a precursor ion mass tolerance of 10 p.p.m., and the fragment ion mass tolerance was set to 1.0 Da Scaffold (version 3.4.9; Proteome Software, Portland, OR) was used to compare spectral counts, validate MS/MS-based peptides and identify matching proteins according to the criteria of 99% minimum protein probability by the Protein Prophet algorithm,18 with 80% minimum peptide probability and one minimum peptide (FDR <0.5%) as specified by the Peptide Prophet algorithm.19 Proteins that contained redundant peptides and multiple isoforms, which could not be differentiated based on MS/MS spectra, were grouped into a primarily assigned protein. Scaffold PTM (version 1.1.3; Proteome Software) was used to annotate Post-Translational Modification (PTM) sites derived from MS/MS sequencing results. The Scaffold PTM assigned possible phosphorylation sites from the Scaffold validated tandem data using the Ascore algorithm developed by Beausoleil et al.20
Quantification and bioinformatic analysis of phosphoproteins
To quantify phosphoproteins, we used spectral counts of phosphopeptides from triplicate samples of SNU216 and SNU216-LR. The data set was entered into the R program (version 2.15) with power law global error model (PLGEM, version 1.28), a statistical analysis software package (www.bioconductor.org) used to determine signal-to-noise ratio and P-value.21 Based on PLGEM, the annotation of protein cellular localization and evaluation of biological function was performed after uploading the signal-to-noise ratio and P-value to the Ingenuity Pathway Analysis (IPA; Ingenuity Systems; Redwood City, CA, USA) database content version 14197757. Hence, the phosphopeptides of the identified proteins, including those in proteins of the MET-mediated MAPK/ERK and PI3K/AKT signaling pathways, were manually quantified by the XCalibur program (Thermo Fisher Scientific). The quantification of precursor ion for the first isotopes of each ion was performed using the area of the peptides present in the raw data set across all samples in extracted ion chromatograms.
Results
Identification of phosphoproteins in SNU216 and SNU216-LR cells
To investigate possible mechanisms responsible for acquiring resistance to HER2-targeted therapy in gastric cancer, we established a LR cell line (SNU216-LR) by chronically exposing HER2-positive SNU216 cells (lapatinib-hypersensitive gastric cancer cells) to increasing concentrations of lapatinib in vitro for 8 months. A LR cell clone (SNU216-LR) was selected and characterized. The half-maximal inhibitory concentration value in SNU216-LR cells was >10 μM, which is 500-fold higher than that of parental cells (0.02 μM).
Next, we performed a large-scale quantitative phosphoproteome analysis to identify differentially phosphorylated proteins (DPPs) between SNU216-LR and SNU216 cells. A total of 6556 unique phosphopeptides representing 1686 phosphoproteins was identified with high confidence by MS analysis of the TiO2-enriched phosphopeptides (protein: probability >99% and FDR <1.5% peptide: probability >80% and FDR <0.5%), and 97.7% of proteins overlapped between SNU216 and SNU216-LR cell lines (Supplementary Figure 1A and Supplementary Tables 1 and 2). We then determined the phosphorylation sites of all identified phosphopeptides using Ascore and localization probability by Scaffold PTM.20 Our approach identified 7791 phosphorylation sites (Ascore >80%) among 11 852 unique phosphorylation sites (Supplementary Table 3). Serine was found to be the most abundantly phosphorylated residue (9745 sites, 82%), followed by threonine (1763 sites, 15%) and tyrosine (344 sites, 3%) (Supplementary Figure 1B). The reproducibility of detecting the same proteins in triplicate analysis by LC-MS/MS exceeded 91% for both cell lines (Supplementary Figures 1C and 1D).
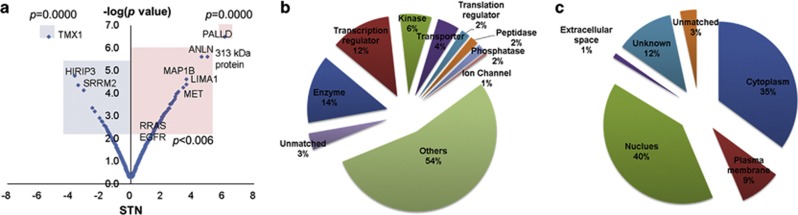
To estimate quantitative changes in the phosphorylation of each phosphopeptide, we selected DPPs in SNU216-LR cells using the spectral counting method in Scaffold (version 3.4.9). The signal-to-noise ratio and P-values for the DPPs were obtained using PLGEM. A list of 95 significantly regulated phosphoproteins (P<0.006) is presented in Table 1. As shown in the volcano plot, 69 proteins were upregulated (red box), and 26 proteins were downregulated (blue box) in SNU216-LR cells (Figure 1a). To understand the functions of the DPPs, we examined cellular processes represented by the 95 regulated proteins using the IPA tool and found that they belong to enzyme (14%), transcription regulator (12%) and kinase (6%) groups of proteins (Figure 1b). We also examined the cellular distribution of these proteins and found that most were associated with either the nucleus (40%) or cytoplasm (35%), with fewer associated with the membrane (9%) (Figure 1c).
