Abstract
Genomewide quantitative-trait locus (QTL) linkage analysis was performed using a continuous measure of relative hand skill (PegQ) in a sample of 195 reading-disabled sibling pairs from the United Kingdom. This was the first genomewide screen for any measure related to handedness. The mean PegQ in the sample was equivalent to that of normative data, and PegQ was not correlated with tests of reading ability (correlations between −0.13 and 0.05). Relative hand skill could therefore be considered normal within the sample. A QTL on chromosome 2p11.2-12 yielded strong evidence for linkage to PegQ (empirical P=.00007), and another suggestive QTL on 17p11-q23 was also identified (empirical P=.002). The 2p11.2-12 locus was further analyzed in an independent sample of 143 reading-disabled sibling pairs, and this analysis yielded an empirical P=.13. Relative hand skill therefore is probably a complex multifactorial phenotype with a heterogeneous background, but nevertheless is amenable to QTL-based gene-mapping approaches.
Family, twin, and adoption studies have provided evidence for a significant genetic contribution to individual differences in human handedness (Carter-Saltzman 1980; McManus and Bryden 1992; Sicotte et al. 1999; McKeever 2000). According to one meta-analysis (McManus and Bryden 1992), the offspring relative risk for left-handedness is ∼2.0. However, rather than attempting to utilize the greater potential power inherent in a continuous description of the trait, most large-scale genetic epidemiological studies have defined left- and right-handedness as categorical states (Annett 1994). The nature of the genetic influence on handedness remains unknown, and various models have been proposed, some of which postulate a single major gene (Annett 1985, 1994; McManus and Bryden 1992; Corballis 1997). Handedness may relate in part to cerebral lateralization (Geschwind and Miller 2001), which is also presumed to underlie the development of much complex human cognition and behavior, including language. Identifying genetic factors involved in handedness therefore may have implications for the understanding of human neurological development in broader terms. The only previous molecular genetic study of handedness was targeted at chromosome X in a sample of male sibling pairs who used their left hands for writing (Laval et al. 1998), and evidence for an Xq21 quantitative-trait locus (QTL) influencing relative hand skill (P<.002) was identified in that study. Until now, no genomewide screen for linkage to any measure related to handedness has been performed.
As part of an ongoing study of reading disability in the United Kingdom, we have collected a sample of 89 nuclear families, each with at least two siblings who show symptoms of dyslexia (Fisher et al. 1999; Marlow et al. 2001). This sample yields 222 siblings, or 195 total sibling pairs, with a mean age of 14.5 years. In addition to tests related to reading ability, a commonly used and well characterized test of relative hand skill (Annett 1985) was administered to all siblings in the sample. The test involved the measurement of the time taken by subjects to move, with each hand, a row of pegs on a board from one location to another. From these data, we derived the measure PegQ for each sibling, which was calculated as the difference between the times for the left hand (L) and the right hand (R), L-R, divided by the average time for both hands, (L+R)/2, to adjust for overall differences in hand skill between subjects. Positive PegQ scores therefore indicate superior relative right-hand skill. In control populations, the peg-board task yields a continuous and normally distributed variable with a positive mean (Annett 1994), consistent with the high prevalence (∼90%) of qualitatively defined right-handedness in modern human populations (Perelle and Ehrman 1994).
