Abstract
Autism is caused, in part, by inheritance of multiple interacting susceptibility alleles. To identify these inherited factors, linkage analysis of multiplex families is being performed on a sample of 105 families with two or more affected sibs. Segregation patterns of short tandem repeat polymorphic markers from four chromosomes revealed null alleles at four marker sites in 12 families that were the result of deletions ranging in size from 5 to >260 kb. In one family, a deletion at marker D7S630 was complex, with two segments deleted (37 kb and 18 kb) and two retained (2,836 bp and 38 bp). Three families had deletions at D7S517, with each family having a different deletion (96 kb, 183 kb, and >69 kb). Another three families had deletions at D8S264, again with each family having a different deletion, ranging in size from <5.9 kb to >260 kb. At a fourth marker, D8S272, a 192-kb deletion was found in five families. Unrelated subjects and additional families without autism were screened for deletions at these four sites. Families screened included 40 families from Centre d'Etude du Polymorphisme Humaine and 28 families affected with learning disabilities. Unrelated samples were 299 elderly control subjects, 121 younger control subjects, and 248 subjects with Alzheimer disease. The deletion allele at D8S272 was found in all populations screened. For the other three sites, no additional deletions were identified in any of the groups without autism. Thus, these deletions appear to be specific to autism kindreds and are potential autism-susceptibility alleles. An alternative hypothesis is that autism-susceptibility alleles elsewhere cause the deletions detected here, possibly by inducing errors during meiosis.
Introduction
Autism (MIM 209850) is a developmental disorder that is recognized in early childhood and persists throughout life. The disorder is characterized by deficits in reciprocal social interaction and in verbal and nonverbal communication and by a restricted stereotyped range of interests and activities. Autism spectrum disorders (ASD) include autism, as the most classic form, and pervasive developmental disorder not otherwise specified (PDD), Asperger syndrome, and childhood disintegrative disorder, as closely related syndromes. Whether ASD represents a continuum of a single disorder or a mixture of clinically related syndromes is unknown.
Autism as a complex of symptoms can occur at elevated rates in individuals with fragile-X syndrome (FMR1 [MIM 309550]), tuberous sclerosis (TS [MIM 191100]), Prader-Willi syndrome (PWS [MIM 176270]), Angelman syndrome (AS [MIM 105830]), and other rare disorders. Duplication of the Prader-Willi region at 15q11-13 is also associated with autism (Gillberg et al. 1991; Robinson et al. 1993; Baker et al. 1994; Cook et al. 1997b). Numerous other case reports of autism associated with cytogenetic abnormalities have appeared with no apparent clustering of the chromosomal regions involved. However, most individuals with ASD do not have one of these known medical disorders or a chromosomal abnormality. For these cases, the etiology of ASD is unknown.
Inheritance contributes substantially to ASD susceptibility. MZ twins are 60% concordant for autism and as much as 95% concordant for less-severe ASD symptoms related to autism (Bailey et al. 1995). The high—but not 100%—concordance rate for MZ twins (Folstein and Rutter 1977a, 1977b; Ritvo et al. 1985; Steffenburg et al. 1989; Le Couteur et al. 1996) indicates that genetic factors are a major contributor to ASD susceptibility but also that environmental factors contribute to risk. DZ twins and siblings have much lower concordance rates (3%–4.5%) (Bolton et al. 1994), indicating that the genetic basis of ASD is complex and that presumably multiple genes contribute in an interactive manner to confer ASD susceptibility. Estimates of the number of genes range from 2–10 (Pickles et al. 1995) to ⩾15 (Risch et al. 1999; Pritchard 2001), and the exact number of genes and the mode of inheritance are unknown.
Genomewide linkage analysis studies of families with autism have yielded positive signals for chromosomes 1, 2q, 7q, 9, 13, 15, and 16p (Barrett et al. 1999; Philippe et al. 1999; Risch et al. 1999; Buxbaum et al. 2001; Liu et al. 2001; IMGSAC 2001a), though no region has been unambiguously confirmed in all studies. The best candidates for regions containing an autism locus are chromosomes 2q and 7q (Barrett et al. 1999; Philippe et al. 1999; IMGSAC 2001a, 2001b; Buxbaum et al. 2001). Specific genes have been tested as candidate autism-susceptibility loci, selected for any of four reasons:
-
1.
Biological plausibility—for example, HOXA1 (MIM 142955), HOXB1 (MIM 142968) (Ingram et al. 2000), EN-2 (MIM 131310) (Petit et al. 1995), and the serotonin transporter SLC6A4 (MIM 182138) (Cook et al. 1997a).
-
2.
Location in a region with a positive linkage signal—for example, reelin (RELN (MIM 600514) (Persico et al. 2001), WNT2 (MIM 147870) (Wassink et al. 2001), and VIPR2 (MIM 601970) (Asano et al. 2001).
-
3.
Disruption by a translocation in a subject with autism—for example, RAY1 (MIM 600833) (Vincent et al. 2000).
-
4.
Location in the PWS-AS deletion region—for example, the GABA receptor (GABRB3 [MIM 137192]) (Cook et al. 1998).
However, no gene has been conclusively identified as an autism-susceptibility locus.
In the course of performing a genome scan for autism-susceptibility genes, we identified four interstitial deletions in individuals from 12 families that had two or more siblings with autism. One of these deletions, initially detected with marker D8S272, is 192 kb and is also found in control samples. Thus, this is an insertion/deletion (in/del) polymorphism with a very-large-deletion allele. For the remaining three sites, deletion alleles, ranging in size from <5.9 kb to >250 kb, were detected only in multiplex families with autism. These deletions did not segregate perfectly with the disease in each case. Thus these deletions may not be causative, but rather suggest that autism is the result of a genetic predisposition to errors in meiosis.
