Abstract
Causative TP63 mutations have been identified in five distinct human developmental disorders that are characterized by various degrees of limb abnormalities, ectodermal dysplasia, and facial clefts. The distribution of mutations over the various p63 protein domains and the structural and functional implications of these mutations establish a clear genotype-phenotype correlation.
Introduction
Different syndromes can result from mutations in a single gene. Quite often, these represent variations in severity, as do the FGFR3 mutations, which cause hypochondroplasia, achondroplasia, and thanatophoric dysplasia. More-divergent phenotypes may reflect the difference between gain-of-function and loss-of-function mutations, as occurs with dominant brachydactyly type B and recessive Robinow syndrome, which are caused by different mutations of the ROR2 gene. Other examples of allelic heterogeneity reflect a selective disruption of some but not all the gene’s functions, as in the contrast, because of an FGFR2 mutation, between Apert and Crouzon syndromes. For some genes there are multiple phenotypes associated with mutations, but a clear genotype-phenotype correlation is lacking. A striking example of such allelic heterogeneity are the GLI3 morphopathies: Greig syndrome, Pallister-Hall syndrome, preaxial polydactyly type IV, and postaxial polydactyly type A/B. We here review the spectrum of p63 mutations that have been found to underlie five human malformation syndromes: ectrodactyly–ectodermal dysplasia–clefting (EEC) syndrome (MIM 604292), ankyloblepharon–ectodermal dysplasia–clefting (AEC) syndrome (MIM 106260), limb-mammary syndrome (LMS [MIM 603543]), acro-dermato-ungual-lacrimal-tooth (ADULT) syndrome (MIM 103285), and nonsyndromic split-hand/split-foot malformation (SHFM). The localization and functional effects of the mutations that underlie these syndromes establish a striking genotype-phenotype correlation. Functional analysis of these mutations has provided valuable new insights into p63 protein structure and function and has provided a basis for further dissection of molecular and cellular pathways involving p63.
p63 Completes the p53/p73 Family
TP53 is the principal tumor-suppressor gene, being mutated in >50% of human cancers (Levine 1997). The p53 protein is a key regulator of the cell cycle and, to prevent damaged cells from becoming uncontrolled, allows completion of genomic-repair processes before the cell cycle proceeds. For the past 20 years, p53 has continued to surprise biomolecular researchers—publications on p53 that were listed in PubMed numbered 23,500 by January 2002. For this reason, the discovery of two genes related to TP53, denoted as “TP73” and “TP63,” created considerable excitement among tumor biologists (Jost et al. 1997; Kaghad et al. 1997; Schmale and Bamberger 1997; Osada et al. 1998; Senoo et al. 1998; Trink et al. 1998; Yang et al. 1998). It is now evident that TP73 and TP63 do not represent classical tumor-suppressor genes. Rather, they act as key regulators in development—p73 in the development of neuronal and pheromonal pathways (Yang et al. 2000) and p63 in limb, epithelial, and craniofacial development (Mills et al. 1999; Yang et al. 1999). Both p63 and p73 exhibit high amino acid identity with p53, especially among their transactivation (TA) domains, DNA-binding domain (DBD), and tetramerization (ISO) domain (fig. 1). Unlike TP53, both TP63 and TP73 encode a number of isoforms (fig. 1). For both TP63 and TP73, two transcription initiation sites were initially described—one that would give rise to proteins containing the TA domain (the TA isotypes) and another that would give rise to proteins lacking this domain (the ΔN isotypes). For TP63, additional transcripts were subsequently uncovered in human and rodents, resulting from both the use of at least four transcription initiation sites and extensive alternative splicing at the 5′ end of the gene (Schmale and Bamberger 1997; Yang et al. 1998; Hagiwara et al. 1999; Bamberger and Schmale 2001). Additionally, extensive alternative splicing is seen at the 3′ end of the gene, resulting in three different C-termini for p63 and six for p73. For TP63, differential splicing of intron 8 creates additional variability in the final polypeptide sequences (either GTKRP or A), but the functional significance of this is not known. The extended 3′ coding sequences of the α isotypes of TP63 and TP73 encode a protein-protein–interaction motif that resembles the sterile-α-motif (SAM) domain (Schultz et al. 1997; Bork and Koonin 1998), which is not contained in p53. SAM domains are small globular protein-protein–interaction modules that are usually involved in homo- and hetero-oligomerization with other SAM domains. It has been demonstrated that the p63 and p73 SAM domains do not oligomerize with one another (Chi et al. 1999), and the interacting proteins still need to be identified. Besides these major structural protein motifs, all members of the p53 family contain, in front of the DBD, proline-rich domains, which are involved in pro-apoptotic activity and in the capacity to activate target sequences (Zhu et al. 1999). A second proline-rich region is located between the ISO and SAM domains in p63 and p73 but not in p53. This proline-rich domain is engaged in a physical association with the YES-associated protein YAP. The binding of YAP to p73α enhances its transcriptional activity at known p73-responsive elements (Strano et al. 2001).
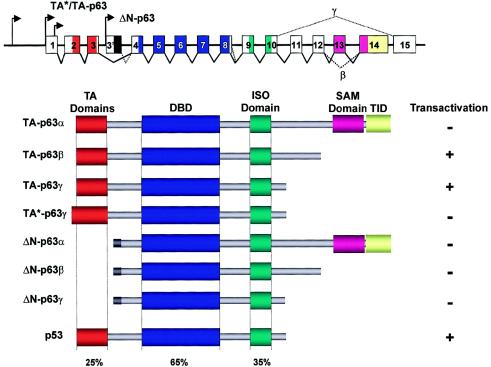
Figure 1.
Structure of both TP63 and major protein isotypes. TP63 uses several transcription initiation sites (arrows) and extensive alternative splicing, to generate a bewildering number of different mRNAs. For clarity, several alternative-splicing routes at the 5′ end of the gene have not been indicated. Several protein domains can be distinguished; of these, the TA domains, the DBD, and the ISO domain are highly homologous to the corresponding domains in p53. The SAM domain and the TID are not contained in the p53 protein. The capacity to transactivate gene expression at a p53-responsive target is given for each of the indicated isotypes.
Differential Properties of the p63 Isotypes
From the primary sequence, one would predict that only the p63 isotypes, which contain the acidic TA domain, have transactivation activity, whereas the ΔN isotypes, which lack this domain, do not have transactivation activity. Although this is generally true, there are still some exceptions to this rule. The largest p63 isotype, TA-p63α, is unable to drive transcription on the optimized p53-responsive element PG13, in contrast to TA-p63β and TA-p63γ (Yang et al. 1998) (fig. 1). This unexpected lack of activity is caused by an inhibitory effect that is contained within the α-specific C-terminal end. This inhibitory activity of the α tail also acts in trans toward TA-p63β/γ transcriptional activation, indicating that the various p63 isotypes can have opposing properties. The repressive activity has been mapped to the region, downstream from the SAM domain, that has been denoted as the “transactivation inhibitory domain” (TID) (Ozaki et al. 1999; V. Doetsch, personal communication). Tentative evidence suggests the presence of other regions within p63 that either promote or repress transactivation activity. For example, the N-terminal TA-isotype variants that contain an additional 39 amino acids (TA*) are unable to drive transcription from the PG13 target sequence (Yang et al. 1998). Interestingly, activation of transcription can be mediated by p63 domains other than the canonical TA domain (Dohn et al. 2001; Strano et al. 2001; Duijf et al. 2002; also, see the “Genotype-Phenotype Correlations: Molecular Dissection of the p63 Gene” section, below). That all known and predicted isotypes contain the tetramerization domain creates an enormous potential for the number of possible different p63 tetrameric complexes that can be formed. Whether all possible complexes occur in vivo remains to be demonstrated.