Table 1. Differentially expressed phosphoproteins in SNU216-LR (P<0.006).
| ID | Protein | Symbol | Location | Type(s) | S/N | P-value |
|---|---|---|---|---|---|---|
| IPI00292009 | Palladin | PALLD | Cytoplasm | Other | 6.09 | 0.00000 |
| IPI00555610 | 313 kDa protein | — | — | — | 4.97 | 0.00000 |
| IPI00657687 | Isoform 1 of Actin-binding protein anillin | ANLN | Cytoplasm | Other | 4.60 | 0.00000 |
| IPI00008868 | Microtubule-associated protein 1B | MAP1B | Cytoplasm | Other | 3.61 | 0.00002 |
| IPI00008918 | Isoform β of LIM domain and actin-binding protein 1 | LIMA1 | Cytoplasm | Other | 3.61 | 0.00004 |
| IPI00291802 | Isoform 3 of LIM domain only protein 7 | LMO7 | Cytoplasm | Enzyme | 3.42 | 0.00005 |
| IPI00304589 | 182 kDa tankyrase 1-binding protein | TNKS1BP1 | Nucleus | Other | 3.10 | 0.00008 |
| IPI00029273 | Isoform 1 of Hepatocyte growth factor receptor precursor | MET | Plasma membrane | Kinase | 3.01 | 0.00010 |
| IPI00009286 | Isoform 1 of zinc finger protein HRX | MLL | Nucleus | Transcription regulator | 2.96 | 0.00010 |
| IPI00219301 | Myristoylated alanine-rich C-kinase substrate | MARCKS | Plasma membrane | Other | 2.90 | 0.00014 |
| IPI00745518 | Microtubule-associated protein 4 isoform 1 variant (Fragment) | — | — | — | 2.88 | 0.00018 |
| IPI00013076 | Isoform 1 of FGFR1 oncogene partner | FGFR1OP | Cytoplasm | Other | 2.77 | 0.00021 |
| IPI00028481 | Ras-related protein Rab-8A | RAB8A | Plasma membrane | Other | 2.68 | 0.00026 |
| IPI00006196 | Isoform 2 of Nuclear mitotic apparatus protein 1 | NUMA1 | Nucleus | Other | 2.65 | 0.00028 |
| IPI00329115 | Isoform 1 of protein C10orf47 | C10orf47 | Unknown | Other | 2.57 | 0.00030 |
| IPI00289334 | Isoform 1 of filamin-B | FLNB | Cytoplasm | Other | 2.55 | 0.00030 |
| IPI00658152 | Tensin-like SH2 domain containing 1 | TNS3 | Unknown | Phosphatase | 2.54 | 0.00030 |
| IPI00237884 | Isoform 1 of A-kinase anchor protein 12 | AKAP12 | Cytoplasm | Transporter | 2.51 | 0.00046 |
| IPI00216219 | Isoform long of tight junction protein ZO-1 | TJP1 (includes EG:21872) | Plasma membrane | Other | 2.48 | 0.00046 |
| IPI00218753 | Isoform 3 of DNA topoisomerase 2-α | TOP2A | Nucleus | Enzyme | 2.47 | 0.00046 |
| IPI00294742 | HDCMA18P protein | LARP7 | Unknown | Other | 2.45 | 0.00047 |
| IPI00020513 | Zyxin | ZYX | Plasma membrane | Other | 2.36 | 0.00060 |
| IPI00298289 | Isoform 2 of reticulon-4 | RTN4 | Cytoplasm | Other | 2.27 | 0.00068 |
| IPI00157144 | GYS1 protein | GYS1 | Cytoplasm | Enzyme | 2.21 | 0.00076 |
| IPI00010471 | Plastin-2 | LCP1 | Cytoplasm | Other | 2.20 | 0.00078 |
| IPI00178440 | Elongation factor 1-β | EEF1B2 | Cytoplasm | Translation regulator | 2.18 | 0.00078 |
| IPI00220740 | Isoform 2 of nucleophosmin | NPM1 | Nucleus | Transcription regulator | 2.16 | 0.00081 |
| IPI00552897 | Isoform 1 of mediator of DNA damage checkpoint protein 1 | MDC1 | Nucleus | Other | 2.16 | 0.00083 |
| IPI00294840 | Absent in melanoma 1 protein | AIM1 (includes EG:11630) | Unknown | Other | 2.15 | 0.00084 |
| IPI00031605 | AHNAK nucleoprotein, isoform 2 | AHNAK | Nucleus | Other | 2.12 | 0.00098 |
| IPI00015029 | Prostaglandin E synthase 3 | PTGES3 | Cytoplasm | Enzyme | 2.11 | 0.00099 |
| IPI00032064 | Isoform A of A-kinase anchor protein 2 | — | — | — | 2.10 | 0.00099 |
| IPI00152011 | PTPL1-associated RhoGAP 1 | ARHGAP29 | Cytoplasm | Other | 2.10 | 0.00101 |
| IPI00015953 | Isoform 1 of nucleolar RNA helicase 2 | DDX21 | Nucleus | Enzyme | 2.09 | 0.00101 |
| IPI00004233 | Isoform long of antigen KI-67 | MKI67 | Nucleus | Other | 2.08 | 0.00103 |
| IPI00027831 | Glutamate-rich WD repeat-containing protein 1 | GRWD1 | Nucleus | Other | 2.02 | 0.00111 |
| IPI00419933 | Pleckstrin homology domain containing, family A member 2 | PLEKHA7 | Cytoplasm | Other | 2.02 | 0.00111 |