Subjects, Material, and Methods
Characterization of Families with Autism and Diagnostic Criteria
Families who identified themselves as having two or more children with autistic disorder and/or PDD, were recruited through the National Institutes of Health Collaborative Programs of Excellence in Autism, a network of sites at the University of Washington (98 families), the University of California at Irvine (families AUO17, AUO27, and AUO39), and the University of Utah (4 families). The University of Washington recruitment network included testing sites in Washington, Oregon, Florida, Tennessee, Arizona, Minnesota, and Texas. When possible, a blood sample was obtained from both parents, each affected sibling, and an unaffected sibling. At least one parent was available from each family. Exclusionary criteria included an age <3 years, the presence of a known genetic condition—for example, neurofibromatosis (NF1 [MIM 162200]), TS, phenylketonuria (PKU [MIM 261600]), FMR1, William-Beuren syndrome (WBS [MIM194050]), or velocardiofacial syndrome (VCFS [MIM 192430])—or a serious head injury.
All affected individuals were administered a diagnostic evaluation consisting of the Autism Diagnostic Interview—Revised (ADI-R) (Lord et al. 1994) and the Autism Diagnostic Observation Schedule—Generic (ADOS-G). Both instruments assess the symptoms of PDD, including autism, as defined in DSM-IV (American Psychiatric Association 1994). In addition, a clinical diagnosis was made by the clinician evaluating the child, on the basis of the presence or absence of DSM-IV symptoms of autism. All children were administered a standardized intellectual evaluation appropriate for their age level (Mullen Scales of Early Learning or Weschler Scales of Intelligence, short form).
Diagnosis of autism was defined as meeting criteria for autism on the ADOS-G and ADI-R and clinical diagnosis. In addition, if a child received a diagnosis of autism on the ADOS-G and clinical diagnosis, and came within 2 points of meeting criteria on the ADI-R, the child was also considered to have autism. Diagnosis of PDD case type 1 was defined as meeting criteria for PDD on the ADOS-G, meeting criteria for autism on the ADI-R or missing criteria on the ADI-R by ⩽2 points, and meeting criteria for PDD or autism on the basis of clinical diagnosis. Diagnosis of PDD case type 2 (PDD2) was defined as meeting criteria for PDD on the basis of clinical diagnosis but failing to meet criteria for PDD on the ADOS-G and/or ADI-R. Individuals who had clinical features of ASD (e.g., clinically significant language and/or social impairment, according to the ADI-R and ADOS-G) but whose symptoms were not numerous or severe enough to receive a clinical diagnosis of autism or PDD were referred to as “broader phenotype.” The studies described here were approved by the University of Washington institutional review board, and appropriate informed consent was obtained from all subjects.
Genetic Markers
For the initial genome screen, STRP markers were from the ABI Prism linkage mapping set, version 2, spaced at ∼10 cM. Chromosome 7 markers were D7S531, D7S517, D7S513, D7S507, D7S493, D7S516, D7S484, D7S510, D7S519, D7S502, D7S669, D7S630, D7S657, D7S515, D7S486, D7S530, D7S640, D7S684, D7S661, D7S636, D7S798, and D7S2465. Other chromosome 7 markers genotyped were D7S2500, D7S1813, and D8S645, which are between D7S502 and D7S669. Chromosome 8 markers were D8S264, D8S277, D8S550, D8S549, D8S258, D8S1771, D8S505, D8S285, D8S260, D8S270, D8S1784, D8S514, D8S284, and D8S272. Chromosome 15 markers were D15S128, D15S1002, D15S165, D15S1007, D15S1012, D15S994, D15S978, D15S117, D15S153, D15S131, D15S205, D15S127, D15S130, and D15S120. Chromosome 16 markers were D16S423, D16S404, D16S3075, D16S500, D16S3046, D16S3068, D16S3136, D16S415, D16S503, D16S515, D16S516, D16S3091, and D16S520. SNP markers were from annotations of genomic sequence contigs found in GenBank; the designations for these loci begin with “rs” (e.g., rs1016807 is an SNP found in the annotation for contig NT_017168.5) (see figs. 1 and 2). New STRP loci were identified by searching genomic sequences for di-, tri-, and tetranucleotide repeat tracks. These loci are designated with a number followed by the repeated nucleotides (e.g., 2127-CT is a dinucleotide CT-repeat polymorphism) (fig. 1).
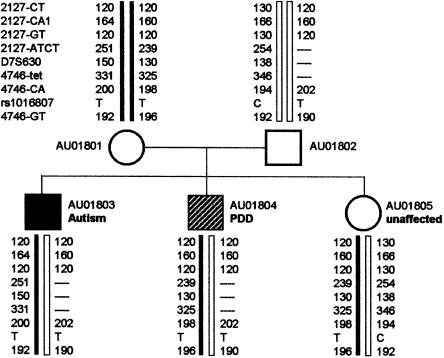
Figure 1.
Family AU018 genotypes for D7S630 and flanking polymorphic markers. Genetic markers are listed next to genotypes for subject AU01801. Markers are listed in their physical order (fig. 2). Null alleles are shown as dashed lines.
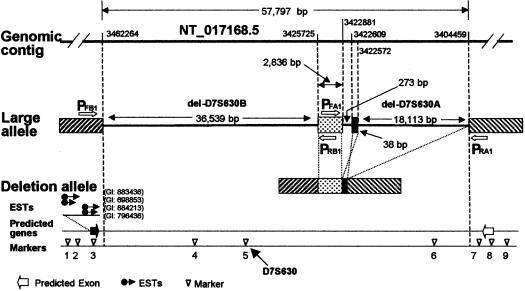
Figure 2.