Regulation of p63 Protein Levels
Activation of the tumor suppressor p53 occurs in response to a number of stressors, including DNA damage, hypoxia, oncogene activation, and UV irradiation (Vogelstein et al. 2000). The main level of activation is mediated by posttranslational protein modifications, which affect protein function and protein stability. UV-B-induced DNA damage decreases levels of ΔN-p63α (a naturally occurring dominant-negative form of the protein), before increasing levels of p53 (Liefer et al. 2000; Yang et al. 2002). Simultaneously, the levels of the transactivating TA-p63 isoforms increase. The down-regulation of dominant-negative ΔN-p63α, as well as the up-regulation that activates TA-p63 isoforms, may be a prerequisite for UV-induced apoptosis in skin (Liefer et al. 2000). This notion is supported by the recent observation that the transactivating TA-p63α (and TA-p73α) isoforms are required for p53-dependent apoptosis induced by DNA damage (Flores et al. 2002). The role that this switch from inhibitory to activating p63 isoforms plays in normal skin development is further discussed below (see “p63 in Mammalian Embryonic Development”).
Little is known about p63 protein activation and turnover. For p53, protein levels are tightly controlled by the ubiquitin ligase MDM2, which promotes the rapid degradation, by the proteasome pathway, of p53 (Haupt et al. 1997; Kubbutat et al. 1997). However, conflicting results have been reported regarding the regulation, by ubiquitination through MDM2 and the related protein MDMX, of p63. For example, in three different studies, MDM2 was shown to have either a repressive effect, a stimulatory effect, or no effect at all on p63-mediated transactivation (Kadakia et al. 2001; Little and Jochemsen 2001; Calabro et al. 2002). Very likely, these conflicting data are the result of variability in both experimental design and the type of cell lines that were used in these studies. One should be extremely cautious in interpreting the results of these and similar studies that are characterized by directive analyses based on knowledge previously obtained for p53.
Upstream and Downstream from p63: Smad and Jagged
A major breakthrough in our knowledge of upstream factors that control TP63 expression was recently established in a screen for genes that are positively regulated by bone morphogenetic proteins (Bmps) during zebrafish gastrulation. A differential display strategy identified ΔN-p63α as a direct transcriptional target of Bmp signaling (Bakkers et al. 2002). The early expression of ΔN-p63α appears to be driven by Smad4 and Smad5, which are common mediators of Bmp signaling. Accordingly, Smad4 and Smad5 binding sites are present in the ΔN-p63 promoter (Bakkers et al. 2002). Bmps promote the specification of ventral cell types in both the mesoderm and the ectoderm, along the dorsoventral axis of vertebrate embryos. In contrast, dorsal cell types, such as the dorsal mesoderm and the neuroectoderm, are formed unless they are instructed by Bmps not to do so. This is known as the “neural default” model. The zebrafish studies revealed that the effect that the Bmps have on the counteraction of this neural default occurs through ΔN-p63α, which acts as the neural repressor.
After the isolation of TP63, it was readily established that the corresponding p63 proteins were able to bind to an engineered p53-responsive element, PG13, and to activate transcription of a downstream reporter gene (Yang et al. 1998). Overexpression of p63 in mammalian cell lines or in yeast can activate or repress the promoters of several p53-responsive genes, including p21, Bax, MDM2, GADD45, 14-3-3σ, and EGFR (Osada et al. 1998; Shimada et al. 1999; Pochampally et al. 2000; Dohn et al. 2001; Nishi et al. 2001; L. Guerrini, personal communication). It is not clear, however, whether p53-responsive genes are also regulated by p63 under physiological conditions. Transfection experiments in keratinocyte cell lines revealed that TA-p63 promotes the expression of loricrin and involucrin, two markers of terminal differentiation of epidermal cells (De Laurenzi et al. 2000). Again, the physiological significance of this observation is not known, although one may speculate that p63 is involved in epidermal differentiation through loricrin and involucrin. The first and only bona fide target genes for p63 are Jagged1 (JAG1) and Jagged2 (JAG2), which encode ligands for Notch receptors (Sasaki et al. 2002). A cDNA microarray analysis showed an increased JAG1 and JAG2 expression in cell lines that were transfected with adenoviral vectors expressing TA-p63γ. The physiological significance of this result was convincingly demonstrated by chromatin-immunoprecipitation experiments, which revealed binding of TA-p63γ to promoter elements of JAG1 in vivo. Also, co-culturing of Notch1-expressing Jurkat cells with p63-transfected cells led to an up-regulation of HES-1, a downstream target of Notch signaling. This indicates that p63 can trigger the Notch pathway in neighboring cells, possibly by induction of JAG1 and JAG2. Although JAG1 mutations cause Alagille syndrome in humans, no human disease has been linked to JAG2 mutations. Interestingly, mice with homozygous inactivating Jag2 mutations have syndactyly and defective craniofacial development, including cleft palate (CP) (Sidow et al. 1997; Jiang et al. 1998). Much work still needs to be done to elucidate other in vivo targets of p63 transactivation and to determine the downstream effects of this transactivation.
p63 in Mammalian Embryonic Development
The spatiotemporal expression of the individual TP63 transcripts has not yet been explored in great detail. Immunohistochemical analyses of mouse embryos show high p63 levels in epithelial cells, especially in progenitor or stem-cell populations of epithelial tissues (Yang et al. 1998, 1999; Mills et al. 1999). The main isotype in these cells is the dominant-negative ΔN-p63α isotype, which likely acts in the maintenance of the proliferative capacity of such cells (Yang et al. 1999). As these cells start to differentiate, their ΔN-p63α levels gradually drop, and the levels of TA-p63 increase. It thus appears that dominant-negative ΔN-p63α is crucial for the maintenance of the capacity of regenerative proliferation of epithelial stem cells. Cells that no longer express ΔN-p63α lose this capacity and are committed to differentiation—and eventually reach terminal differentiation (Yang and McKeon 2002). Indeed, application of retinoic acid, which prevents degradation of ΔN-p63α, effectively blocks the differentiation of skin epithelial stem cells (i.e., keratinocytes) in culture (Bamberger et al. 2002).