| IPI00017450 | Transcription initiation factor IIF α subunit | GTF2F1 | Nucleus | Transcription regulator | 2.01 | 0.00132 |
| IPI00219841 | DNA ligase 1 | LIG1 | Nucleus | Enzyme | 1.97 | 0.00144 |
| IPI00218136 | 137 kDa protein | CGN | Plasma Membrane | Other | 1.95 | 0.00149 |
| IPI00020021 | Protein DEK | DEK | nucleus | Transcription regulator | 1.92 | 0.00161 |
| IPI00031698 | Hypothetical protein MGC5509 | C2orf49 | Unknown | Other | 1.90 | 0.00177 |
| IPI00033054 | CTD small phosphatase like 2 | CTDSPL2 | Unknown | Other | 1.89 | 0.00179 |
| IPI00376199 | Interferon regulatory factor 2 binding protein 2 | IRF2BP2 | Nucleus | Transcription regulator | 1.85 | 0.00195 |
| IPI00411635 | KIAA0802 protein | — | — | — | 1.83 | 0.00202 |
| IPI00005688 | Zinc finger protein 185 | ZNF185 | Nucleus | Other | 1.82 | 0.00206 |
| IPI00470483 | WD repeat domain 62 | WDR62 | Nucleus | Other | 1.77 | 0.00219 |
| IPI00152881 | Shroom-related protein | SHROOM3 | Cytoplasm | Other | 1.76 | 0.00225 |
| IPI00337426 | Isoform 1 of BMP-2-inducible protein kinase | BMP2K | Nucleus | Kinase | 1.76 | 0.00225 |
| IPI00025512 | Heat-shock protein β-1 | HSPB1 | Cytoplasm | Other | 1.73 | 0.00265 |
| IPI00456887 | Scaffold attachment factor A2 | HNRNPUL2 | Nucleus | Other | 1.71 | 0.00272 |
| IPI00020418 | Ras-related protein R-Ras | RRAS | Cytoplasm | Enzyme | 1.70 | 0.00277 |
| IPI00291175 | Isoform 1 of vinculin | VCL | Plasma membrane | Enzyme | 1.66 | 0.00291 |
| IPI00010746 | Phosphatidylserine synthase 1 | PTDSS1 | Cytoplasm | Enzyme | 1.61 | 0.00336 |
| IPI00216654 | Isoform β of nucleolar phosphoprotein p130 | NOLC1 | Nucleus | Transcription regulator | 1.59 | 0.00365 |
| IPI00645814 | KIAA1187 protein | MAP7D1 | Unknown | Other | 1.58 | 0.00370 |
| IPI00019996 | modulator of estrogen induced transcription isoform b | SLTM | Nucleus | Other | 1.56 | 0.00388 |
| IPI00013743 | Hypothetical protein BUD13 | BUD13 (includes EG:215051) | Nucleus | Other | 1.54 | 0.00400 |
| IPI00024579 | Postreplication repair protein RAD18 | RAD18 | Nucleus | Other | 1.52 | 0.00406 |
| IPI00025087 | Isoform 1 of Cellular tumor antigen p53 | TP53 (includes EG:22059) | Nucleus | Transcription regulator | 1.52 | 0.00406 |
| IPI00008708 | Ribosomal L1 domain-containing protein 1 | RSL1D1 | Cytoplasm | Other | 1.51 | 0.00448 |
| IPI00295457 | Myosin phosphatase-Rho interacting protein isoform 1 | MPRIP | Cytoplasm | Other | 1.50 | 0.00449 |
| IPI00002135 | Transforming acidic coiled-coil-containing protein 3 | TACC3 | Nucleus | Other | 1.49 | 0.00458 |
| IPI00184317 | Isoform short of transcription intermediary factor 1-α | TRIM24 | Nucleus | Transcription regulator | 1.49 | 0.00508 |
| IPI00394829 | Hypothetical protein LOC286077 | FAM83H | Unknown | Other | 1.48 | 0.00512 |
| IPI00294618 | Proline-rich protein PRCC | PRCC | Nucleus | Other | 1.45 | 0.00534 |
| IPI00181905 | Isoform 1 of protein KIAA1914 | AFAP1L2 | Cytoplasm | Other | 1.44 | 0.00560 |
| IPI00302829 | Retinoblastoma-associated protein | RB1 | Nucleus | Transcription regulator | 1.43 | 0.00569 |
| IPI00018274 | Isoform 1 of epidermal growth factor receptor precursor | EGFR | Plasma membrane | Kinase | 1.40 | 0.00598 |
| IPI00289819 | Cation-independent mannose-6-phosphate receptor precursor | IGF2R | Plasma membrane | Transmembrane receptor | −1.39 | 0.00592 |
| IPI00019870 | Isoform α of caveolin-2 | CAV2 | Plasma membrane | Other | −1.43 | 0.00543 |
| IPI00301294 | Protein C2orf17 | FAM134A | Unknown | Other | −1.43 | 0.00543 |
| IPI00100151 | 5′–3′ exoribonuclease 2 | XRN2 | Nucleus | Enzyme | −1.44 | 0.00528 |
| IPI00010080 | Serine/threonine-protein kinase OSR1 | OXSR1 | Nucleus | Kinase | −1.45 | 0.00520 |