Physical map of D7S630 region and structure of the deletion allele. Locations of deletion boundaries are given on the top genomic contig line as positions in NT_017168.5 (accession number GI:15298273). Arrows labeled PFB1 and PRB1 indicate the location of primers used to amplify del-D7S630. Arrows labeled PFA1 and PRA1 indicate the location of primers used to amplify del-D7S630-A (table 1 and fig. 3D). Genetic markers, with locations in NT_017168.5 given in parentheses, are as follows: 1, 2127-CT (3534931); 2, 2127-CA1 (3484630); 3, 2127-GT (3466041); 4, 2127-ATCT (3457424); 5, D7S630 (3452308); 6, 4746-TET (3412376); 7, 4746-CA (3398684); 8, rs1016807 (3370218); and 9, 4746-GT (3324233). GenScan-predicted genes are shown as unblackened arrows, and an EST-predicted gene is shown as a blackened arrow.
Genotype Analysis
For genotyping, DNA samples from families with autism were extracted from blood. Genotypes were determined by PCR amplification of polymorphic loci, performed with primers labeled with fluorescent probes. DNA fragments were resolved by use of an ABI 377 DNA sequencer. The computer program Loki was used to identify genotypes incompatible with Mendelian inheritance. SNPs were genotyped by single-nucleotide extension assays performed with SnapShot reagents (PE Biosystems), and the resulting products were analyzed by use of an ABI 377 Sequencer. Primers used to assay specific deletions alleles are given in table 1. Primer sequences used for SNPs, STRP loci, and DNA sequencing are in the online-only appendixonline-only appendix.
Table 1.
PCR Primers Used to Genotype Deletion Alleles[Note]
| Locus Primer/Contig | Primer Sequence(5′→3′) | Product Size(bp)a | Location in Contig |
| D7S630/NT_017168.5: | |||
| 2127-LG2-F | AGATCCCCATGACTATGTTTAGTACAGGG | 3466226 | |
| 2127-LG4-F | CATTGAGGGTAGAACTGAAACCATGGG | 3462955 | |
| 4746-LG1-R | AGTATAACCAGCAGTTAACAATTGAGGG | 3398803 | |
| 4746-LG2-R | ATACCTCCATCATTATTCCTAGGCTGG | 3404326 | |
| PFA1 | ATCTACAAAGGGGTGACTTTAGG | 400 | 3404174 |
| PRA1 | AAATGAAGACAATGTCAAACTTATTG | 3422995 | |
| PFB1 | GGCCAAATTATGTGCTATACAAGG | 161 | 3425643 |
| PRB1 | GTCAGCTTCTATACAGAGATTTGG | 3462342 | |
| D7S517/NT_007784.5: | |||
| PFA2 | CCCAAAACTCGGAAACGTGAAG | 374 | 122158 |
| PRA2 | AGTTTGGCACAGCCCATATAGG | 25811 | |
| PFB2 | CCTAGCTGCATAAAAACTCCTGG | 1687 | 219891 |
| PRB2 | GGACTGCGTGGAAGAGCTGG | 35217 | |
| D8S272/NT_008104.5: | |||
| PFA3 | GTGTAGTGGAGCCACTATGCTC | 181 | 1086974 |
| PRA3 | GATCAAGGGATGATGAGTATCTC | 1269241 |
Note.— Primer sequences for genotyping STRP and SNP loci are available in the online-only appendixthe online-only appendix.
Product sizes are for amplification of the deletion allele. For the nondeletion allele, no product is amplified.
Long-Range PCR Amplification.
DNA fragments spanning deletion junctions were amplified by use of long-range PCR. Amplification was performed by use of the Takara LA Taq DNA Polymerase Kit (Takara Shuzo), with primers at 0.2 μM each and 200 ng of template genomic DNA in a final volume of 50 μl. PCR profile consists of 1 cycle for 1 min at 94°C, 30 cycles for 20 s at 98°C, and 10 min at 68°C, and a final extension at 72°C for 10 min. The amplified products were analyzed by agarose gel electrophoresis, and fragments were visualized by use of ethidium bromide.
DNA Sequence Analysis
For DNA-sequence analysis, PCRs were carried out by mixing 20 pmol of primers and 40 ng of genomic DNA with HotStarTaq DNA Polymerase master mixture (Qiagen) in a final volume of 20-μl reactions. After 30 cycles, 2 μl of ExoSAP-IT (USB) was added to the PCR products and was digested for 1 h to remove the residual primers and dNTPs. For sequencing of long-range PCR products, fragments were purified by agarose gel electrophoresis, and bands were recovered by use of the Gel Band Purification Kit (Amersham Pharmacia Biotech). Sequencing reactions were performed with the BigDye Terminator cycle sequencing kit, using 4 μl of the processed PCR fragments in a final volume of 10 μl. Fragments were resolved using an ABI 377 DNA sequencer. Repeat sequences were identified by use of the RepeatMaster (version 4/4/2000), ProcessRepeats (version 4/4/2000), and Repbase (version 3/31/200), and the results were integrated by use of the Seqhelp search interface.
Generation of Somatic Cell Hybrids
GM459A cells—an HPRT-negative Chinese hamster ovary (CHO) cell line—were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco). A lymphoblastoid cell line (LCL) from family AU018, subject AU01803, was maintained in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FBS (Gibco). GM459A cells were seeded at a concentration of 107 cells per 100-mm tissue-culture dish. After 24 h, cells were washed with DMEM without serum, and 5 × 107 LCLs were added in 5 ml of DMEM without serum containing 125 ml of a 1 mg/ml stock of phytohemagglutin-P (Sigma). Dishes were incubated at 37°C for 15 min. The media were gently aspirated, and 2 ml of 50% polyethylene glycol (1,000 MW) (Sigma) was layered over each dish. After treatment for 1 min, cells were washed three times with DMEM and were incubated in 10 ml of serum-free DMEM at 37°C for 30 min. Cells were allowed to recover overnight in 10 ml DMEM, 10% FBS. The next day cells from each 100-mm dish were trypsinized and seeded 1:5 into fresh 100-mm dishes containing selection media composed of DMEM with 20% FBS and a 1× solution of hypoxanthine-aminopterin-thymidine (HAT) (Sigma). Cells were fed fresh selection media every 2–3 d. After 2–3 wk, individual colonies were transferred to a 16-mm well (24-well plate) and were expanded in duplicate 35-mm wells (6-well plates). One well was used for genomic DNA isolation and genotyping, while the other was used for further expansion and freezing of the clone. Clones were maintained in HAT selection media throughout the experiment.