In mouse embryos, TP63 expression is first evident in nuclei of cells in the basal layer, which develop into the progenitor cells of the epidermis and related derivatives, such as hair and sweat glands. Basal cells of the cervix, tongue, esophagus, mammary glands, prostate, and urothelium also show high levels of p63. Early TP63 expression is further evident in ectodermal cells of the limb buds and tail bud, branchial arches, and the oral epithelium. In the developing limb bud, TP63 expression is restricted to the apical ectodermal ridge (AER), a key determinant of limb-bud emergence and progression. Proper signaling along the antero-posterior axis between the AER and the underlying mesoderm is crucial for normal formation of the distal limb. The sites of TP63 expression are well in line with the phenotypic consequences of homozygous TP63 inactivation in mice. These p63-deficient newborns exhibit striking limb defects. The forelimbs are severely truncated, and the hindlimbs are lacking altogether. The skin of the knockout animals is absent, and newborn animals die from dehydration shortly after birth. Other skin derivatives, such as hair shafts and follicles are not present. Finally, p63-deficient animals lack tooth primordia and eyelids. Both the maxilla and the mandible are truncated, and the secondary palate fails to close. Taken together, the defects in p63-deficient mice present as severe ectodermal dysplasia, abnormal limb development, and facial dysmorphism.
A Family of EEC-like Syndromes
A group of multiple-congenital-anomaly syndromes is characterized by EEC. The prototypic EEC syndrome has this triad of features (Rudiger et al. 1970). EEC syndrome frequently presents with other associated anomalies, such as lacrimal-tract anomalies, urogenital anomalies, anal atresia, and conductive hearing loss (Rudiger et al. 1970; Rodini and Richieri-Costa 1990; Roelfsema and Cobben 1996). EEC syndrome is relatively common, with >200 cases having been reported in the literature, and is well known for having both variable expressivity and reduced penetrance. A comparison of interfamilial and intrafamilial variability in expressivity found significantly greater interfamilial variability, suggesting that more than one gene or allele might be involved (Roelfsema and Cobben 1996). Several autosomal dominant syndromes have been described that share features with EEC (table 1), including lacrimo-auricular-dental-digital (LADD) syndrome (MIM 149730) and LMS. Bamshad et al. (2000) proposed the combination of the aforementioned four syndromes as “LEAD syndrome” (named for limb, lacrimal, ectodermal, and apocrine dysplasia). Other dominant syndromes resemble the EEC syndrome in only one or two of the cardinal features; for example, AEC syndrome (also known as “Hay-Wells syndrome”) and Rapp-Hodgkin syndrome (RHS [MIM 129400]) lack ectrodactyly, the ectrodactyly–cleft palate (ECP) syndrome (MIM 129830) lacks ectodermal dysplasia, ADULT syndrome and the ectrodactyly–ectodermal dysplasia (EE) syndrome (MIM 129810) lack cleft lip with or without cleft palate (CL/P), and isolated SHFM is characterized only by ectrodactyly.
Table 1.
Phenotypic Characteristics of Human Syndromes[Note]
|
Human Syndrome |
|||||||||
| EECSyndrome | EESyndrome | AECSyndrome | RHS | LMS | SHFM | ADULTSyndrome | LADDSyndrome | ECPSyndrome | |
| Limb: | |||||||||
| Split hand/split foot | ++ | +++ | − | − | ++ | +++ | ++ | − | +++ |
| Fused digits, other | ++ | − | +/− | − | ++ | ++ | ++ | ++ | + |
| Ectodermal: | |||||||||
| Hair | + | ++ | +++ | +++ | − | − | + | − | − |
| Skin | + | − | +++ | + | − | − | ++ | − | − |
| Nail | + | − | ++ | ++ | +/− | − | + | − | − |
| Teeth | + | ++ | ++ | ++ | +/− | − | ++ | ++ | − |
| Lacrimal ducts | + | − | + | ++ | + | − | + | ++ | − |
| Sweat glands | + | − | ++ | ++ | + | − | − | − | − |
| Breast/nipple hypoplasia | +/− | − | − | − | +++ | − | + | − | − |
| Facial: | |||||||||
| CL/P | +++ | − | + | + | − | − | − | − | − |
| CP | − | − | + | ++ | + | − | − | − | +++ |
| Microretrognathia | + | − | + | − | − | − | − | ++ | − |
| Other: | |||||||||
| Fused eyelid | − | − | +++ | − | − | − | − | − | − |
| Supernumerary nipple | +/− | − | + | − | − | − | − | − | − |
| Poor saliva production | − | − | − | − | − | − | − | ++ | − |
| Deformed ears/ear canals | + | − | − | + | − | − | − | +++ | − |
| Hearing loss | + | − | + | − | − | − | − | + | − |
| Extensive freckling | − | − | − | − | − | − | +++ | − | − |
| Kidney hypoplasia | + | − | + | − | − | − | − | + | − |
| Urethra/bladder | + | − | − | − | − | − | − | − | − |
| Hypospadias | − | − | − | + | − | − | − | + | − |
Note.— +++ = Consistent feature; ++ = frequently observed; + = occasionally observed; +/− = rarely observed; − = never observed.
TP63 Mutations: All in the Family
In 1999, linkage mapping of human EEC-like syndromes identified a locus on 3q27, coinciding with the localization of TP63. This result, combined with the data generated by the basic biological research on p63, suggested that TP63 was a strong positional candidate gene, which led to the rapid identification of causative TP63 gene mutations in patients with EEC syndrome (Celli et al. 1999). At the same time, these results established that germline mutations in p63 are not associated with a cancer-prone phenotype, as is the case for p53/Li-Fraumeni syndrome. Moreover, the implication of p63 in EEC syndrome paved the way to testing of the TP63 gene in the EEC-like syndromes, and by that, provided insight into the molecular mechanisms underlying this group of disorders (table 2).
Table 2.
