Abstract
Eating disorders, such as anorexia nervosa (AN), have a significant genetic component. In the current study, a genomewide linkage analysis of 192 families with at least one affected relative pair with AN and related eating disorders, including bulimia nervosa, was performed, resulting in only modest evidence for linkage, with the highest nonparametric linkage (NPL) score, 1.80, at marker D4S2367 on chromosome 4. Since the reduction of sample heterogeneity would increase power to detect linkage, we performed linkage analysis in a subset (n=37) of families in which at least two affected relatives had diagnoses of restricting AN, a clinically defined subtype of AN characterized by severe limitation of food intake without the presence of binge-eating or purging behavior. When we limited the linkage analysis to this clinically more homogeneous subgroup, the highest multipoint NPL score observed was 3.03, at marker D1S3721 on chromosome 1p. The genotyping of additional markers in this region led to a peak multipoint NPL score of 3.45, thereby providing suggestive evidence for the presence of an AN-susceptibility locus on chromosome 1p.
Eating disorders—psychiatric disorders characterized by severe disturbances in eating behavior that typically have onset during late adolescence and young adulthood—are divided into three major types: anorexia nervosa (AN), bulimia nervosa (BN), and eating disorder not otherwise specified (EDNOS). AN is characterized by obsessive fear of weight gain, severely restricted eating, and low body weight, and, in women, AN has the highest mortality among the psychiatric disorders (Sullivan 1995); AN is divided into two clinical subtypes: restricting anorexia nervosa (RAN) and binge-eating/purging anorexia nervosa (BPAN). A related disorder (Hsu et al. 1979; Casper et al. 1980; Strober et al. 1982; Bulik et al. 1997), BN, can occur at any body weight and is characterized by binge-eating and compensatory weight-loss behaviors. Family studies have shown significantly increased prevalence of eating disorders in relatives of probands with AN, compared to relatives of control probands (Gershon et al. 1983; Strober et al. 1985; Hudson et al. 1987; Lilenfeld et al. 1998), thereby providing evidence for the familiality of eating disorders. Twin studies of AN have included both clinically derived samples (Garfinkel and Garner 1982; Holland et al. 1984, 1988) and population-based investigations (Wade et al. 2000). In these studies, concordance rates for MZ twins with AN have been estimated to be 52%–56%, whereas concordance rates for DZ twins with AN have been estimated to be 5%–11% (Garfinkel and Garner 1982; Holland et al. 1984, 1988; Wade et al. 2000). These studies strongly suggest that the familiality observed in family studies is primarily due to genetic causes.
We undertook a multicenter study with an allele-sharing linkage strategy to identify loci that may contribute to AN. Clinical and diagnostic information on the sample is summarized in table 1. This sample was composed of 192 families with at least two affected relatives, all of which were ascertained through a proband who met DSM-IV criteria (American Psychiatric Association 1994, pp. 544–545) for >3 years when criterion D, amenorrhea, for AN was waived (because some subjects were menstruating owing to treatment with exogenous hormone replacement and because some subjects were male) and who reported a family history of eating disorders in a first-degree relative (for a detailed description of this sample, see Kaye et al. [2000]). Affected relatives met DSM-IV (American Psychiatric Association 1994, pp. 544–545) criteria for AN, BN, or EDNOS (Kaye et al. 2000). Full assessments were completed on the proband and affected relative(s) (for details, see Kaye et al. 2000). Eating-disorder diagnoses were confirmed by trained raters with the Structured Interview for Anorexia Nervosa and Bulimic Syndromes (Fichter et al. 1998). Of the 230 relative pairs, 88.8% were female-female, 10.8% were female-male, and 0.4% was male-male, consistent with the increased prevalence of eating disorders in females. Mean ± SD age of the probands was 27.8±9.4 years. More than 98% of the families were of European ancestry, with 138 families from North America, 37 families from Germany, and 21 families from England. Informed consent was obtained from all study participants, and all sites received approval from their local institutional review board.
Table 1.