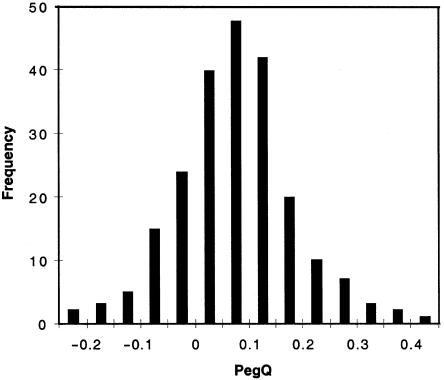
A phenotypic association between left-handedness/ambidexterity and dyslexia has been reported (Geschwind and Behan 1982; Tønnesen et al. 1993), but other investigations have not upheld this association (Pennington et al. 1987; Gilger et al. 1992). Our data do not support a straightforward relationship between relative hand skill and dyslexia. As for normative data, the distribution of PegQ in the sibling sample appeared roughly normally distributed, with a positive mean (fig. 1). Despite having ascertained the sibling sample through evidence of reading disability, the mean PegQ in the sample, 0.072, was very similar to the value (0.073) derived from age-matched normative data of Kilshaw and Annett (1983). Furthermore, all Pearson correlation coefficients between PegQ and reading-related measures were low, between −0.13 and 0.05 (all were two-tailed P values >.15, with the exception of a measure of phonological decoding ability [P=.047], with which the correlation was −0.13 [i.e., the opposite direction to that predicted by the reported phenotypic association]). We therefore reasoned that relative hand skill could be considered to be normal in the siblings and, consequently, that ascertainment history would not have an obvious impact on linkage analysis of PegQ.
Figure 1.
Distribution of PegQ, a quantitative measure of relative hand skill, in the genome-screen sample (222 siblings). Mean = 0.072; SD = 0.104; range −0.24 to 0.42; skewness = 0.154; kurtosis = 0.876. The distribution is unimodal, continuous, and roughly normal, suggesting that linkage analysis can be performed within a classical oligogenic framework (see text). A positive mean (i.e., superior right-hand skill) is characteristic of unselected populations. The Pearson correlation of PegQ with self-reported “writing hand,” a dichotomous variable, was 0.51 (one-tailed P=6.3×10-17). Twenty-two (9.9%) of the siblings wrote with their left hands; all 22 left-handed writers had negative PegQ scores, as did 27 right-handed writers.
The distributional properties of PegQ in the sibling sample (fig. 1) meant that linkage analysis could be performed using a standard biometric model (Falconer et al. 1989) in which phenotype scores are defined by three effects causing deviation from a grand mean:
-
1.
A locus-specific additive genetic effect, for which sibling covariance is conditional on identity-by-descent (IBD) sharing.
-
2.
A familial effect, for which sibling covariance includes the effects of shared environment and half the residual additive polygenic variance.
-
3.
A residual environmental component.
To perform this analysis, we used the variance-components (VC) quantitative-linkage method of Amos (1994), as implemented in the software GENEHUNTER 2.0 (Kruglyak Lab Home Page) (Pratt et al. 2000). The method involves maximum-likelihood estimation of the mean and VC for each locus under investigation. The likelihood of the full model is compared with the likelihood under the null hypothesis in which the locus-specific effect is constrained to equal zero, yielding a LOD score as a test of linkage. The null model also yields a direct estimate of trait familiality, which sets an upper bound for trait heritability. The familiality estimate for PegQ in the genome-screen sample, under the VC approach, was 0.35 (standard error 0.17 by use of SOLAR; Almasy and Blangero 1998). This was comparable to the familiality estimate of 0.36 reported by Laval et al. (1998) for their sample of brothers, by use of a measure similar to PegQ that was derived from the same peg-board test.
Genotyping of children and parents from each nuclear family was performed using 401 highly polymorphic markers spanning the genome (Fisher et al. 2002). The majority of markers came from the ABI PRISM LMS2-MD10 panels (Applied Biosystems). Sex-averaged marker maps were taken from the Cooperative Human Linkage Centre and were supplemented with data from Généthon then verified by comparison to maps estimated from our genome-screen marker data. Semi-automated fluorescent genotyping, as well as data checking and handling, was performed using techniques described by Fisher et al. (1999). The mean marker success rate was 92.5%, and the mean multipoint information content (Kruglyak and Lander 1995), by use of data at 1-cM intervals across the genome, was 75%.