TP63 Mutations in EEC Syndrome, LMS, AEC Syndrome, ADULT Syndrome, and Isolated SHFM
| Amino Acid Change | No. ofFamilies(n=78) | CpG Isleta | Disorder(s) | Reference(s) |
| Exon 3′: | ||||
| N6H | 1 | − | ADULT syndrome | Amiel et al. 2001 |
| Exon 4: | ||||
| G76W | 1 | − | LMS | Authors' unpublished data |
| Exon 5: | ||||
| IVS4-2A→Cb | 1 | − | SHFM | van Bokhoven et al. 2001 |
| Y163C | 1 | − | EEC syndrome | Authors' unpublished data |
| Y192C | 2 | − | EE syndrome | Authors' unpublished data |
| K193E | 1 | − | SHFM | van Bokhoven et al. 2001 |
| K194E | 1 | − | SHFM | Ianakiev et al. 2000 |
| V202M | 1 | − | EE syndrome | Authors' unpublished data |
| Exon 6: | ||||
| R204W | 6 | + | EEC syndrome | Celli et al. 1999, van Bokhoven et al. 2001 |
| R204Q | 4 | + | EEC syndrome | van Bokhoven et al. 2001, authors' unpublished data |
| R227Q | 4 | + | EEC syndrome | Authors' unpublished data |
| Exon 7: | ||||
| C269Y | 1 | − | EEC syndrome | van Bokhoven et al. 2001 |
| S272N | 1 | − | EEC syndrome | Celli et al. 1999 |
| C273Y | 1 | − | EEC syndrome | Authors' unpublished data |
| R279H | 8 | + | EEC syndrome | Celli et al. 1999, Ianakiev et al. 2000, van Bokhoven et al. 2001, authors' unpublished data |
| R279C | 3 | + | EEC syndrome | Kosaki et al. 2001, van Bokhoven et al. 2001 |
| R279Q | 1 | − | EEC syndrome | van Bokhoven et al. 2001 |
| R280C | 5 | + | EEC syndrome, SHFM | Ianakiev et al. 2000, van Bokhoven et al. 2001, authors' unpublished data |
| R280S | 1 | + | EEC syndrome | van Bokhoven et al. 2001 |
| R280H | 2 | + | EEC syndrome, SHFM | van Bokhoven et al. 2001 |
| R298Q | 2 | + | ADULT syndrome | Duijf et al. 2002, authors' unpublished data |
| Exon 8: | ||||
| R304W | 4 | + | EEC syndrome | Celli et al. 1999, Wessagowit et al. 2000, van Bokhoven et al. 2001, authors' unpublished data |
| R304Q | 6 | + | EEC syndrome | Ianakiev et al. 2000, van Bokhoven et al. 2001 |
| C306R | 1 | − | EEC syndrome | Celli et al. 1999 |
| C308S | 1 | − | EEC syndrome | van Bokhoven et al. 2001 |
| P309S | 1 | − | EEC syndrome | van Bokhoven et al. 2001 |
| D312H | 1 | − | EEC syndrome | van Bokhoven et al. 2001 |
| Exon 11: | ||||
| IVS10-2A→Gb | 1 | − | AEC syndrome | Barrow et al., in press |
| Exon 13: | ||||
| L518V | 1 | − | AEC syndrome | McGrath et al. 2001 |
| L518F | 1 | − | AEC syndrome | McGrath et al. 2001 |
| C526G | 1 | − | AEC syndrome | McGrath et al. 2001 |
| C526W | 1 | − | AEC syndrome | McGrath et al. 2001 |
| 1689InsA | 1 | − | EEC syndrome | Celli et al. 1999 |
| 1693-1694DelTT | 1 | − | LMS | van Bokhoven et al. 2001 |
| G534V | 1 | − | AEC syndrome | McGrath et al. 2001 |
| T537P | 1 | − | AEC syndrome | McGrath et al. 2001 |
| Q540L | 1 | − | AEC syndrome | McGrath et al. 2001 |
| I541T | 2 | − | AEC syndrome | McGrath et al. 2001, authors' unpublished data |
| Exon 14: | ||||
| 1859DelC | 1 | − | AEC syndrome | Authors' unpublished data |
| 1860-1861DelAA | 1 | − | LMS | van Bokhoven et al. 2001 |
| Q634X | 1 | − | SHFM | van Bokhoven et al. 2001 |
| E639X | 1 | − | SHFM | Authors' unpublished data |
+ = Mutation at a CpG site (n=45); − = mutation not at a CpG site (n=33).
Intron mutation detected on analysis of the indicated exons.
EEC Syndrome
To date, 20 different heterozygous p63 mutations in 53 families with EEC syndrome are known (reported by Celli et al. [1999], Ianakiev et al. [2000], Wessagowit et al. [2000], Kosaki et al. [2001], and van Bokhoven et al. [2001], as well as in the present article). Of the 50 families for which the entire TP63-coding region was tested in our laboratory, mutations were found in 49, indicating that the p63 gene accounts for many, if not all, patients with this syndrome. The patients with EEC syndrome who had chromosomal abnormalities on 7q21-q22, overlapping the SHFM1 locus (MIM 183600), may be rare exceptions; note that such patients rarely have the full EEC syndrome phenotype (Scherer et al. 1994). Also note that many patients and small families with p63 mutations have all of the clinical hallmarks of EEC syndrome except for CL/P, suggesting that EE syndrome (Wallis 1988) is a variable expression of the EEC syndrome. Two mutations, Y192C and V202M, were observed only in patients with an EE syndrome phenotype. All except one of the mutations in families with EEC syndrome give rise to amino acid substitutions in the DBD that is common to all known p63 isoforms. The arginine codons 204, 227, 279, 280, and 304 were mutated in several unrelated patients. These amino acids are crucially important for direct interactions with DNA target sequences, and their mutation is highly detrimental to DNA binding and transactivation activity.
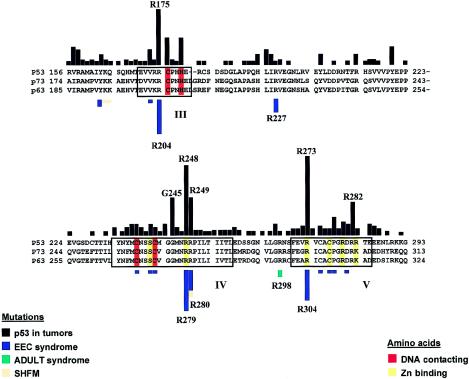
Strikingly, most of the p63 germline mutations correspond precisely to the somatic mutational hotspots in the p53 gene in tumors (fig. 2). An exception is R227 in p63 (corresponding to R196 in p53, which is infrequently mutated in tumors), and the lack of mutations at amino acids G276 and R313, which correspond to the p53 mutational hotspots G245 and R282, respectively (fig. 2). Two explanations, which are not mutually exclusive, may account for the highly specific distribution of p63 mutations.
Figure 2.
Distribution of missense mutations in the DBDs of p53 and p63. Amino acid–sequence comparison of the DBDs of p53, p63, and p73 reveals a high conservation, especially in the core domains (boxed). Within these core domains, amino acids that directly interact with the DNA and amino acids that form the Zn-binding pocket of the protein are highlighted. The black bars on top represent the incidence of somatic p53 mutations in tumors at the corresponding amino acids; the colored bars beneath the sequences represent the incidence of p63 mutations in the indicated human disorders. The p53-mutation spectrum is based on the IARC TP53 Mutation Database.
One explanation is the high mutability of the corresponding codons. Indeed, 46 of the 51 mutations in families with EEC syndrome are C→T transitions at CpG sites. Hence, the high mutability of 5-methylcytosine at CpG sites is a likely explanation for the high proportion of recurrent mutations in EEC syndrome. This would fit with the notion that the codons of the p53 mutational hotspots G245 and R282 contain a CpG, whereas the codons for the nonmutated p63 counterparts G276 and R313 do not have a CpG site.
Another explanation, however, is that these recurrent mutations reflect a specific pathogenetic mechanism. This possibility is supported by the finding that a number of different missense mutations occurred at amino acids R204W/Q, R279C/H/Q, R280C/H/S, and R304W/Q, all of which give rise to EEC syndrome. In contrast, mutations elsewhere in the p63 protein yield phenotypically distinguishable syndromes.
These two explanations are not mutually exclusive, since disease-causing mutations may also be those of DNA sites that are readily mutable. Regardless, it is now clear that mutations in domains other than the DBD give rise to related but clinically distinguishable syndromes, suggesting that there is a specific relationship between TP63 alleles and disease.