Characteristics of Probands Partitioned by AN Subtype[Note]
|
Sample with AN |
|||
| Entire Sample | Subgroup with RANor BPAN | Subgroup with RAN | |
| Mean ± SD age (years) | 27.76 ± 9.38 | 28.82 ± 9.84 | 26.73 ± 8.84 |
| Gender (% males) | 5.6 | 5.5 | 5.4 |
| Mean ± SD body-mass index (kg/m2): | |||
| Current | 18.62 ± 2.55 | 18.87 ± 2.74 | 18.38 ± 2.33 |
| Minimum (past) | 14.27 ± 2.28 | 14.29 ± 2.17 | 14.25 ± 2.39 |
| Maximum (past) | 21.27 ± 2.88 | 21.49 ± 3.24 | 21.07 ± 2.47 |
| No. of families: | |||
| With one affected sibling paira | 147 | 92 | 26 |
| With three or four affected siblingsa | 16 | 15 | 8 |
| With other affected relative pairsb | 29 |
20 |
3 |
| Total | 192 | 127 | 37 |
Note.— The original sample was composed of 196 families (4 of which were not used in the genetic analysis, as detailed in the note to table 2), each including both a proband with a diagnosis of either the RAN or BPAN subtype and one or more family members affected with AN, BN, or EDNOS. For the families in the subgroup with RAN, two or more individuals had a diagnosis of RAN. For the families in the subgroup with AN, two or more individuals had a diagnosis of AN of either subtype. For both the RAN and AN subgroups, any additional family members with BPAN, BN, or EDNOS were identified as affected, in this table and subsequent analyses.
Families may have had additional affected relatives.
Families included, for example, cousin-cousin relative pairs, aunt-niece relative pairs, or half sibs.
In the entire sample of families, the only multipoint NPL score ⩾1.5 was observed at marker D4S2367 on chromosome 4 (see boldface italic values under the heading “Entire Sample” in table 2). Values from multipoint LOD analysis that allows for heterogeneity (HLOD), under both dominant and recessive inheritance are shown. The estimated α values (i.e., estimates of the fraction of families showing linkage at that locus) under dominant inheritance was 22% at this position. Multipoint NPL scores >1.0 were observed at marker D4S403 on chromosome 4 (NPL score 1.42; P=.07), at marker D11S1392 on chromosome 11 (NPL score 1.20; P=.11), at marker D13S796 on chromosome 13 (NPL score 1.03; P=.15), and at marker D15S657 on chromosome 15 (NPL score 1.23; P=.11). This modest evidence for linkage could reflect either multiple interacting genes, each with weak to moderate effect, and/or sample heterogeneity.
Table 2.
Summary of Highest Multipoint NPL Scores[Note]
|
Entire Sample |
Sample with RAN |
||||||
| HLOD |
HLOD |
||||||
| Chromosome (Marker) | Distance(cM) | MultipointNPL z (P) | Recessive | Dominant | MultipointNPL z (P) | Recessive | Dominant |
| 1 (D1S3721) | 72.37 | .84 (.20) | .19 | .17 | 3.03 (.001) | 2.0 | 1.5 |
| 2 (D2S1391) | 186.45 | .12 (.45) | .00 | −.08 | 1.70 (.05) | .43 | .70 |
| 4 (D4S403) | 25.97 | 1.42 (.08) | .26 | .79 | 1.82 (.03) | .59 | 1.54 |
| 4 (D4S2367) | 78.50 | 1.80 (.03) | .23 | 1.05 | 2.44 (.008) | .65 | 2.23 |
| 5 (D5S1501) | 85.25 | −.24 (.59) | .00 | .00 | 1.63 (.05) | .02 | 1.06 |
| 8 (D8S264) | 4.04 | .11 (.45) | .00 | .03 | 1.90 (.03) | 1.36 | .70 |
| 9 (D9S922) | 80.08 | .78 (.22) | .09 | .21 | 1.51 (.07) | .28 | .78 |
| 16 (D16S748) | 22.19 | −.78 (.78) | .00 | .00 | 1.70 (.05) | .55 | .34 |
| 22 (D22S683) | 36.16 | −.63 (.73) | −.01 | .00 | 1.50 (.07) | .44 | .13 |
Note.— A peripheral-blood sample was collected from each proband, affected relative, and participating biological parent, and DNA was extracted according to standard methods (Lahiri and Nurnberger 1991). The Weber screening set, version 9 (Center for Medical Genetics, Marshfield Medical Research Foundation), which is a screening panel of 386 fluorescently tagged markers dispersed at a density slightly less than 10 cM, was used for the genomewide scan. Mean ± SD marker heterozygosity was 0.77 ± 0.06. Genotype information and reported family structures were compared for inconsistencies (including paternity relationships, twin status, etc.). DNA from four families yielded genotypes that were inconsistent with the stated family structure and were excluded from subsequent genetic analysis. Multipoint NPL scores were determined using GENEHUNTER (Kruglyak et al. 1996). The z score and associated P value for markers with peak NPL scores ⩾1.5 for each sample are indicated in boldface italic. In addition, peak scores and from multipoint HLOD analysis are shown (Smith 1963), under dominant and recessive models and with penetrance set at 50% (Vieland et al. 1992; Greenberg et al. 1998; Durner et al. 1999). For these parametric analyses, disease-allele frequency was arbitrarily set at 0.1 for recessive inheritance and .006 for dominant inheritance.