Results
Families with two or more subjects affected with autism, PDD (case types 1 and 2), or broader-phenotype autism were identified for use in genetic linkage studies (table 2). Chromosomes 7 and 15 were initially targeted because previous studies indicated the presence of autism loci on these chromosomes (Barrett et al. 1999; Philippe et al. 1999). Chromosomes 8 and 16 were also analyzed as part of accompanying marker panels to be used in a complete genome screen. Genotypes for 66 dinucleotide STRP markers spaced at ∼10-cM intervals across these four chromosomes were checked for compatibility with reported family relationships. Apparent non-Mendelian inheritance was observed for six markers (D7S630, D7S517, D8S264, D8S272, D8S258, and D8S1784) in 18 families (table 3 and fig. 1). In these families, one or more children were homozygous and failed to inherit an allele from a homozygous parent. The incompatible genotypes could not be explained by mutations adding or deleting a single dinucleotide repeat unit (Weber and Wong 1993). These same families did not exhibit incompatible genotypes with other markers, thus eliminating inaccurate paternity information and sampling errors as the source of incompatibilities. The discrepancies in the families could be rectified if the incompatible parent-child pairs were assumed to have a null allele. Because the rate of apparently null alleles appeared higher than in our other studies, we determined the cause of these null-allele phenotypes.
Table 2.
Families with Autism[Note]
| Diagnosis of Sibs | No. ofFamilies |
| Autism/autism | 44 |
| Autism/PDD | 22 |
| Autism/PDD2 | 18 |
| Autism/BPH | 7 |
| PDD/PDD | 3 |
| PDD/PDD2 | 6 |
| PDD/BPH | 4 |
| PDD2/PDD2 | 1 |
| Total | 105 |
Note.— The average age of affected subjects at assessment is 8.95 years (SD = 4.76 years). The ethnicity is predominantly (84%) white. The mean full-scale IQ values for affected subjects, unaffected sibs, and parents are 74.9 (SD = 26.6), 107.9 (SD = 16.2), and 113.5 (SD = 13.0), respectively.
Table 3.
Deletions in Families with Autism[Note]
|
No. of Sibs (Diagnosis) |
||||
| Markerand Family | Sex of Parent with Deletion | With a Deletion | Without a Deletion | Deletion Size(kb) |
| D7S630: | ||||
| AU018 | Male | 2 (PDD) | 1 (unaffected) | 57 |
| D7S517: | ||||
| AU026 | Female | 1 (autism), 1 (PDD2) | 1 (unaffected) | >69 |
| AU044 | Male | 2 (autism), 1 (unaffected) | 0 | 183 |
| AU098 | Male | 1 (autism) | 1 (autism) | 96 |
| D8S264: | ||||
| AU025 | Female | 0 | 2 (autism) | <5.9 |
| AU058 | Female | 2 (autism) | 0 | 260–283a |
| AU113 | Female | 1 (autism) | 1 (PDD2), 1 (unaffected) | 252–266a |
| D8S272: | ||||
| AU002 | Male | 1 (autism), 1 (unaffected) | 1 (autism) | 192 |
| AU085 | Female | 3 (PDD2) | 0 | 192 |
| AU141 | Female | 2 (autism) | 0 | 192 |
| AU142 | Female | 1 (autism), 1 (PDD2) | 0 | 192 |
| UO2 | Female | 2 (autism), 2 (unaffected) | 1 (unaffected) | 192 |
Note.— Deletions were initially detected in DNA samples isolated directly from blood.
Sizes may not be correct, because of potential errors in the genomic DNA contig covering this region.
Null alleles can result from failure to amplify an allele caused by a polymorphic base in the sequence corresponding to the primers used to amplify the locus, a common source of genotype errors (Ewen et al. 2000). Therefore, for the six sites with potential null alleles, new primers were designed that were either both internal or both external to the original primer pairs. For D8S258 and D8S1784, previously unobserved alleles amplified with the new primers resulting in genotypes compatible with the family structures. However, for the remaining four loci, the incompatibilities were not resolved. These four loci were characterized as follows.
D7S630 Deletion
To characterize the D7S630 null allele, SNP and STRP loci flanking D7S630 were genotyped in family AU018 (figs. 1 and 2). For the two flanking STRP sites 4746-tet and 2127-ATCT, non-Mendelian inheritance compatible with a null allele was observed (fig. 1). These results are compatible with a deletion with a minimum size of 45.2 kb (fig. 2). In contrast, genotypes for the more distantly flanking STRP sites 2127-GT and 4746-CA were compatible with the family structure, defining a maximum deleted region of 67.3 kb.