Analysis of the primary amino acid sequence (fig. 2) and modeling of the structure of the p63 DBD predicted, for the mutant p63 proteins in EEC syndrome, a general disruptive effect on the DNA-binding capacity (Celli et al. 1999). Support for this prediction was provided by functional studies with mutant proteins in EEC syndrome. Mutant TA-p63γ isotypes were no longer able to promote the expression of a reporter gene, in contrast to their wild-type counterpart. Also, mutant ΔN-p63α was unable to compete with p53 for binding to this target site. These data established that missense mutations in EEC syndrome disrupt DNA binding for all p63 isotypes. The effects on transactivation will differ, however, depending on the sum of the transactivating TA-p63γ and the dominant-negative ΔN-p63α activities, thereby making it difficult to predict the net result on transactivation in vivo. A single frameshift mutation found in a patient with EEC syndrome did not disrupt the DNA-binding capacity. Strikingly, this frameshift mutation, which affects the p63α isotypes only, conferred a gain of transactivation on the otherwise repressive ΔN-p63α isotype (Celli et al. 1999).
AEC (Hay-Wells) Syndrome
AEC syndrome, which is also known as “Hay-Wells syndrome,” has little or no limb involvement but instead includes ankyloblepharon, which is a partial or complete fusion of the eyelids that is very rare in other EEC-like syndromes (Hay and Wells 1976). Also, the ectodermal dysplasia is much more pronounced in AEC than in the other EEC-like syndromes. Severe infections of the scalp are common during the first years of life. Mutations in 12 unrelated patients with AEC have been detected, and 10 of these mutations are missense mutations within the SAM domain of p63 (McGrath et al. 2001). These mutations are predicted to disrupt protein-protein interactions, by either destroying the compact globular structure of the SAM domain or substituting amino acids that are crucial for such interactions (McGrath et al. 2001). Further interpretation of the consequences of the SAM-domain mutations is obscured by our ignorance of the normal biological role of this domain. The missense mutations in AEC syndrome affect only the α isotypes of p63, which behave as inhibitors of transactivation. Transactivation studies did not reveal any increase in activity for mutant TA-p63γ, indicating that the SAM-domain mutations do not relieve the intramolecular repressive activity. In contrast, cotransfections of mutant ΔN-p63α with wild-type p53 or TA-p63γ revealed a clear decrease of intermolecular repressive activity. Tentative evidence indicates that the effects of the SAM-domain mutations varies for different isotypes and at different DNA target sites (L. Guerrini, personal communication). For the functional and developmental consequences of these mutations to be better understood, it will be necessary to identify the protein(s) interacting with the SAM domain.
ADULT Syndrome
ADULT syndrome differs from EEC syndrome by the absence of facial clefting in patients with the former (Propping and Zerres 1993). Instead, these patients show neurodermitic signs—namely, exfoliative dermatitis of the digits—and excessive freckling. Linkage studies in one large German family indicated allelism with EEC syndrome, and, indeed, a pathogenetic mutation, R298Q, was found in the p63 gene (Propping et al. 2000; Duijf et al. 2002). The same R298Q mutation was recently found in an unrelated Italian family with ADULT syndrome. Another missense mutation was reported in an isolated patient with features of ADULT syndrome (Amiel et al. 2001). This mutation lies in exon 3′ and results in a substitution (N6H) that is specific to the ΔN-p63 isotypes.
LMS
LMS differs from EEC syndrome in at least three respects. First, mammary-gland and nipple hypoplasia are consistent features of LMS but are only occasionally seen in EEC syndrome. Second, patients with LMS do not have the hair and skin defects that are seen in EEC syndrome. Third, whereas patients with LMS have CP (van Bokhoven et al. 1999), those with EEC syndrome have CL/P but never have CP only. Phenotypically, LMS is most similar to ADULT syndrome (Propping et al. 2000). Three different mutations have been detected in patients with LMS. Two isolated patients with an LMS phenotype have, in exons 13 and 14, frameshift mutations that result in truncations of the p63α protein. Therefore, the abundant p63 product in epithelial cells would be missing the TID. The third mutation was identified in the large Dutch family with LMS (van Bokhoven et al. 1999). The mutation is in exon 4 and creates a substitution (G76W) just upstream from the TA domain (P. Duijf, personal communication).
SHFM
SHFM is genetically heterogeneous, and three loci have previously been identified by linkage analysis and study of SHFM1, on 7q21-q22; SHFM2 (MIM 313350), on Xq26; and SHFM3 (MIM 600095), on 10q24. In two affected families, SHFM chromosomal abnormalities did not map to any of these established loci but instead mapped to 3q27-q28, thereby indicating the existence of a fourth locus (SHFM4 [MIM 605289]) (Ianakiev et al. 2000). Causative TP63 mutations were found in both families. A subsequent analysis of a group of ∼50 unrelated patients with SHFM revealed five mutations, suggesting that p63 mutations account for ∼10% of these cases (van Bokhoven et al. 2001). Five of the seven p63 mutations seen in patients with SHFM are unique to this syndrome—namely, missense mutations K193E and K194E, nonsense mutations Q634X and E639X, and splice-site mutation IVS4-2A→C (which causes the insertion of a proline residue at position 233). The two aforementioned nonsense mutations create truncations of eight and three amino acids, respectively, in the C-terminal end of the α isotypes. This C-terminal domain contains the repressive domain, and removal of the last eight amino acids partially abolishes this repression (V. Doetsch, personal communication). In addition, the last five amino acids, KEEGE, may form an endoplasmic retention signal, suggesting that protein routing may also be impaired. Two other mutations, both at the same codon, have been found in both SHFM and EEC syndrome—namely, R280C and R280H. This arginine, like the lysines at positions 193 and 194, is not in direct contact with the DNA, and mutation of these residues probably induces more-subtle effects on the DNA-binding capacity of p63.
Involvement of p63 in Other EEC-like Syndromes
We and others have tested a number of patients with LADD syndrome (Hollister et al. 1973; Amiel et al. 2001; authors' unpublished data). A nucleotide change that creates a P472T substitution was identified in an isolated patient with LADD syndrome. Although this change was not detected in >250 individuals (185 healthy controls and 78 patients with an EEC-like syndrome), it is likely to reflect a polymorphism, since the healthy father was also carrying the same change. Hence, it appears that p63 mutations do not account for LADD syndrome. The anticipated involvement in other related conditions, such as ECP syndrome and RHS (Rapp and Hodgkin 1968; Opitz et al. 1980), still needs to be established.
Genotype-Phenotype Correlations: Molecular Dissection of the p63 Gene
The pattern of mutations in the five human disorders linked to p63 reveals a remarkable specificity of the molecular defects in this gene and clinical consequences. The clustering of mutations—in the DBD, for EEC syndrome, and in the SAM domain, for AEC syndrome—establishes a clear genotype-phenotype correlation. Furthermore, the mutations in ADULT syndrome, as well as most of the mutations in LMS and SHFM, are distinctive to these syndromes.