| IPI00719818 | Nucleolar protein 5A | NOP56 | Nucleus | Other | −1.46 | 0.00496 |
| IPI00011635 | Isoform 2 of Bcl-2-like 13 protein | BCL2L13 | Cytoplasm | Other | −1.47 | 0.00482 |
| IPI00006987 | ATP-dependent RNA helicase DDX24 | DDX24 | Nucleus | Enzyme | −1.49 | 0.00474 |
| IPI00032107 | Sulfate transporter | SLC26A2 | Plasma membrane | Transporter | −1.49 | 0.00465 |
| IPI00297859 | Isoform 1 of myeloid/lymphoid or mixed-lineage leukemia protein 2 | MLL2 | Nucleus | Transcription regulator | −1.49 | 0.00465 |
| IPI00019502 | Myosin-9 | MYH9 | Cytoplasm | Enzyme | −1.54 | 0.00377 |
| IPI00004273 | RNA binding motif protein 25 | RBM25 | Nucleus | Other | −1.54 | 0.00377 |
| IPI00009146 | FLN29 gene product | TRAFD1 | Unknown | Other | −1.62 | 0.00291 |
| IPI00719759 | Akt substrate AS250 | RALGAPA2 | Cytoplasm | Other | −1.62 | 0.00291 |
| IPI00396634 | OTTHUMP00000028561 | KIAA1671 | Unknown | Other | −1.65 | 0.00274 |
| IPI00217709 | Isoform β-1 of DNA topoisomerase 2-β | TOP2B | Nucleus | Enzyme | −1.72 | 0.00251 |
| IPI00025156 | Isoform 1 of STIP1 homology and U box-containing protein 1 | STUB1 | Cytoplasm | Enzyme | −1.84 | 0.00180 |
| IPI00554788 | Keratin, type I cytoskeletal 18 | KRT18 | Cytoplasm | Other | −1.84 | 0.00179 |
| IPI00061758 | PRR8 protein | RBM33 | Unknown | Other | −1.89 | 0.00168 |
| IPI00304587 | Dispatched B | DISP2 | Unknown | Other | −2.02 | 0.00120 |
| IPI00021275 | Isoform 1 of ephrin type-B receptor 2 precursor | EPHB2 | Plasma membrane | Kinase | −2.33 | 0.00063 |
| IPI00024062 | Band 4.1-like protein 1 | EPB41L1 | Plasma membrane | Other | −2.49 | 0.00044 |
| IPI00744036 | Tight junction protein 3 | TJP3 | Plasma membrane | Other | −3.04 | 0.00007 |
| IPI00748920 | Splicing coactivator subunit SRm300 | SRRM2 | Nucleus | Other | −3.40 | 0.00004 |
| IPI00304875 | Isoform 1 of HIRA-interacting protein 3 | HIRIP3 | Nucleus | Other | −3.62 | 0.00002 |
| IPI00395887 | Thioredoxin domain-containing protein 1 precursor | TMX1 | Cytoplasm | Enzyme | −5.34 | 0.00000 |
Abbreviation: S/N, signal-to-noise ratio.
Up- or down-regulated phosphoproteins in SNU216-LR compared with SNU216 cells. S/N and P-values were generated by the power law global error model (PLGEM) statistical analysis tool.
Figure 1.
Differentially expressed phosphoproteins in SNU216-LR and their functional annotations. (a) Volcano plot showing the P-value (−log10) versus signal-to-noise ratio (S/N) of phosphoproteins determined by power law global error model (PLGEM) statistical analysis for label-free quantification. (b) Molecular functions of identified phosphoproteins by the Ingenuity Pathway Analysis (IPA) tool. (c) Subcellular localization of identified phosphoproteins by the IPA tool.
We further evaluated quantitative changes in phosphorylation of proteins using label-free quantification methods (MS/MS spectral count and extraction ion chromatography) that are generally accepted in phosphoproteomic studies.22, 23, 24 More MS/MS spectral counts of phosphorylated EGFR and HER2 peptides were identified in SNU216-LR than SNU216 cells (Supplementary Figures 2A and 2B). These data were consistent with the peptide extracted ion chromatogram, which showed significantly greater signal intensities of phosphorylated peptides in SNU216-LR cells (Supplementary Figures 2C and 2D). Using the same quantification approaches, we were able to measure the phosphorylation levels of key molecules (MET, RAS, C-RAF, ERK1/2, PI3K and AKT; see Supplementary Figures 2E–J). In addition, we found a new phosphorylation site in HER2 (S1073) based on the phosphopeptide reference database phosphoSitePlus (www.phosphosite.org). This phosphorylation site was confirmed by the matched tandem mass spectra shown in Supplementary Figure 3.