The deletion allele was characterized by initially designing four PCR primer pairs flanking the obligate deleted segment. These primers were used in long-range PCR experiments to amplify across the junctions of the deletion, using DNA from deletion carrier AU01804. The nondeleted allele was not amplified because the distance between the primers was too great. Three different primer pairs produced fragments that were ∼9, ∼6.9, and ∼3.7 kb in size (fig. 3A). The 3.7-kb fragment could be amplified from family AU018 subjects predicted to have the null allele but not from family members heterozygous for D7S630 or from unaffected control subjects (fig. 3B). This 3.7-kb fragment was sequenced to define the breakpoints of the deletion. The structure of the deleted allele is complex. Three segments (36.5 kb, 18 kb, and 273 bp) are missing, and two segments (2,836 bp and 37 bp) are retained. The orientation of the shorter retained segment is reversed from its normal orientation (fig. 2). Thus the null allele in family AU018 is a 54,925-bp complex deletion.
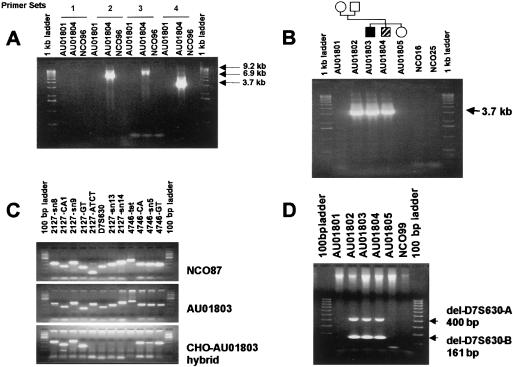
Figure 3.
Long-range PCR amplification of the D7S630 deletion allele. Genomic DNA samples are from the AU018 family (fig. 1) or unrelated unaffected control subjects (NCO samples). Amplified fragments were resolved by use of agarose gel electrophoresis and were detected with ethidium bromide staining. A, Long-range PCR performed to identify primer pairs that can amplify across the D7S630 deletion-allele junction. Primer combinations were as follows: 1, 2127-LG2-F and 4746-LG1-R; 2, 2127-LG2-F and 4746-LG2-R; 3, 2127-LG4-F and 4746-LG1-R; and 4, 2127-LG4-F and 4746-LG2-R (table 1). B, Primer pair D (2127-LG4-F and 4746-LG2-R) was used to assay for the D7S630 deletion allele in the entire AU018 family and in unaffected control subjects NCO16 and NCO25. C, Genetic markers were used as STSs in an unaffected control subject NCO087, in subject AU01803, who has the AU018 deletion allele, and in a CHO-AU01803 hybrid containing a single copy of human chromosome 7 that has the D7S630 deletion allele. D, Deleted segments del-D7S630-A and del-D7S630-B were amplified in a single PCR reaction, using primer pairs PFA1/PRA1 and PFB1/PRB1, respectively (table 1 and fig. 2). This combination of primer pairs was used to directly screen for the D7S630 complex deletion (table 4).
To confirm the presence and structure of the deletion, cells from deletion-carrier AU01803 were fused with CHO cells to obtain somatic-cell hybrid lines that contain a single human chromosome 7. Two lines were obtained, 1 with the deletion allele chromosome 7 and the second with the nondeletion allele chromosome 7. DNA from the hybrids was genotyped for sequence-tagged sites (STSs) located within and flanking the deletion. The STS content of each line was compatible with the predicted deletion (fig. 3C).
No known gene, EST, or GenScan-predicted genes were in the deletion. There is a cluster of EST sites immediately upstream of the deletion (fig. 2). Although these ESTs matched the genomic sequence, there was no evidence of intron-exon splicing, and this region was not part of a GenScan-predicted gene. A gene was predicted by GenScan, immediately downstream of the deletion that did not match any known genes or ESTs.
To facilitate large-population screening for the D7S630 deletion, primer sets PFA1/PRA1 and PFB1/PRB1 (table 1) were designed to separately amplify the segment of the deletion allele missing del-D7S630A and the segment missing del-D7S630B, respectively (fig. 2). The PCR products were 400 bp and 161 bp, corresponding to alleles missing 18 kb and 36.5 kb, respectively (fig. 3D). The same primers did not produce a product from the nondeletion allele.
D7S517 Deletion
Characterization of the D7S517 null alleles was performed by the same approach as used for the D7S630 deletion. Six STRP and 21 SNP sites flanking D7S517 were genotyped in the three families showing a D7S517 null allele. While not all sites were informative, the results indicated that each family had a different deletion (fig. 4 and table 3). For families AU044 and AU098, the deletions were entirely within genomic contig Celera-1R. For family AU026, the deletion extended past the end of the available sequence. Marker genotypes from the next contig (NT_007747) were compatible with the known family structure for AU026. Thus, one end of this deletion was not present in available genomic sequence. The three deletions have a 24-kb region in common that contains D7S517 (fig. 4). Three GenScan-predicted genes were in the deletion region, but only one was missing in all three types of deletions. These predicted genes did not match known genes or ESTs.
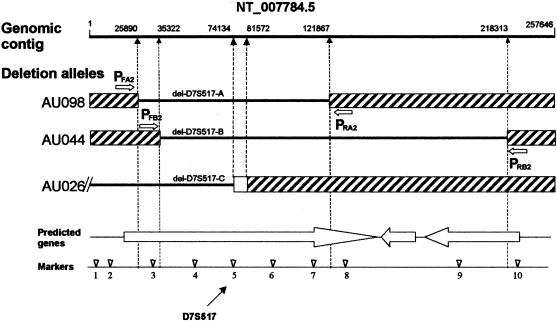
Figure 4.
D7S517 genomic structure and deletion alleles. Deletion boundary positions listed above the top line (genomic contig) are based on numbering from contig NT_007784.5 (accession number GI:15299808). Arrows PFA2 and PRA2 show primer locations for amplifying del-D7S517A. Arrows PFB2 and PRB2 show primer locations for amplifying del-D7S517B. Genetic markers with locations in NT_007784.5 given in parentheses are as follows: 1, rs975926 (4421); 2, CE1R-AGGG (8891); 3, CE1R-CA (29444); 4, 16898-AT (53633); 5, D7S517 (74190); 6, CE1R-AGAT (93628); 7, CE1R-GT (115042); 8, D7S472 (132296); 9, rs730813 (195200); and 10, rs2005745 (219539). Genes predicted by GenScan are shown as large unblackened arrows.