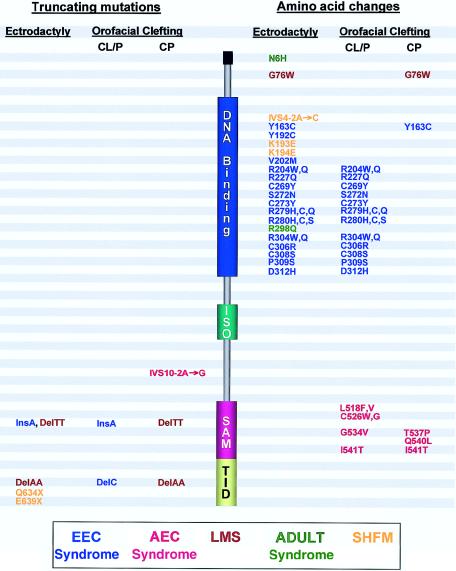
A number of observations can be made from more-detailed analysis of the pattern of TP63 mutations (fig. 3). First, it is notable that the truncating mutations are all located in the C-terminal part of the protein. Hence, all p63 mutations known to date leave the ISO domain intact. This allows the formation of tetrameric complexes between wild-type and mutant p63 proteins, which offers an explanation for the dominant effect of p63 mutations. We have previously rejected haploinsufficiency as an explanation for the disease mechanism, because patients with a heterozygous deletion on 3q27-q29, encompassing the p63 gene, do not display signs of EEC syndrome (Chitayat et al. 1996; van Bokhoven et al. 2001).
Figure 3.
Distribution of mutations in p63, revealing a genotype-phenotype correlation. The approximate positions of truncating mutations (left) and amino acid changes (right) are indicated, together with the associated phenotype, with respect to the occurrence of ectrodactyly and the type of facial clefting. (For discussion, see text.) DNA binding = DBD; ISO = ISO domain; SAM = SAM domain.
The genotype-phenotype comparison suggests that there are further associations with regard to the occurrence of ectrodactyly and the type of facial clefting. Ectrodactyly is only seen in combination with missense mutations before or in the DBD of p63 or in combination with truncating mutations in the C-terminal part of the protein. In contrast, missense mutations in the C-terminal part are never associated with ectrodactyly. A second remarkable phenomenon involves the type of facial clefting. The facial clefts seen in conjunction with missense mutations in the conserved part of the DBD always involve the primary palate (CL/P), whereas mutations toward the C-terminal end of p63 may either involve the primary palate or the secondary palate (CP). Also, the most N-terminal amino acid substitutions, which are located outside the conserved regions of the DBD, give rise to either CP or no clefting at all but never CL/P. The distinction between CL/P and CP is relevant, from both genetic and developmental points of view (Fraser 1970; Ferguson 1988). Anatomically, the primary palate and the secondary palate form independently of one another. Genetically, CL/P and CP also appear to be distinct, since the mixture of these types of facial clefting is rarely observed within families. The Van der Woude and AEC syndromes are rare examples of mixture of embryological and genetic types of facial clefting (CL/P vs. CP) (Schinzel and Klausler 1986). The pattern of mutations associated with ectrodactyly and with the type of facial clefting provides a basis for our conceptual thinking about the normal and abnormal activities, in developmental cascades, of p63.
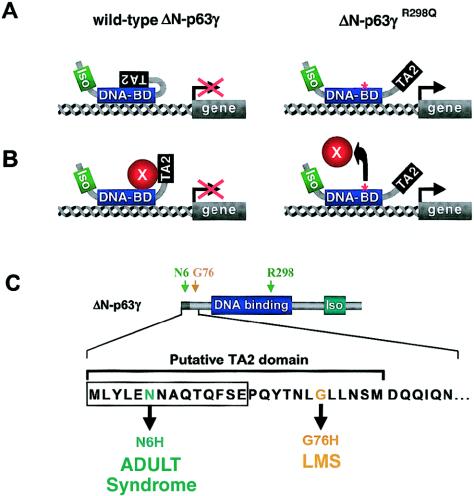
The genotype-phenotype correlations are extended by the mutations found in ADULT syndrome. Like most mutations in EEC syndrome, the R298Q mutation in ADULT syndrome causes an amino acid substitution in the DBD (Duijf et al. 2002). Yet, this mutation is strikingly different from the mutations in EEC syndrome, since, on the basis of the structural model of the DBD, the arginine at position 298 itself does not participate in DNA binding. This prediction was supported by transactivation assays, which revealed no significant loss of activity for TA-p63γR298Q, in sharp contrast to EEC-associated mutations of the same p63 isotype (Duijf et al. 2002). Although this result was predicted, an unexpected finding was the exceptional high transactivation activity for mutant ΔN-p63γR298Q, because this isoform is usually inert in these assays, since it lacks the canonical TA domain. We offer two explanations for this gain of transactivation activity: (1) The mutation creates a novel site for binding of a transcriptional coactivator. (2) The mutation releases a second TA domain (denoted as “TA2”) that is normally kept in an inactive state. Although the first option is already less likely from a mechanistic point of view, there is additional support for the second possibility from studies in cell lines with inducible p63 expression (Dohn et al. 2001). In the present study, we provide evidence that there is, in the N-terminal end of p63, a TA2 domain, encompassing 14 amino acids from the ΔN-specific end and 12 amino acids common to all isotypes (fig. 4). Thus, we suggest that the R298Q mutation constitutively activates this TA2 domain, either by the release of intramolecular protein-protein interactions (fig. 4A) or by the abolition of an interaction with a repressor molecule (fig. 4B). Interestingly, the N6H mutation, found in another family with ADULT syndrome, and the G76W mutation, found in a family with LMS, map precisely within this postulated TA2 domain (fig. 4C). The close juxtaposition of these two residues in the putative TA2 domain of the ΔN isotypes may be relevant, in light of the phenotypic similarity between LMS and ADULT syndrome.
Figure 4.
A gain-of-function mutation in ADULT syndrome reveals a second TA domain (TA2) in p63. A and B, Models explaining the gain-of-function effect that the R298Q mutation has on the ΔN-p63γ isotype, which normally does not posses transactivation. The TA2 domain is normally kept in an inactive state, either because of intramolecular interaction (A) or because of binding of another protein (B). This inhibition is proposed to be released in patients with ADULT syndrome because the R298Q mutation abolishes this protein-protein interaction. DNA-BD = DBD; Iso = ISO domain; TA2 = TA2 domain. C, Position of TA2 domain, as determined by Dohn et al. (2001). The TA2 domain consists of 14 amino acids specific to the ΔN isotypes and 12 amino acids common to all p63 isotypes. Interestingly, another ADULT-syndrome mutation and an LMS mutation both give rise to amino acid substitutions within the TA2 domain. DNA binding = DBD; Iso = ISO domain.
The finding that most of the mutations in SHFM are unique for this condition further extends the genotype-phenotype correlation. Notable exceptions to this relationship are the R280H and R280C mutations, which are found in both SHFM and EEC syndrome. Interestingly, these two missense mutations and the two other SHFM missense mutations, K193E and K194E, involve amino acids that are not in direct contact with the DNA (Ianakiev et al. 2000; P. Duijf, personal communication). Another exception to genotype-phenotype correlation is provided by the frameshift mutations in the α tail of p63. For example, the two frameshift mutations in exon 13 predict similar truncations, but one of these is associated with EEC syndrome, whereas the other gives rise to LMS. Likewise, the exon 14 frameshift mutations are almost identical at the molecular level, but one is seen in LMS, whereas the other is seen in AEC syndrome. The variable outcome of these mutations suggests the influence of additional genetic or environmental factors. Interestingly, within families, mutation of the arginine at position 280 always has the same phenotypic outcome—namely, either SHFM or EEC syndrome—supporting the notion that genetic modifiers or epigenetic factors have a modulatory effect.