The VC framework provides a powerful test of linkage that uses most of the available trait information (Amos 1994). However, since VC analysis can be sensitive to deviations from trait multivariate normality (Allison et al. 1999), we performed simulations within our sample, to determine unbiased, empirical significance levels for our linkage results. The essence of the approach was to simulate genotype data (see Documentation to the SIMULATE Program, originally written by J. Terwilliger) for a single unlinked marker with four equally frequent alleles (75% heterozygosity), while maintaining the family structures and phenotypic scores of the real sample. One hundred thousand replicates were simulated and analyzed for QTL linkage by the VC method. We have shown elsewhere (Fisher et al. 2002) that the resulting empirical LOD significance distributions are suitable for interpreting multipoint linkage results, for which 70%–80% of IBD information is typically extracted at each locus.
The VC approach has not been implemented for X-linked analysis, and we therefore tested for chromosome X linkage by means of the more traditional Haseman-Elston (HE) method (Haseman and Elston 1972), a regression-based method that is also framed around a locus-specific additive genetic effect. We used the “unweighted, all pairs” option of MAPMAKER/SIBS (Kruglyak and Lander 1995) to perform this analysis, and simulations were again employed, this time primarily to correct for statistical dependence within multiple sibships. MAPMAKER/SIBS uses only male-male pairings for HE regression.
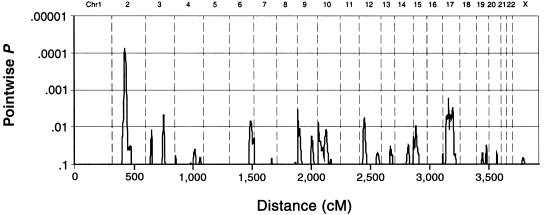
In genomewide multipoint VC linkage analysis of PegQ (fig. 2), one locus at 2p11.2-12, between markers D2S2333 and D2S2216, yielded a pointwise empirical P=.00007 (LOD=2.3), thus approaching the conservative value P=.00002, which was proposed by Lander and Kruglyak (1995) and corresponds to a genomewide significance level α=0.05. Although the approximate magnitudes of P values <.0001 are likely to be correctly specified using 100,000 simulations, their precise specification is not possible without performing millions of simulations, which would be impractical. Therefore, the question of whether or not the empirical P=.00007 surpasses any threshold is not relevant in this context. To verify the 2p11.2-12 linkage, a targeted simulation study was performed, in which all chromosome 2 markers were simulated 100,000 times according to the allelic numbers and frequencies of the real data, patterns of missing genotype data, and genetic distances between markers, again without regard to phenotypic data. VC linkage was then assessed at the same 2p11.2-12 location that showed the peak multipoint linkage in the real analysis. The resulting empirical LOD significance distribution was effectively identical to the single-marker situation, thereby demonstrating that the 2p11.2-12 locus had no unusual marker/map properties and also that the single-marker simulation approach was indeed applicable in a general multipoint context. According to the targeted simulations of chromosome 2, the pointwise empirical P value associated with the 2p11.2-12 multipoint linkage was .00005.
Figure 2.
Genomewide multipoint linkage analysis of PegQ, a measure of relative hand skill. Cumulative distance (in Haldane centimorgans) is shown along the X-axis, with chromosome identities along the top. Evidence for linkage (Y-axis) is given as pointwise empirically derived significance, calculated under a VC framework (see text). The QTL on chromosome 2p11.2-12 yields a pointwise peak P=.00007, suggesting a positive relationship between sibling phenotypic and genetic similarity at this locus.
The second-most-significant multipoint linkage was to a locus on 17p11-q23, between markers D17S1857 and D17S944, with pointwise empirical P=.002. The strongest multipoint linkage on chromosome X was to Xp11, between DXS993 and DXS991, and overlaps the result of Laval et al. (1998) but has a pointwise empirical P of only .07. Single-point VC linkage analysis across the genome (and HE analysis for chromosome X) agreed well with the multipoint results (table 1). Finally, as another check on our autosomal VC analysis, we employed less powerful HE analysis genomewide, using all siblings and the “unweighted, all pairs” options of GENEHUNTER 2.0 (Kruglyak Lab Home Page), this time without performing simulations. The peak HE linkage on 2p11.2-12 yielded a nominal (i.e., not empirical) P=.0003, and the peak HE linkage on 17p11-q23 yielded a nominal P=.0017.