The reduction of sample heterogeneity should increase the likelihood of identification of susceptibility loci for eating disorders. In this study, all families were ascertained through a proband who met criteria for AN; however, the criteria for affection status were broader for affected relatives. With the goal of examining the effect that reduction of clinical heterogeneity has on the genetic linkage analysis of this sample, we used additional clinical data to define a subgroup (n=37) from the original sample of families in which two or more affected individuals had the RAN subtype (table 1). We focused on the RAN subtype because of its clinical homogeneity (Kaye et al. 1984; Herzog et al. 1996), which may represent a sample with greater genetic homogeneity. The multipoint linkage analysis in the subgroup of 37 families with RAN identified nine peaks with NPL scores ⩾1.5 (see boldface italic values under the heading “Sample with RAN” in table 2; fig. 1). Two of these peaks (at markers D4S403 and D4S2367 on chromosome 4) were included in the set of five peaks, in the entire sample, that surpassed an NPL threshold of 1.0. The highest peak that we observed in the sample with RAN was a multipoint NPL score of 3.03 at marker D1S3721 on chromosome 1. Multipoint HLOD scores under dominant and recessive models were 1.5 and 2.0, respectively. Estimates of the fraction of families linked at this position (α) were 0.45 under the dominant model and 0.41 under the recessive model. This contrasts with estimated α values, in the entire sample, of 0.17 (dominant) and 0.18 (recessive) at this locus. Although the sample with RAN included eight families with three or more siblings, these families did not disproportionately contribute to the linkage signal.
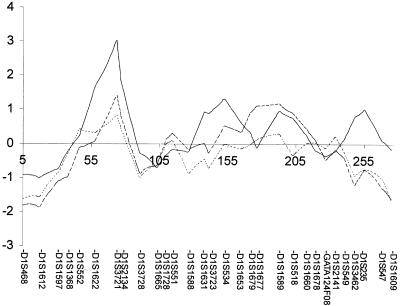
Figure 1.
Evidence for linkage on chromosome 1 in the subgroup with AN. Multipoint NPL z scores (Kruglyak et al. 1996) are plotted, as a function of chromosomal position, for the three categories of families defined in table 1: the subgroup with RAN (n=37 families; solid line), the subgroup with AN (n=127 families; dashed line), and the entire eating-disorder sample (n=192 families; dotted line).
In a follow-up exploratory study, we defined a further subgroup of the entire sample; this subgroup was composed of 127 families in which two or more affected individuals had AN of either the RAN or BPAN subtype, and it thus excluded the 65 families with both a proband with a diagnosis of AN and one or more relatives with BN or EDNOS. The subgroup with AN is more inclusive than the subgroup with RAN, since the former is composed of all members of the subgroup with RAN in addition to all members of the subgroup with BPAN. Sample characteristics for the subgroup with AN are shown in table 1. In the subgroup with AN, the NPL score at marker D1S3721 on chromosome 1 was 1.42, whereas, in the entire sample, the NPL score was 0.83 (fig. 1). Thus, as the affection model was narrowed, the peak NPL scores at this location increased. This increase in evidence for linkage is consistent with there being decreasing genetic heterogeneity as the phenotype is refined. At the second-highest peak (on chromosome 4 at marker D4S2367) in the original sample, there was a more modest increase in NPL score as the affection model was narrowed (table 2).
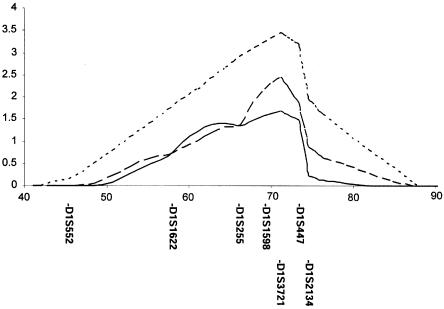
To increase the genetic information concerning the observed peak on chromosome 1, we genotyped three additional markers (D1S255, D1S1598, and D1S447) in the families with RAN. Additional markers in this region resulted in a peak multipoint NPL score of 3.45 (P=.00038) at marker D1S3721 in the subgroup with RAN (fig. 2). Multipoint HLOD scores were 1.68 under the dominant model and 2.43 under the recessive model (fig. 2). Estimated α values were 0.45 under the dominant model and 0.47 under the recessive model.