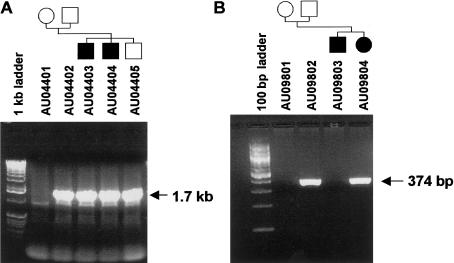
The DNA sequences for the deletion junctions for families AU044 and AU098 were determined as described above for D7S630. Primer sets PFA2/PRA2 and PFB2/PRB2, which flank the deletion junctions, produced 1.7-kb and 374-bp PCR products from null-allele subjects from families AU044 and AU098, respectively (fig. 5). These fragments were sequenced to identify the deletion junctions.
Figure 5.
Amplification of D7S517 deletion alleles. Deletion alleles were amplified with primer pairs PFA2/PRA2 and PFB2/PRB2 for del-D7S517A and D7S517B, respectively (table 1 and fig. 4). PCR products were resolved by agarose gel electrophoresis and were stained with ethidium bromide. Samples used and family structures are shown at the top.
D8S272 Deletion
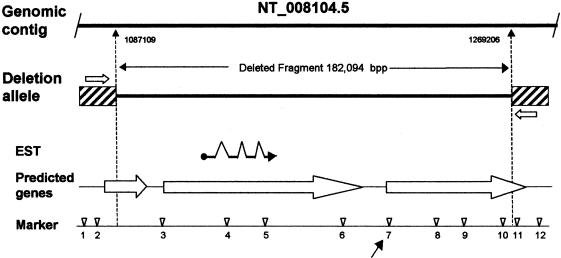
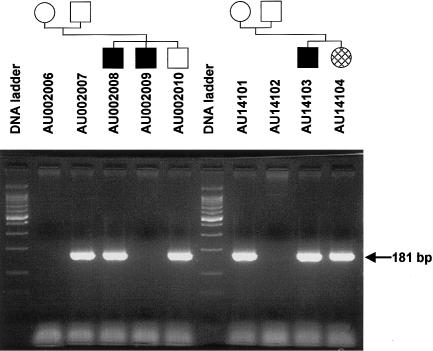
To characterize the D8S272 deletion, 11 STRP and SNP sites were genotyped in the five autism kindreds with null alleles at this site (fig. 6). The results were compatible with a deletion of minimum and maximum sizes of 157.1 kb and 207.2 kb, respectively. Long-range PCR was performed with primer pairs flanking the deletion site, using DNA from subjects with a D8S272 null allele. One primer set produced a 1.7-kb fragment in subjects with the null allele but no product in subjects without a null allele. Sequence from this junction fragment indicates the deletion allele is missing 192 kb. For all five families with this deletion, identical-sized fragments were produced when the deletion allele was amplified by use of the flanking primers. Thus the deletion appears to be identical in each family. To facilitate screening for this mutation, a second primer pair (PFA3/PRA3) was designed to produce a 181-bp product spanning the deletion (fig. 7).
Figure 6.
D8S272 region genomic structure and deletion alleles. Deletion-boundary positions given beneath the top line (genomic contig) are from contig NT_008104.5 (accession number GI:15300207). Arrows mark the location of primers PFA3 and PRA3 that were used to directly amplify the deletion allele (fig. 7). Genetic markers with locations in NT_008104.5 given in parentheses are as follows: 1, 26261-CT (1044194); 2, 8104-CCCT (1082018); 3, 8104-TG (1084178); 4, 13646-GTT (1138446); 5, 13646-CTTT (1155366); 6, 13646-ATTT (1193790); 7, D8S272 (1214065); 8, 13646-TA (1235221); 9, 13707-ATCT (1249914); 10, 13707-AAAT (1267145); 11, 12410-GT (1373533); and 12, 12410-CA (1410210). Genes predicted by GenScan are shown as large unblackened arrows.
Figure 7.
Amplification of the D7S272 deletion allele. Samples are from families with autism AU002 (left) and AU141 (right). Deletion alleles were amplified with primer pair PFA3/PRA3. PCR products were resolved by agarose gel electrophoresis and were stained with ethidium bromide. Samples used and family structures are shown at the top. Subjects showing a 181-bp PCR product were heterozygous for the D7S272 deletion allele and subjects showing no product were homozygous for the nondeletion long allele.
There were three GenScan predicted genes in the deletion region. The flanking two predicted genes did not match known genes or ESTs. The center predicted gene that is completely deleted matched an EST from a random activation of gene expression (RAGE) library (Harrington et al. 2001). Comparison of the EST sequence to the genomic sequence indicates the EST sequence is compatible with intron removal by RNA splicing (fig. 6).