Table 1.
Empirical P Values <.01 in the Genome Screen, for Single-Point VC Linkage
| Chromosome and Marker | CytogeneticBand | Positiona(cM) | MarkerHeterozygosityb(%) | Empirical Pc |
| Chromosome 2: | ||||
| D2S286 | 2p12 | 94 | 61 | .0026 |
| D2S2333 | 2p11.2 | 108 | 82 | .0036 |
| D2S2216 | 2p11.2 | 116 | 73 | .0069 |
| Chromosome 3: | ||||
| D3S1266 | 3p24.1 | 46 | 81 | .0015 |
| D3S1267 | 3q13.32 | 146 | 89 | .0024 |
| Chromosome 9: | ||||
| D9S288 | 9p24.1 | 0 | 85 | .0017 |
| D9S271 | 9q31.2 | 115 | 67 | .0076 |
| Chromosome 10: | ||||
| D10S197 | 10p12.1 | 58 | 73 | .0015 |
| Chromosome 17: | ||||
| D17S1852 | 17p12 | 28 | 85 | .0029 |
| D17S921 | 17p12 | 48 | 69 | .0025 |
| D17S1857 | 17p11.2 | 51 | 64 | .0026 |
| D17S250 | 17q11.2 | 72 | 83 | .0042 |
| D17S787 | 17q22 | 86 | 81 | .0033 |
Distance (in Haldane centimorgans) from the most p-telomeric marker.
In the genome-screen sample.
Pointwise empirical P values are shown for single-point linkages, derived from simulations of a marker with four equally frequent alleles as described in the text. Single-point linkage results can be influenced by marker-specific patterns of missing genotype data and cannot be precisely corrected by simulations that were performed for the entire sample. The P values shown in this table therefore are approximate, but nonetheless are likely to be more accurate than nominal P values, which are not at all corrected for deviations from trait multivariate normality.
The putative 2p11.2-12 QTL for relative hand skill is near a locus on 2p12-16 that has been implicated in dyslexia susceptibility by two linkage studies (i.e., the study by Fagerheim et al. [1999] and the U.S. sample studied by Fisher et al. [2002]). There is no suggestive evidence for linkage of 2p11-16 to reading-related measures in the present study sample (Fisher et al. 2002), which is not surprising given the low correlations between PegQ and reading-related measures in this sample. (Note that Fisher et al. [2002] found suggestive evidence for reading-related linkage 60 cM distally, to 2p25, in the present study sample). The proximity of the 2p11.2-12 relative-hand-skill QTL identified in the present study and the reported 2p12-16 dyslexia QTL may represent coincidence, although the possibility that these QTLs are related remains open and awaits investigation in independent samples.