Figure 2.
Evidence for linkage on chromosome 1p, with additional markers in the subgroup with RAN. Multipoint NPL scores (dotted line) were calculated across the indicated interval, as were multipoint HLOD analyses performed under dominant (solid line) and recessive (dashed line) models, as detailed in the note to table 2. Across this interval, markers were D1S552, D1S1622, D1S255, D1S1598, D1S3721, D1S447, and D1S2134. Positions of markers are indicated. The highest score was over the position of marker D1S3721.
In the current study, we looked for susceptibility loci for eating disorders. This study represents the first genomewide linkage analysis of eating disorders. To date, population-based association studies have been the primary approach to the study of the genetics of eating disorders. Thus far, these candidate-gene analyses have demonstrated only modest success in the identification of genes that may be related to AN susceptibility.
The peak scores that we identified in the entire sample of affected families may indicate the presence of susceptibility loci at these positions but may also represent false positives. The absence of higher peaks in this large sample could be due to a number of potential factors:
-
1.
Although eating disorders are substantially heritable, the genetic susceptibility may be due to a number of interacting loci of small-to-modest effect. If eating disorders arise from a number of interacting genes, then susceptibility loci may reflect variants of small effect that are fairly common in the general population. These variants therefore are difficult to identify by linkage analysis.
-
2.
As discussed above, there may be substantial genetic heterogeneity in eating disorders—a possibility that is supported by the low α values observed parametric linkage analysis in the entire sample.
The reduction of sample heterogeneity by narrowing the affection-status model resulted in a peak NPL score of 3.03, at marker D1S3721 on chromosome 1, which increased to 3.45 when additional markers were considered. Evidence for linkage in the subgroup with RAN was also seen on chromosomes 2, 4, 5, 8, 9, 16, and 22. The evidence for linkage in the subgroup with RAN must be tempered by our study design, which included multiple tests. Therefore, until replicated in an independent sample, these results can be taken as only suggestive linkage.
That a more restrictive phenotype increases NPL scores and HLODs at marker D1S3721 on chromosome 1 suggests that there is decreased heterogeneity in the subgroup with RAN, which is consistent with the clinical presentation of RAN. Decreased heterogeneity is also supported by the higher α values observed in the sample with RAN, compared to both the sample with AN and the entire sample. To evaluate whether the evidence for linkage at marker D1S3721 was derived primarily from the subgroup with RAN, we performed linkage analysis in the subset of all families except the 37 families with RAN (n=155). By Morton’s predivided sample test, these comparisons provided significant evidence for heterogeneity, for both the peak NPL score and HLOD (with χ2=10.13, df=1, and P=.0015 and χ2=8.43, df=1, and P=.0037, respectively), and support genetic heterogeneity in eating disorders (Morton 1956). Interestingly, <25% of relative-pair families with RAN had additional family members in whom other eating disorders had been diagnosed. This suggests that the genetic vulnerability in these families may lead to manifestation of eating disorders beyond RAN. In summary, our data support the presence, on chromosome 1, of a gene for AN and further suggest that the use of restrictive diagnoses can decrease heterogeneity and thereby facilitate the identification of such genes.
Acknowledgments
The authors wish to thank the Price Foundation for the support of the clinical collection of subjects, genotyping, and data analysis. Genotypic markers, allele frequencies, and genetic maps were generated at the Center for Medical Genetics, Marshfield Medical Research Foundation, with support from the National Heart, Lung and Blood Institute. The authors are indebted to the participating families for their contributions of time and effort in support of this study.