D8S264 Deletion
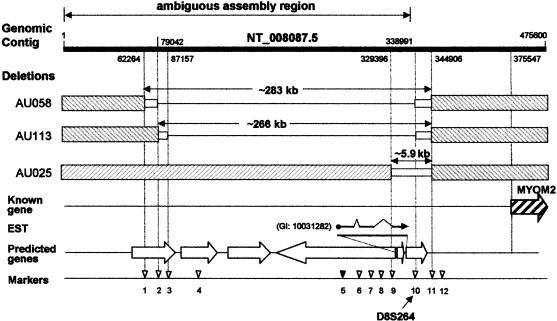
To characterize the D8S264 deletion, 65 STRP and SNP sites flanking D8S264 were genotyped in the three families with null alleles at this locus. Genotype results for some of these markers are shown in table 4 and locations for a subset of markers is shown in figure 8. Annotation of genomic contig NT_008087.5 for this region indicates that the sequence assembly may contain errors due to repeated sequences (fig. 8). Thus, the marker order for site 1–9 in figure 8 may be incorrect. When these polymorphic markers were genotyped in the three families with null alleles at D8S264, different patterns emerged for each family. For all three families, the deletion alleles appear to end at approximately the same site, defined by marker 11 in fig. 8. In each of the three families, at least one subject with a D8S264 null allele was heterozygous at SNP 11. Genotype patterns indicated that the other ends of the deletions were different in each family. For family AU025, marker 9 in figure 8 was also heterozygous in the parent and child with the null allele. The only marker showing a null allele in family AU025 was D8S264. Thus, the deletion in this family is <5.9 kb, and it may be considerably smaller. For family AU058, marker 2 in figure 8 was consistent with a null allele, whereas marker 1 was heterozygous and thus is outside the deleted region. The minimum and maximum sizes for this deletion are 260 and 283 kb, respectively. Family AU113 has a somewhat smaller deletion defined by markers 2 and 3 with a size range of ∼252–266 kb (fig. 8). Of the markers shown, all except site 5 were consistent with the deletion pattern shown in fig. 8. Marker 5 was heterozygous in the AU058 family parent who had the null allele. This inconsistency suggests that the contig assembly (as indicated by the GenBank annotation) is incorrect or that these deletions are complex, as was observed with D7S630. Because the genomic sequence was not reliable, we were unable to identify and clone fragments spanning these deletions.
Table 4.
Frequency of Deletions in Different Populations[Note]
|
Deletion Allele Frequencies (Chromosomes with Deletion/Total Chromosomes Sampled)a |
||||||
| Marker | Autism | Learning Disability | CEPH | Younger Control Subjectsb | Elderly Control Subjectsc | LOAD |
| D7S630 | .003 (1/388) | .0 (0/164) | .0 (0/238) | .0 (0/188) | .0 (0/574) | .0 (0/474) |
| D7S517 | .008 (3/388) | .0 (0/192) | .0 (0/253) | NA | NA | NA |
| del-D7S517A | .003 (1/388) | ND | ND | .0 (0/242) | .0 (0/504) | .0 (0/396) |
| del-D7S517B | .003 (1/388) | ND | ND | .0 (0/180) | .0 (0/498) | .0 (0/374) |
| del-D7S517C | .003 (1/388) | ND | ND | NA | NA | NA |
| D8S264 | .008 (3/388) | .0 (0/191) | .0 (0/270) | ND | ND | ND |
| D8S272 | .013 (5/388) | .005 (1/192)d | .104 (25/241) | .082 (14/170) | .035 (21/598) | .026 (13/494) |
Note.— ND = not determined. NA = not applicable. It is not possible to screen directly for del-D7S517C by a PCR assay. Therefore, the complete set of D7S517 deletions can be examined only by segregation of the D7S517 STRP in families with one or both parents sampled.
Autism, CEPH, and LD groups were initially screened for null alleles by examination of segregation patterns of marker genotypes in the families. All subjects in the families with autism were screened for deletions D7S630, del-D7S517A, del-D7S517B, and D8S272, by use of the direct assays described in figures 3, 5, and 7. These same deletion assays were used to screen the CEPH grandparents (in three-generation families) or parents (in two-generation families), and the control and LOAD populations, but not the families with LD. For the autism sample, the younger control subjects, elderly control subjects, and subjects with LOAD, the DNA samples were isolated directly from blood. For the subjects with LD, most of the DNA samples were isolated directly from blood with the rest from lymphoblastoid cell lines. All CEPH DNA samples were drawn from lymphoblastoid cell lines.
Younger control subjects were laboratory workers and subjects recruited by an advertisement for control subjects. The mean age was 36.9 years (SD = 12.7).
Elderly control subjects were spouses and caregivers of subjects with dementia who were seen by the University of Washington Alzheimer’s Disease Research Center. The mean age at the time of sampling was 73.5 years (SD = 8.56). Subjects with LOAD seen at the same clinic had a mean age at sampling of 73.3 years (SD = 9.34).
The families with LD were screened for the deletions only by use of segregation analysis of marker genotypes. The direct PCR assays for the deletions were not performed on these families.
Figure 8.
D8S264 region genomic and deletion-allele structures. Deletion-boundary positions given above and beneath the top line (genomic contig) are based on contig NT_008087.5 (accession number GI:15300158). The region labeled “ambiguous assembly region” potentially has contig-assembly errors due to repeat sequences (on the basis of the annotation provided for NT_008087.5). Genetic markers with locations in NT_008087.5 given in parentheses are as follows: 1, rs897445 (62264); 2, rs1824863 (79042); 3, rs922501 (87157); 4, rs899410 (87837); 5, rs1455638 (257680); 6, rs1382608 (304379); 7, rs958660 (309132); 8, 22257-ATCT (316326); 9, 8152018-CA (329355); 10, D8S264 (338805); 11, rs2119237 (344906); and 12, rs936553 (347075). Predicted genes are shown as unblackened arrows, and the known gene MYOM2 is shown as a hatched arrow.
Six GenScan-predicted genes were in the region covered by the deletions. Two are in the region potentially deleted in all three families. One of the predicted genes was homologous to a single EST that showed evidence of splicing (fig. 8). The closest known gene is the endosarcomeric cytoskeletal M-protein gene (MYOM2 [MIM 603509]).