Network analysis and quantification of differentially phosphorylated proteins
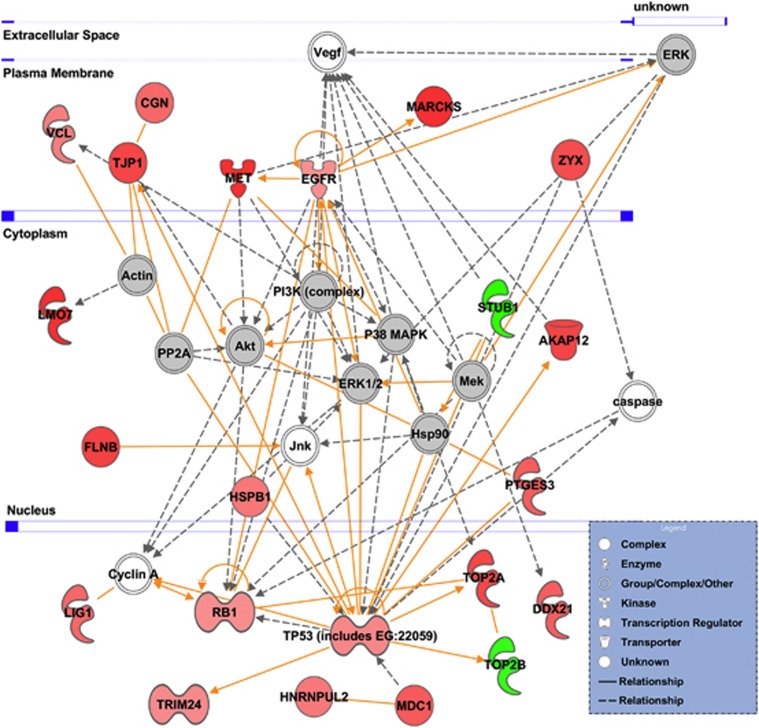
To investigate the extent to which activation of signaling pathways contributes to EGFR/HER2-targted therapy, we integrated the DPPs (P<0.006) into the protein interaction network using the IPA tool to interpret the DPPs in the context of biological processes. The IPA tool generated the top three networks that are associated with cancer, cell morphology and cellular and tissue development (Supplementary Figure 4). Considering this network analysis, our upregulated phosphoproteins, including EGFR and MET, appeared to be closely related to PI3K, AKT and ERK1/2 in SNU 216-LR (Figure 2).
Figure 2.
Biological network analysis of phosphoproteins. Associations among phosphoproteins are shown by solid or dashed lines, which represent direct or indirect interactions, respectively. Upregulated proteins are shown in red, and downregulated proteins are shown in green.
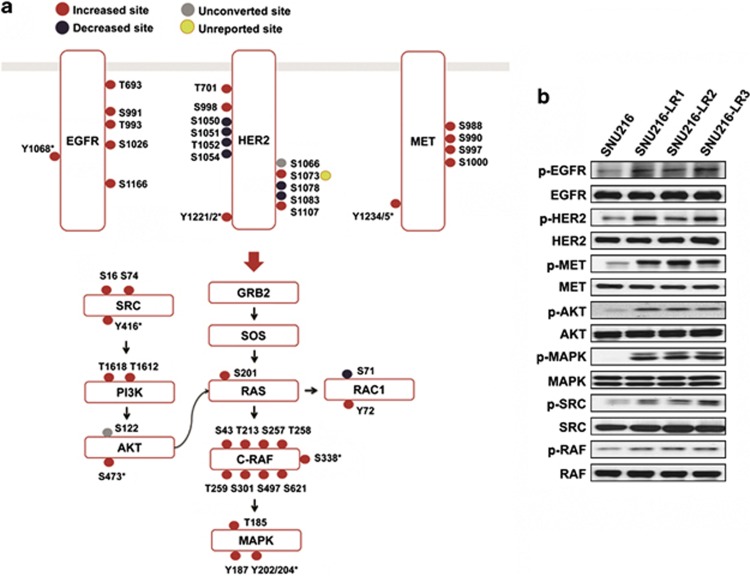
Our analysis of results from the IPA tool also revealed that EGFR/HER2, MET and ERK/MAPK signaling pathways were enriched (Supplementary Figure 5). Based on the results from the network and canonical signaling pathway analyses, we constructed the signaling network for EGFR/HER2 and MET with the two major downstream signaling pathways (PI3K/AKT and MAPK/ERK) including SRC (Figure 3). As shown in Figure 3a, categorization of expression levels of specific phosphorylated sites revealed that the majority of signaling molecules were highly expressed in SNU216-LR. We further compared SNU216 and SNU216 LR, as well as three other resistant clones (SNU216-LR1, 2 and 3), using immunoblot analysis with phospho-specific antibodies. Consistent with the quantitative MS results, the phosphorylation level of selected proteins (EGFR, HER2, MET, AKT, MAPK, SRC, and RAF) was increased in SNU216-LR (Figure 3b).
Figure 3.
Merged signaling pathways in SNU216-LR. (a) Reconstructed EGFR/HER2 and MET signaling pathways with two downstream signaling pathways (PI3K/AKT and MAPK/ERK). Red, blue and yellow closed circles on each protein represent increased, decreased and newly identified phosphorylation sites in SNU216-LR, respectively. Specific phosphorylated sites (marked with an asterisk) of HER2, EGFR, MET, SRC, C-RAF and MAPK were also identified from immunoblot analysis with antibodies against p-EGFR (pY1068), p-HER2 (pY1221/1222), p-MET (pY1234/1235), p-AKT (pS473), p-MAPK (pY202/pY204), p-SRC (pY416) and p-RAF (pS338). Protein names for symbols are as follows: AKT, α-serine/threonine-protein kinase; C-RAF, RAF proto-oncogene serine/threonine-protein kinase; EGFR, epidermal growth factor receptor; ERK, extracellular signal-regulated kinase; HER2, human epidermal growth factor receptor 2; MAPK, mitogen-activated protein kinase 1; GRB2, growth factor receptor-bound protein 2; MET, hepatocyte growth factor/mesenchymal–epithelial transition factor; PI3K, phosphatidylinositide 3-kinase; RAC1, Ras-related C3 botulinum toxin substrate 1; RAS, Ras-related protein R-Ras; SOS, son of sevenless homolog; SRC, proto-oncogene tyrosine protein kinase Src. (b) Immunoblot analysis of SNU216 and three different resistant clones (SNU216-LR 1, 2 and 3) with phospho-tyrosine or serine-specific antibodies. α-Tubulin was used as a loading control. Data are representative of three independent experiments.