Electronic-Database Information
The URL for data in this report is as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for Weber screening set, version 9)
References
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC [Google Scholar]
- Bulik CM, Sullivan PF, Fear J, Pickering A (1997) Predictors of the development of bulimia nervosa in women with anorexia nervosa. J Nerv Ment Dis 185:704–707 [DOI] [PubMed] [Google Scholar]
- Casper RC, Eckert ED, Halmi KA, Goldberg SC, Davis JM (1980) Bulimia: its incidence in patients with anorexia nervosa. Arch Gen Psychiatry 37:1030–1035 [DOI] [PubMed] [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichter MM, Herpertz S, Quadflieg N, Herpertz-Dahlmann B (1998) Structured interview for anorexic and bulimic disorders for DSM-IV and ICD-10: updated (third) revision. Int J Eat Disord 24:227–249 [DOI] [PubMed] [Google Scholar]
- Garfinkel PE, Garner DM (eds) (1982) Anorexia nervosa: a multidimensional perspective. Brunner/Mazel, New York [Google Scholar]
- Gershon ES, Hamovit JR, Schreiber JL, Dibble ED, Kaye W, Nurnberger JI Jr, Anderson A, Ebert M (1983) Anorexia nervosa and major affective disorders associated in families: a preliminary report. In: Guze SB, Earls FJ, Barrett JE (eds) Childhood psychopathology and development. Raven Press, New York, pp 279–291 [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog DB, Field AE, Martin KB, West JC, Robbins WM, Staley J, Colditz GA (1996) Subtyping eating disorders: is it justified? J Am Acad Child Adolesc Psychiatry 35:928–936 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Hall A, Murray R, Crisp AN (1984) Anorexia nervosa: a study of 34 twin pairs and one set of triplets. Br J Psychiatry 145:414–419 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Sicotte N, Treasure J (1988) Anorexia nervosa: evidence for a genetic basis. J Psychosom Res 32:561–571 [DOI] [PubMed] [Google Scholar]
- Hsu LKG, Crisp AH, Harding B (1979) Outcome of anorexia nervosa. Lancet 1:61–65 [DOI] [PubMed] [Google Scholar]
- Hudson JI, Pope HG, Jonas JM, Yurgelun-Todd D, Frankenburg FR (1987) A controlled family history study of bulimia. Psychol Med 17:883–890 [DOI] [PubMed] [Google Scholar]
- Kaye WH, Ebert MH, Gwirstman HE, Weiss SR (1984) Differences in brain serotonergic metabolism between nonbulimic and bulimic patients with anorexia nervosa. Am J Psychiatry 141:1598–1601 [DOI] [PubMed] [Google Scholar]
- Kaye WH, Lilenfeld LR, Berrettini WH, Strober M, Devlin B, Klump K, Goldman D, Bulik CM, Halmi KA, Fichter MM, Kaplan A, Woodside DB, Treasure J, Plotnicov KH, Pollice C, Rao R, McConaha C (2000) A search for susceptibility loci for anorexia nervosa: methods and sample description. Biol Psychiatry 47:794–803 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MJ, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Nurnberger JI Jr (1991) A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acid Res 19:5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilenfeld LR, Kaye WH, Greeno C, Merikangas KR, Plotnikov K, Pollice C, Rao R, Strober M (1998) A controlled family study of anorexia nervosa and bulimia nervosa. Arch Gen Psychiatry 55:603–610 [DOI] [PubMed] [Google Scholar]
- Morton NE (1956) The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am J Hum Genet 8:80–96 [PMC free article] [PubMed] [Google Scholar]
- Smith CAB (1963) Testing for heterogeneity of the recombination fraction values in human genetics. Ann Hum Genet 27:175–182 [DOI] [PubMed] [Google Scholar]
- Strober M, Morrell W, Burroughs J, Salkin B, Jacobs C (1985) A controlled family study of anorexia nervosa. J Psychiatr Res 19:239–246 [DOI] [PubMed] [Google Scholar]
- Strober M, Salkin B, Burroughs J, Morrell W (1982) Validity of the bulimia-restricter distinction in anorexia nervosa. J Nerv Ment Dis 170:345–351 [DOI] [PubMed] [Google Scholar]
- Sullivan PF (1995) Mortality in anorexia nervosa. Am J Psychiatry 152:1073–1074 [DOI] [PubMed] [Google Scholar]
- Vieland VJ, Hodge SE, Greenberg DA (1992) Adequacy of single locus approximations for linkage analysis of oligogenic traits. Genet Epidemiol 9:45–49 [DOI] [PubMed] [Google Scholar]
- Wade TD, Bulik CM, Neale MC, Kendler KS (2000) Anorexia and major depression: shared genetic and environmental risk factors. Am J Psychiatry 157:469–471 [DOI] [PubMed] [Google Scholar]