We further analyzed the 2p11.2-12 relative-hand-skill QTL by genotyping 11 microsatellite markers in a second, independent sample of reading-disabled siblings. The markers (D2S2259, D2S391, D2S337, D2S2368, D2S286, D2S2114, D2S329, D2S2333, D2S2216, D2S160, and D2S347) spanned an interval extending >28 cM on either side of the peak of multipoint linkage identified in the genome screen. This second sample contained 84 nuclear families from the United Kingdom and yielded 143 total sibling pairs, who were ascertained using the same criteria as the genome-screen sample (Fisher et al. 1999; Marlow et al. 2001). The mean PegQ in the second sample was 0.063, slightly lower than that for the genome-screen sample but within the range of the age-matched normative subsets of Kilshaw and Annett (1983) (range of means 0.044–0.085). The correlations between PegQ and reading-related measures in the second sample were again low, between −0.04 and 0.00 (all two-tailed P values >.5). The VC familiality estimate for PegQ in the second sample was 0.26 (standard error 0.19), which was lower than that for the genome-screen sample, and, therefore, the power to replicate the 2p11.2-12 QTL in the second sample may have been somewhat reduced. Note, however, that the VC estimate of familiality, like VC LOD scores, can be affected by sample-specific deviations from trait multivariate normality. Multipoint and single-point VC linkage analysis of 2p11.2-12 in the second sample failed to provide significant support for replication of the QTL influencing relative hand skill. The pointwise empirical P was .18 for multipoint analysis of the location corresponding to the peak of multipoint linkage in the genome screen. The minimum pointwise empirical P was .13 in the second sample, at a location 25 cM proximal to the genome-screen peak, which yielded P=.041 in the screen.
Three explanations for the lack of replication of the 2p11.2-12 QTL in the second sibling sample can be considered:
-
1.
Although the genome-screen linkage was strong (P=.00007), the result may represent a type I (i.e., false-positive) error.
-
2.
The proportion of PegQ variance determined by the 2p11.2-12 QTL may differ substantially between the two samples. This would imply that relative hand skill, as assessed by the peg-board method, is a complex phenotype with a multifactorial background that includes heterogeneous environmental and/or genetic influences. No large-scale twin or adoption analysis of PegQ has been performed to help address this possibility.
-
3.
Even if the 2p11.2-12 QTL determines similar proportions of variance in both samples, it is nevertheless possible, in samples of this size, that stochastic IBD sharing, superimposed on the QTL effect, biased toward detection in the genome-screen sample and toward nonreplication in the second sample (Suarez et al. 1994; Göring et al. 2001).
In conclusion, we have performed the first genomewide screen for a measure related to handedness in humans and have identified a putative QTL influencing relative hand skill on chromosome 2p11.2-12, demonstrating the amenability of the trait to a quantitative genetic-mapping approach in sibling pairs. The QTL was not detected at a significant level in a second sibling sample, suggesting a complex genetic etiology for relative hand skill. The 2p11.2-12 QTL awaits investigation in independent samples.
Acknowledgments
We are very grateful to all the families who participated in this study. We thank Janet Walter, Pam Southcott, Sue Fowler, and Chris Clisby, for the collection and testing of families; Dianne Newbury, Yumiko Ishikawa-Brush, Helene Rees, and Joanne Smith, for assistance with genotyping; Kathleen Taylor, for assistance with handling of phenotype data; and Joel Talcott, Eva Cyhlarova, and Catherine Stoodley, for providing useful background information. Collection of families and all genotyping were funded by the Wellcome Trust. I.L.M was funded by the British Council and the Natural Sciences and Engineering Research Council of Canada. C.F. was a Wellcome Trust Prize Student. A.P.M. is a Wellcome Trust Principal Research Fellow.