Effects of targeted anticancer agents on SNU216-LR cells
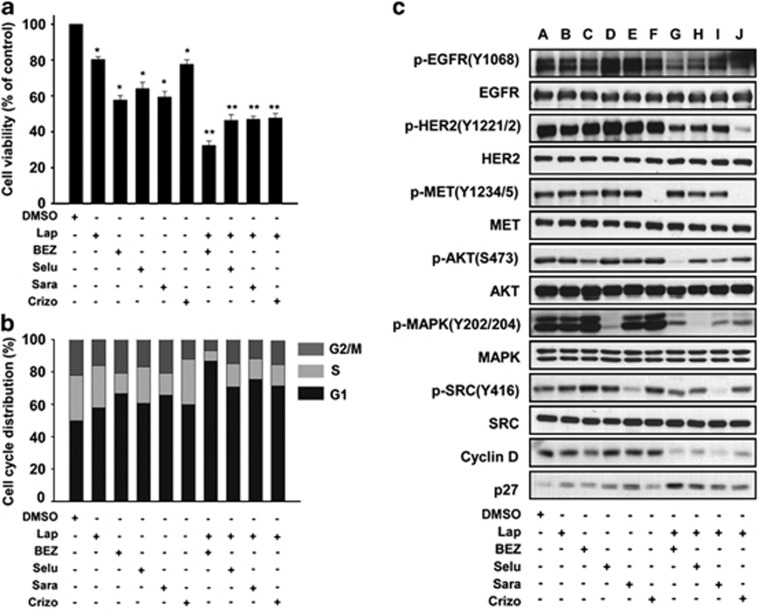
To verify whether constitutive activation of the PI3K/AKT and MAPK/ERK signaling pathways is the MET-mediated cascade that confers an acquired lapatinib resistance, we investigated the effects of selected kinase inhibitors, including lapatinib, NVP-BEZ235 (PI3K/mTOR inhibitor), selumetinib (MEK inhibitor), saracatinib (SRC inhibitor) and crizotinib (MET inhibitor), on the growth of SNU216-LR cells. As shown in Figure 4a, combination treatment with these drugs had a greater inhibitory effect on cell growth in SNU216-LR cells compared with single-agent treatment. The combination of lapatinib and NVP-BEZ235 produced the best growth inhibition effect. To confirm our data, we investigated cell cycle arrest by staining the cells with the DNA-intercalating dye propidium iodide. Combination treatment with lapatinib and NVP-BEZ235, selumetinib, saracatinib or crizotinib induced an increase in the percentage of SNU216-LR cells in the G1 phase compared with single-agent treatment (Figure 4b). Next, we examined the levels of total and phosphorylated EGFR, HER2, MET, AKT, MAPK, SRC, cyclin D and p27 in SNU216-LR cells (Figure 4c). As expected, treatment with NVP-BEZ235, selumetinib, sacaratinib or crizotinib alone reduced the phosphorylation level of each target or of the downstream effectors (AKT, MAPK, SRC and MET, respectively). The phosphorylation levels of the selected signaling molecules remained the same after treatment with dimethyl sulfoxide (control) or lapatinib. Treatment with a single kinase inhibitor had no effect on the expression of cell cycle regulatory molecules, including cyclin D. The combination treatment of lapatinib with a kinase inhibitor showed markedly decreased phosphorylation levels in each of the target molecules, such as EGFR and HER2, as well as in the downstream effectors, compared with single treatment. Furthermore, the levels of cell proliferation-related cyclin D were reduced, and those of the cell cycle inhibitor p27 were increased, after combination treatment. The levels of p-SRC in SNU216-LR were not affected by individual TKI treatment or the combination treatment except the SRC inhibitor (saracatinib) when p-SRC was monitored with the specific tyrosine antibody (Y416). As SRC is known to be an essential communicator between EGFR and MET in LR cancer cells,25 maintaining SRC activation may be necessary for efficient compensatory TKI activation for acquired TKI resistance.
Figure 4.
Antiproliferative effects of selected tyrosine kinase inhibitors on SNU216-LR. SNU216-LR cells were treated via single or combined treatment of lapatinib (Lap; 1 μM), NVP-BEZ235 (BEZ; 100 nM), selumetinib (Selu; 1 μM), saracatinib (Sara; 1 μM) and crizotinib (Crizo; 1 μM). (a) Viable cells were measured after 72 h. The percentage of viable cells is shown relative to untreated controls. Data represent the mean±s.d. (*P<0.05 for single vs dimethyl sulfoxide (DMSO); **P<0.05 for combination vs single) from three independent experiments. (b) Cells were fixed with 70% (v/v) ethanol, stained with propidium iodide and analyzed using flow cytometry. The proportion of cells in the G1, S and G2/M phases were quantified using the ModFit LT program (Verity Software House). Columns represent the means of three independent experiments. (c) SNU216-LR cells were treated with the indicated drug for 48 h. Whole-cell extracts were analyzed by western blot with antibodies recognizing phosphorylated and total EGFR, HER2, MET, AKT, MAPK, SRC, cyclin D and p27. Data are representative of three independent experiments. AKT, α-serine/threonine-protein kinase; EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor receptor 2; MAPK, mitogen-activated protein kinase 1; MET, hepatocyte growth factor/mesenchymal–epithelial transition factor; SRC, proto-oncogene tyrosine protein kinase Src.