Evidence for genetic modifiers is found in mice with mutations in genes that are likely to be involved in p63 pathways. The limb phenotype of the dactylaplasia (Dac) mouse, a model for human SHFM3, not only requires mutation of the dactylin gene but also requires homozygosity for an as-yet-unknown modifier allele that has been denoted as “mDac” (Sidow et al. 1999). Another fascinating example, in the syndactylism (sm) mouse, is caused by a disruption of the p63 target gene Jag2 (Sidow et al. 1997; Sasaki et al. 2002). The sm phenotype is strongly modified by genetic background, and several loci, acting as either enhancers or suppressors, have been mapped (Sidow et al. 1997). One of these, the suppressor locus on mouse chromosome 16, is syntenic to human chromosome 3q27-q29 and encompasses the TP63 gene. TP63 may be a modifier of the mutant JAG2 phenotype, and, by analogy, JAG2 may be a modifier of the mutant p63 phenotype. The hypothesis that there are specific modifier genes can be further pursued by molecular studies of large families with a single TP63 mutation. Other candidate modifiers include (a) genes that are known to be mutated in human syndromes with features that overlap those of the EEC syndrome or (b) genes that are active in genetic programs that are governed by p63. For full comprehension of the normal and disrupted properties of the complex array of p63 isotypes, it will be necessary to identify those genes that act together with or in response to p63. It is to be expected that some of these will be found either to be modifiers of the spectrum of EEC-like disorders or to underlie LADD syndrome or the 90% of cases of SHFM that lack TP63 mutations.
Acknowledgments
We are indebted to the members of the Nijmegen Human Genetics laboratory—in particular, Pascal Duijf, Jacopo Celli, and Ellen van Beusekom. The continuous support from and discussions with our collaborators Frank McKeon, Annie Yang, Volker Doetsch, and John McGrath is greatly appreciated. Special thanks to Ben Hamel and Arie Smits for their studies of the family with LMS. Finally, we would like to thank Hans Scheffer, the many clinicians from other Institutes, and, most of all, the patients, for their participation in the studies described. Work in our laboratory is supported by grants from the Dutch Foundation for Scientific Research (NWO 903-42-190).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- IARC TP53 Mutation Database, http://www.iarc.fr/p53/ (for mutation frequencies in the TP53 gene)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for EEC syndrome [MIM 604292], LADD syndrome [MIM 149730], ADULT syndrome [MIM 103285], LMS [MIM 603543], AEC syndrome [MIM 106260], RHS [MIM 129400], ECP syndrome [MIM 129830], EE syndrome [MIM 129810], SHFM1 [MIM 183600], SHFM2 [MIM 313350], SHFM3 [MIM 600095], and SHFM4 [MIM 605289])
References
- Amiel J, Bougeard G, Francannet C, Raclin V, Munnich A, Lyonnet S, Frebourg T (2001) TP63 gene mutation in ADULT syndrome. Eur J Hum Genet 9:642–645 [DOI] [PubMed] [Google Scholar]
- Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M (2002) Zebrafish ΔNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell 6:617–627 [DOI] [PubMed] [Google Scholar]
- Bamberger C, Pollet D, Schmale H (2002) Retinoic acid inhibits downregulation of ΔNp63α expression during terminal differentiation of human primary keratinocytes. J Invest Dermatol 118:133–138 [DOI] [PubMed] [Google Scholar]
- Bamberger C, Schmale H (2001) Identification and tissue distribution of novel KET/p63 splice variants. FEBS Lett 501:121–126 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Jorde LB, Carey JC (2000) Getting a LEAD on EEC. Am J Med Genet 90:183–184 [DOI] [PubMed] [Google Scholar]
- Barrow LL, van Bokhoven H, Daack-Hirsch S, Andersen T, van Beersum SEC, Gorlin R, Murray JC. Analysis of the p63 gene in classic EEC syndrome, related syndromes, and nonsyndromic orofacial clefts. Am J Med Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Koonin EV (1998) Predicting functions from protein sequences—where are the bottlenecks? Nat Genet 18:313–318 [DOI] [PubMed] [Google Scholar]
- Calabro V, Mansueto G, Parisi T, Vivo M, Calogero RA, La Mantia G (2002) The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J Biol Chem 277:2674–2681 [DOI] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G, van Haeringen A, Woods CG, van Essen AJ, de Waal R, Vriend G, Haber DA, Yang A, McKeon F, Brunner HG, van Bokhoven H (1999) Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99:143–153 [DOI] [PubMed] [Google Scholar]
- Chi SW, Ayed A, Arrowsmith CH (1999) Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J 18:4438–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitayat D, Babul R, Silver MM, Jay V, Teshima IE, Babyn P, Becker LE (1996) Terminal deletion of the long arm of chromosome 3 [46,XX,del(3)(q27→qter)]. Am J Med Genet 61:45–48 [DOI] [PubMed] [Google Scholar]
- De Laurenzi V, Rossi A, Terrinoni A, Barcaroli D, Levrero M, Costanzo A, Knight RA, Guerrieri P, Melino G (2000) p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem Biophys Res Commun 273:342–346 [DOI] [PubMed] [Google Scholar]
- Dohn M, Zhang S, Chen X (2001) p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20:3193–3205 [DOI] [PubMed] [Google Scholar]
- Duijf PHG, Vanmolkot KRJ, Propping P, Friedl W, Krieger E, McKeon F, Dötsch V, Brunner HG, van Bokhoven H (2002) Gain-of-function mutation in ADULT syndrome reveals the presence of a second transactivation domain in p63. Hum Mol Genet 11:799–804 [DOI] [PubMed] [Google Scholar]
- Ferguson MW (1988) Palate development. Development Suppl 103:41–60 [DOI] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T (2002) p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560–564 [DOI] [PubMed] [Google Scholar]
- Fraser FC (1970) The genetics of cleft lip and cleft palate. Am J Hum Genet 22:336–352 [PMC free article] [PubMed] [Google Scholar]
- Hagiwara K, McMenamin MG, Miura K, Harris CC (1999) Mutational analysis of the p63/p73L/p51/p40/CUSP/KET gene in human cancer cell lines using intronic primers. Cancer Res 59:4165–4169 [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387:296–299 [DOI] [PubMed] [Google Scholar]
- Hay RJ, Wells RS (1976) The syndrome of ankyloblepharon, ectodermal defects and cleft lip and palate: an autosomal dominant condition. Br J Dermatol 94:277–289 [DOI] [PubMed] [Google Scholar]
- Hollister DW, Klein SH, De Jager HJ, Lachman RS, Rimoin DL (1973) The lacrimo-auriculo-dento-digital syndrome. J Pediatr 83:438–444 [DOI] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P (2000) Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet 67:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T (1998) Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev 12:1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost CA, Marin MC, Kaelin WG Jr (1997) p73 is a human p53-related protein that can induce apoptosis. Nature 389:191–194 (erratum 399:817 [1999]) [DOI] [PubMed] [Google Scholar]
- Kadakia M, Slader C, Berberich SJ (2001) Regulation of p63 function by Mdm2 and MdmX. DNA Cell Biol 20:321–330 [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D (1997) Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809–819 [DOI] [PubMed] [Google Scholar]