Electronic-Database Information
URLs for data in this report are as follows:
- Cooperative Human Linkage Center, The, http://lpg.nci.nih.gov/ABI/ (for ABI PRISM Reference Maps)
- Documentation to the SIMULATE Program, http://linkage.rockefeller.edu/ott/simulate.htm
- Généthon, ftp://ftp.genethon.fr/pub/Gmap/Nature-1995/data/
- Kruglyak Lab Home Page, http://www.fhcrc.org/labs/kruglyak/Downloads/ (for GENEHUNTER 2.0)
- MAPMAKER/SIBS, http://www-genome.wi.mit.edu/ftp/distribution/software/sibs/
- SOLAR, http://ns1.sfbr.org/sfbr/public/software/solar/
References
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance component quantitative-trait loci–mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI (1994) Robust variance components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Annett M (1985) Left, right, hand and brain: the right shift theory. Laurence Erlbaum, London [Google Scholar]
- ——— (1994) Handedness as a continuous variable with dextral shift: sex, generation, and family handedness in subgroups of left- and right-handers. Behav Genet 24:51–63 [DOI] [PubMed] [Google Scholar]
- Carter-Saltzmann L (1980) Biological and sociocultural effects on handedness: comparison between biological and adoptive families. Science 209:1263–1265 [DOI] [PubMed] [Google Scholar]
- Corballis MC (1997) The genetics and evolution of handedness. Psychol Rev 104:714–727 [DOI] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tønnessen FE, Pedersen M, Tranebjaerg L, Lubs HA (1999) A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet 36:664–669 [PMC free article] [PubMed] [Google Scholar]
- Falconer DS (1989) Introduction to quantitative genetics. Longman Scientific and Technical, London [Google Scholar]
- Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayán J, Olson RK, Pennington BF, Smith SD, DeFries JC, Stein JF, Monaco AP (2002) Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet 30:86–91 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Miller BL (2001) Molecular approaches to cerebral laterality: development and neurodegeneration. Am J Med Genet 101:370–381 [PubMed] [Google Scholar]
- Geschwind N, Behan P (1982) Left-handedness: association with immune disorder, migraine, and developmental learning disorder. Proc Natl Acad Sci USA 79:5097–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilger JW, Pennington BF, Green P, Smith SM, Smith SD (1992) Reading disability, immune disorders and non-right-handedness: twin and family studies of their relations. Neuropsychologia 30:209–227 [DOI] [PubMed] [Google Scholar]
- Göring HHH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Kilshaw D, Annett M (1983) Right- and left-hand skill. I. Effects of age, sex, and hand preference showing superior skill in left-handers. Br J Psychol 74:253–268 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Laval SH, Dann JC, Butler RJ, Loftus J, Rue J, Leask SJ, Bass N, Comazzi M, Vita A, Nanko S, Shaw S, Peterson P, Shields G, Smith AB, Stewart J, DeLisi LE, Crow TJ (1998) Evidence for linkage to psychosis and cerebral asymmetry (relative hand skill) on the X chromosome. Am J Med Genet 81:420–427 [DOI] [PubMed] [Google Scholar]
- Marlow AJ, Fisher SE, Richardson AJ, Francks C, Talcott JB, Monaco AP, Stein JF, Cardon LR (2001) Investigation of quantitative measures related to reading disability in a large sample of sib-pairs from the UK. Behav Genet 31:219–230 [DOI] [PubMed] [Google Scholar]
- McKeever WF (2000) A new family handedness sample with findings consistent with X-linked transmission. Br J Psychol 91:21–39 [DOI] [PubMed] [Google Scholar]
- McManus IC, Bryden MP (1992) The genetics of handedness, cerebral dominance and lateralization. In: Rapin I, Segalowitz SJ (eds) Handbook of neuropsychology, section 10: developmental neuropsychology. Elsevier, Amsterdam [Google Scholar]
- Pennington BF, Smith SD, Kimberling WJ, Green PA, Haith MM (1987) Left-handedness and immune disorders in familial dyslexics. Arch Neurol 44:634–639 [DOI] [PubMed] [Google Scholar]
- Perelle IB, Ehrman L (1994) An international study of human handedness: the data. Behav Genet 24:217–227 [DOI] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicotte NL, Woods RP, Mazziotta JC (1999) Handedness in twins: a meta-analysis. Laterality 4:265–286 [DOI] [PubMed] [Google Scholar]
- Suarez BK, Hampe CL, Van Eerdewegh P (1994) Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR (eds) Genetic approaches to mental disorders. American Pyschiatric, Washington, DC, pp 23–46 [Google Scholar]
- Tønnesen FE, Løkken MA, Høien T, Lundberg I (1993) Dyslexia, left-handedness, and immune disorders. Arch Neurol 50:411–416 [DOI] [PubMed] [Google Scholar]