Discussion
Lapatinib inhibits growth of HER2-positive gastric cancer cell lines.9 Presently, a phase III clinical trial in patients with advanced gastric cancer is on-going, studying the effects of lapatinib in combination with capecitabine plus oxaliplatin.26 Interest in this trial reflects lapatinib being regarded as one of the promising chemotherapeutic drugs for gastric cancer treatment. On the other hand, the medium duration of response to lapatinib is <1 year, and almost 80% of trastuzumab-pretreated patients fail to respond to lapatinib.27 Hence, understanding the mechanisms of acquired lapatinib resistance may help to improve the therapeutic efficacy of lapatinib in HER2-positive cancers.
To identify molecular changes conferring resistance to lapatinib at the system level, we performed a quantitative phosphoprotein profiling of the isogenic gastric cancer cell lines SNU216-LR and SNU216, in which we identified a total of 11 852 phosphorylation sites on 1686 phosphoproteins. Integrating the DPPs into cellular signaling pathways using the IPA network analysis tool provided potential signaling pathways that are regulated in response to lapatinib-induced resistance in SNU216-LR. Among the predominant signaling pathways generated from the IPA analysis, we highlighted the following: three receptor tyrosine kinases (EGFR, HER2 and MET); the two major downstream signaling pathways (PI3K/AKT and MAPK/ERK); and SRC. We found that the PI3K/AKT and MAPK/ERK signaling pathways and SRC were constitutively activated in SNU216-LR cells. This observation is similar to those of Rexer et al.,28 who observed active PI3K/AKT and MAPK pathways, including increased phosphorylation of SRC, in LR HER2-overexpressing human breast cancer cells. Activation of the SRC pathway is essential for communication between EGFR and MET.25 For example, lateral signaling in the EGFR/SRC/MET axis was indicated by a delay in the EGFR activation of MET in non-small-cell lung cancer produced by an inhibitor of SRC. Interestingly, our data in SNU216-LR cells show a similar pattern of lateral activation of EGFR/SRC/MET, suggesting that cross-talk between EGFR and MET may play a significant role in the regulation of SNU216-LR cells. Our conclusions are also supported by the fact that functional cross-talk between EGFR and MET has been extensively documented as playing an important role in drug resistance. Engelman et al.14 have demonstrated that the compensatory activation of MET is responsible for resistance to EGFR-targeting drugs.14 Puri et al.29 also observed that inhibition of MET in non-small-cell lung cancer is synergistic with EGFR inhibition.
Having found the constitutive activation in LR gastric cancer cells of the PI3K/AKT and MAPK/ERK signaling pathways, as well as activation of SRC, we hypothesized that the constitutive activation of PI3K/AKT and MAPK/ERK confers resistance to lapatinib and may be mediated by MET-axis signaling along the two major downstream pathways. Our data are consistent with the notion of compensatory activation of MET-mediated downstream cellular signaling pathway in response to EGFR blockade.30, 31 Results from several studies have indicated that MET activation is associated with both primary and acquired resistance to EGFR inhibitors in non-small-cell lung cancer.14, 15, 32, 33 Therefore, we postulate that constitutive activation of the MET-axis PI3K/AKT and MAPK/ERK signaling pathways, including the lateral activation of SRC, is a compensatory signaling cascade counteracting the selective pressure on EGFR/HER2 in LR human gastric cancer cells.
In the present study, we found that cotreatment with lapatinib and crizotinib is able to induce lapatinib sensitivity. Thus, the active PI3K/AKT and MAPK/ERK signaling pathways induced by the MET axis may be a critical compensatory intracellular event for the survival of LR cells.28 To the best of our knowledge, the present study is the first large-scale profiling of peptide/protein phosphorylation in gastric cancer cell lines to study lapatinib resistance. Our site-specific and semiquantitative approach demonstrates a useful bioinformatics application, and phosphoproteomic data from the study provide information directly relevant to the mechanisms of lapatinib resistance.
In conclusion, we demonstrated that network-based global phosphoproteomic analysis is a reliable approach for predicting cellular signaling events in LR gastric cancer cells. Our results suggest that the MET-directed PI3K/AKT and MAPK/ERK cellular signaling pathways contribute to lapatinib resistance, and the constituent signaling molecules may be potential targets for the development of complementary therapeutic options. Therefore, the validity of MET-axis PI3K/AKT and MAPK/ERK signaling pathways should be evaluated using clinical samples and will be the subject of further investigations.