- Kosaki R, Ohashi H, Yoshihashi H, Suzuki T, Kosaki K (2001) A de novo mutation (R279C) in the P63 gene in a patient with EEC syndrome. Clin Genet 60:314–315 [DOI] [PubMed] [Google Scholar]
- Kubbutat M, Jones S, Vousden K (1997) Regulation of p53 stability by Mdm2. Nature 387:299–303 [DOI] [PubMed] [Google Scholar]
- Levine AJ (1997) p53, the cellular gatekeeper for growth and division. Cell 88:323–331 [DOI] [PubMed] [Google Scholar]
- Liefer KM, Koster MI, Wang XJ, Yang A, McKeon F, Roop DR (2000) Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res 60:4016–4020 [PubMed] [Google Scholar]
- Little NA, Jochemsen AG (2001) Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene 20:4576–4580 [DOI] [PubMed] [Google Scholar]
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, Orlow SJ, van Haeringen A, Ausems MG, Yang A, McKeon F, Bamshad MA, Brunner HG, Hamel BC, van Bokhoven H (2001) Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 10:221–229 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708–713 [DOI] [PubMed] [Google Scholar]
- Nishi H, Senoo M, Nishi KH, Murphy B, Rikiyama T, Matsumura Y, Habu S, Johnson AC (2001) p53 homologue p63 represses epidermal growth factor receptor expression. J Biol Chem 276:41717–41724 [DOI] [PubMed] [Google Scholar]
- Opitz JM, Frias JL, Cohen MM Jr (1980) The ECP syndrome, another autosomal dominant cause of monodactylous ectrodactyly. Eur J Pediatr 133:217–220 [DOI] [PubMed] [Google Scholar]
- Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, Ikawa S (1998) Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med 4:839–843 [DOI] [PubMed] [Google Scholar]
- Ozaki T, Naka M, Takada N, Tada M, Sakiyama S, Nakagawara A (1999) Deletion of the COOH-terminal region of p73α enhances both its transactivation function and DNA-binding activity but inhibits induction of apoptosis in mammalian cells. Cancer Res 59:5902–5907 [PubMed] [Google Scholar]
- Pochampally R, Li C, Lu W, Chen L, Luftig R, Lin J, Chen J (2000) Temperature-sensitive mutants of p53 homologs. Biochem Biophys Res Commun 279:1001–1010 [DOI] [PubMed] [Google Scholar]
- Propping P, Friedl W, Wienker TF, Uhlhaas S, Zerres K (2000) ADULT syndrome allelic to limb mammary syndrome (LMS)? Am J Med Genet 90:179–182 [PubMed] [Google Scholar]
- Propping P, Zerres K (1993) ADULT-syndrome: an autosomal-dominant disorder with pigment anomalies, ectrodactyly, nail dysplasia, and hypodontia. Am J Med Genet 45:642–648 [DOI] [PubMed] [Google Scholar]
- Rapp RS, Hodgkin WE (1968) Anhidrotic ectodermal dysplasia: autosomal dominant inheritance with palate and lip anomalies. J Med Genet 5:269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodini ES, Richieri-Costa A (1990) EEC syndrome: report on 20 new patients, clinical and genetic considerations. Am J Med Genet 37:42–53 [DOI] [PubMed] [Google Scholar]
- Roelfsema NM, Cobben JM (1996) The EEC syndrome: a literature study. Clin Dysmorphol 5:115–127 [DOI] [PubMed] [Google Scholar]
- Rudiger RA, Haase W, Passarge E (1970) Association of ectrodactyly, ectodermal dysplasia, and cleft lip-palate. Am J Dis Child 120:160–163 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T (2002) The p53 family member genes are involved in the Notch signal pathway. J Biol Chem 277:719–724 [DOI] [PubMed] [Google Scholar]
- Scherer SW, Poorkaj P, Massa H, Soder S, Allen T, Nunes M, Geshuri D, Wong E, Belloni E, Little S (1994) Physical mapping of the split hand/split foot locus on chromosome 7 and implication in syndromic ectrodactyly. Hum Mol Genet 3:1345–1354 [DOI] [PubMed] [Google Scholar]
- Schinzel A, Klausler M (1986) The Van der Woude syndrome (dominantly inherited lip pits and clefts). J Med Genet 23:291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmale H, Bamberger C (1997) A novel protein with strong homology to the tumor suppressor p53. Oncogene 15:1363–1367 [DOI] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci 6:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Seki N, Ohira M, Sugano S, Watanabe M, Inuzuka S, Okamoto T, Tachibana M, Tanaka T, Shinkai Y, Kato H (1998) A second p53-related protein, p73L, with high homology to p73. Biochem Biophys Res Commun 248:603–607 [DOI] [PubMed] [Google Scholar]
- Shimada A, Kato S, Enjo K, Osada M, Ikawa Y, Kohno K, Obinata M, Kanamaru R, Ikawa S, Ishioka C (1999) The transcriptional activities of p53 and its homologue p51/p63: similarities and differences. Cancer Res 59:2781–2786 [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, Birren BW, Altshuler D, Jaenisch R, Johnson KR, Lander ES (1999) A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet 23:104–107 [DOI] [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, Bronson RT, Daly MJ, Reeve MP, Hawkins TL, Birren BW, Jaenisch R, Lander ES (1997) Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature 389:722–725 [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G (2001) Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem 276:15164–15173 [DOI] [PubMed] [Google Scholar]
- Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D (1998) A new human p53 homologue. Nat Med 4:747–748 [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Hamel BC, Bamshad M, Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E, van Beersum SE, Celli J, Merkx GF, Tenconi R, Fryns JP, Verloes A, Newbury-Ecob RA, Raas-Rotschild A, Majewski F, Beemer FA, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates JR, Neri G, Brunner HG (2001) p63 gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet 69:481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, Jung M, Smits AP, van Beersum S, Schendorf R, van Steensel M, Veenstra M, Tuerlings JH, Mariman EC, Brunner HG, Wienker TF, Reis A, Ropers HH, Hamel BC (1999) Limb mammary syndrome: a new genetic disorder with mammary hypoplasia, ectrodactyly, and other hand/foot anomalies maps to human chromosome 3q27. Am J Hum Genet 64:538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408:307–310 [DOI] [PubMed] [Google Scholar]
- Wallis CE (1988) Ectrodactyly (split-hand/split-foot) and ectodermal dysplasia with normal lip and palate in a four-generation kindred. Clin Genet 34:252–257 [DOI] [PubMed] [Google Scholar]
- Wessagowit V, Mellerio JE, Pembroke AC, McGrath JA (2000) Heterozygous germline missense mutation in the p63 gene underlying EEC syndrome. Clin Exp Dermatol 25:441–443 [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Caput D, McKeon F (2002) On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 18:90–95 [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F (1998) p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2:305–316 [DOI] [PubMed] [Google Scholar]
- Yang A, McKeon F (2002) P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol 1:199–207 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714–718 [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D (2000) p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99–103 [DOI] [PubMed] [Google Scholar]
- Zhu J, Jiang J, Zhou W, Zhu K, Chen X (1999) Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene 18:2149–2155 [DOI] [PubMed] [Google Scholar]