Acknowledgments
This research was supported by Grant No. R31-2008-000-10103-0 from the World Class University program of the Ministry of Education, Science, and Technology, the National Research Foundation of Korea and the Proteogenomic Research Program through the National Research Foundation of Korea.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Monsey J, Shen W, Schlesinger P, Bose R. Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system. J Biol Chem. 2010;285:7035–7044. doi: 10.1074/jbc.M109.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa GM, Abraham J. Lapatinib in the treatment of breast cancer. Expert Rev Anticancer Ther. 2007;7:1183–1192. doi: 10.1586/14737140.7.9.1183. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- De Vita F, Giuliani F, Silvestris N, Catalano G, Ciardiello F, Orditura M. Human epidermal growth factor receptor 2 (HER2) in gastric cancer: a new therapeutic target. Cancer Treat Rev. 2010;36 (Suppl 3:S11–S15. doi: 10.1016/S0305-7372(10)70014-1. [DOI] [PubMed] [Google Scholar]
- Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N, et al. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther. 2001;1:85–94. [PubMed] [Google Scholar]
- Cameron DA, Stein S. Drug insight: intracellular inhibitors of HER2--clinical development of lapatinib in breast cancer. Nature clinical practice. Oncology. 2008;5:512–520. doi: 10.1038/ncponc1156. [DOI] [PubMed] [Google Scholar]
- Ryan Q, Ibrahim A, Cohen MH, Johnson J, Ko CW, Sridhara R, et al. FDA drug approval summary: lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2. Oncologist. 2008;13:1114–1119. doi: 10.1634/theoncologist.2008-0816. [DOI] [PubMed] [Google Scholar]
- Kim JW, Kim HP, Im SA, Kang S, Hur HS, Yoon YK, et al. The growth inhibitory effect of lapatinib, a dual inhibitor of EGFR and HER2 tyrosine kinase, in gastric cancer cell lines. Cancer Lett. 2008;272:296–306. doi: 10.1016/j.canlet.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Kim HP, Yoon YK, Kim JW, Han SW, Hur HS, Park J, et al. Lapatinib, a dual EGFR and HER2 tyrosine kinase inhibitor, downregulates thymidylate synthase by inhibiting the nuclear translocation of EGFR and HER2. PLoS One. 2009;4:e5933. doi: 10.1371/journal.pone.0005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GlaxoSmithKline. Lapatinib optimization study in ErbB2 (HER2) positive gastric cancer: a phase III global, blinded study designed to evaluate clinical endpoints and safety of chemotherapy plus lapatinib http://clinicaltrials.gov/ct2/show/NCT006809012008
- Nahta R, Yuan LXH, Du Y, Esteva FJ. Lapatinib induces apoptosis in trastuzumab-resistant breast cancer cells: effects on insulin-like growth factor I signaling. Mol Cancer Ther. 2007;6:667–674. doi: 10.1158/1535-7163.MCT-06-0423. [DOI] [PubMed] [Google Scholar]
- Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin Cancer Res. 2008;14:6730–6734. doi: 10.1158/1078-0432.CCR-08-0581. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- Chen CT, Kim H, Liska D, Gao S, Christensen JG, Weiser MR. Met activation mediates resistance to lapatinib inhibition of HER2-amplified gastric cancer cells. Mol Cancer Ther. 2012;11:660–669. doi: 10.1158/1535-7163.MCT-11-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one-or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Lee YY, McKinney KQ, Ghosh S, Iannitti DA, Martinie JB, Caballes FR, et al. Subcellular tissue proteomics of hepatocellular carcinoma for molecular signature discovery. J Proteome Res. 2011;10:5070–5083. doi: 10.1021/pr2005204. [DOI] [PubMed] [Google Scholar]
- Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. ra46. 2009;2 doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- Casado P, Cutillas PR. A self-validating quantitative mass spectrometry method for assessing the accuracy of high-content phosphoproteomic experiments. Mol Cell Proteomics. 2011;10:M110 003079. doi: 10.1074/mcp.M110.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcolea MP, Casado P, Rodriguez-Prados JC, Vanhaesebroeck B, Cutillas PR. Phosphoproteomic analysis of leukemia cells under basal and drug-treated conditions identifies markers of kinase pathway activation and mechanisms of resistance. Mol Cell Proteomics. 2012;11:453–466. doi: 10.1074/mcp.M112.017483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Gubish CT, Stabile LP, Henry C, Siegfried JM. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011;30:3625–3635. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulig J, Kolodziejczyk P, Kulig P, Legutko J. Targeted therapy for gastric cancer--current status. J Oncol Pharm Pract. 2013;19:75–81. doi: 10.1177/1078155212449030. [DOI] [PubMed] [Google Scholar]
- Nahta R, Shabaya S, Ozbay T, Rowe DL. Personalizing HER2-targeted therapy in metastatic breast cancer beyond HER2 status: what we have learned from clinical specimens. Curr Pharmacogenomics Person Med. 2009;7:263–274. doi: 10.2174/187569209790112337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer BN, Ham AJ, Rinehart C, Hill S, Granja-Ingram Nde M, Gonzalez-Angulo AM, et al. Phosphoproteomic mass spectrometry profiling links Src family kinases to escape from HER2 tyrosine kinase inhibition. Oncogene. 2011;30:4163–4174. doi: 10.1038/onc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Salgia R. Synergism of EGFR and c-Met pathways, cross-talk and inhibition, in non-small cell lung cancer. J Carcinog. 2008;7:9. doi: 10.4103/1477-3163.44372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Stabile LP, Gubish CT, Gooding WE, Grandis JR, Siegfried JM. Dual blockade of EGFR and c-Met abrogates redundant signaling and proliferation in head and neck carcinoma cells. Clin Cancer Res. 2011;17:4425–4438. doi: 10.1158/1078-0432.CCR-10-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabile LP, Rothstein ME, Keohavong P, Lenzner D, Land SR, Gaither-Davis AL, et al. Targeting of both the c-Met and EGFR pathways results in additive inhibition of lung tumorigenesis in transgenic mice. Cancers. 2010;2:2153–2170. doi: 10.3390/cancers2042153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Zerillo C, Kolmakova J, Christensen JG, Harris LN, Rimm DL, et al. Association of constitutively activated hepatocyte growth factor receptor (Met) with resistance to a dual EGFR/Her2 inhibitor in non-small-cell lung cancer cells. Br J Cancer. 2009;100:941–949. doi: 10.1038/sj.bjc.6604937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.