Deletion Alleles in Subjects Not Affected with Autism
To determine whether the deletions observed are specific to families with autism, families affected with learning disability (LD) and families from CEPH were genotyped for D7S630, D7S517, D8S264, and D8S272 to detect null alleles. The families from CEPH consisted of 40 families with two or three generations sampled and 253 founder chromosomes. Founder chromosomes are defined as independent chromosomes that could potentially contribute a deletion allele. The families with LD were 28 kindreds sampled for 2–4 generations with 164–194 founder chromosomes sampled (depending on the marker used). These samples are comparable to the families with autism, which had 388 founder chromosomes sampled. No null alleles were detected in the CEPH and LD families for markers D7S630, D7S517, or D8S264. For D8S272, one family with LD and one family from CEPH (Z1416) had genotypes consistent with a D8S272 deletion allele.
Deletion Alleles in Unrelated Subjects
Because the D7S630, del-D7S517A, del-D7S517B, and D8S272 deletion alleles can be directly detected (figs. 3, 5, and 7), control populations were screened for these alleles (table 4). In addition, the CEPH family founders, were directly assayed for the deletion alleles, as were the autism family members. These families were rescreened, because, in some families, null alleles could be missed if genotypes were uninformative. Unrelated adult control subjects were also screened for these deletion alleles. A cohort of subjects with late-onset Alzheimer disease (LOAD [MIM 104300]) and age-matched control subjects were screened as representative of another neurologic disease, to determine specificity of the deletion alleles. Because the D7S630 deletion was complex, primer pairs were used that amplify independently across the two deleted segments (fig. 3D). For D7S630, del-D7S517A, and del-D7S517B, no examples of deletion alleles were observed in subjects from CEPH, elderly control subjects, younger control subjects, or subjects with LOAD. In contrast, for D8S272, deletion alleles were detected in all the non-autism groups tested (table 4). To detect potential D8S272-deletion homozygotes, samples with deletions were also genotyped for D8S272. An appropriately sized fragment was observed in all subjects with a deletion, from all samples used, indicating that none were homozygous for the deletion allele.
The deletion allele frequencies in subjects with autism were compared to the deletion allele frequencies in control subjects for each of the four deletion sites. Values from the autism sample were compared to each individual control group (LD, CEPH, elderly control subjects, younger control subjects, and subjects with LOAD) and to all control subjects for whom data were available pooled as one group. (Note that the deletion allele frequencies for D7S517, D7S630, and D8S264 were 0 for all control groups tested [table 4].) Contingency-table analysis was carried out with a Fisher exact test, estimated by Markov chain–Monte Carlo methods (Guo and Thompson 1992). For D7S630, D7S517, or D8S264, when the frequencies from the autism sample were compared with those of each control subject or those of the pooled control subjects, there was no significant difference. However, for D8S272, the difference between the autism group and the individual control groups or the pooled control subjects was highly significant (P<.0001 and P=.0018, respectively), with the deletion allele being less common in the subjects with autism, compared with control subjects, except for the LD group. There were also differences among control groups. The healthy control subjects, who were young, were different (P<.0001) from the elderly control subjects and subjects with LOAD (table 4). Also, the subjects from CEPH were different from the elderly control subjects and subjects with LOAD but not the younger control group. The autism sample differs from both the young (P<.0001) and the elderly control subjects (P=.04). In light of the significant results for D8S272, stratified exact contingency-table analysis was performed using either the three nonpolymorphic deletions (D7S630, D7S517, and D8S264) or all four deletion sites (+D8S272) together. For both the three- and the four-marker analyses, there was a significant difference between the autism sample and the pooled control sample (P<.0001).
Discussion
In the process of performing genetic linkage studies of autism, we identified, in 12 of 105 families, large-deletion alleles at four different sites on chromosomes 7 and 8. For three of these sites (D7S630, D7S517, and D7S264), deletion alleles were found in kindreds with autism but not in families with LD or in other control populations. For these three sites, each family with a deletion allele had a different segment deleted. For a fourth site, at D8S272, the deletion allele identified in families with autism was also observed in subjects not affected with autism, including healthy control subjects. For this site, each example of the deletion appeared to be identical.
Because deletions at D7S630, D7S517, and D8S264 were observed only in families with autism or PDD and not in control families (table 4), these deletions are potentially involved in genetic susceptibility to autism. However, interpretation of the results presented here is complicated by the fact that the characteristics of autism-susceptibility alleles are difficult to predict because the mode of inheritance for the disorder is unknown. The difference between concordance rates for MZ twins and those for DZ twins and nontwin sibs, along with the low risk to more-distant relatives, is highly suggestive of a multigenic mechanism for autism susceptibility, involving simultaneous defects at multiple loci. However, the actual number of different contributing loci is unknown, with the lower limit being three to five genes. A much larger number could be involved, particularly if some are small-effect loci. Also, genetic heterogeneity could add to the number of genes contributing to the autism phenotype studied here.
One interpretation of the results for D7S630, D7S517, and D8S264 is that the deletions themselves are autism-susceptibility alleles and can, in combination with other as-yet-unidentified loci, cause the disorder. This is a particularly intriguing possibility for the D7S630 deletion, because this site, at 98 cM, is close to the regions of positive linkage reported by several groups (maximum evidence for linkage is at 103–129 cM) (Ashley-Koch et al. 1999; Barrett et al. 1999; IMGSAC 2001b). Also, D7S630 is the closest marker to the proximal breakpoint for an interstitial 7q inversion reported in a subject with autism, though this breakpoint has not been precisely defined (Ashley-Koch et al. 1999). There is a statistically nonsignificant trend supporting a role for these deletions in the disease: 10 of 14 affected offspring but only 1 of 4 unaffected offspring had one of the deletions. The fact that the deletions at the three sites do not perfectly cosegregate with autism in families is consistent with a multigenic model in which different combinations of genes cause the same phenotype and specific susceptibility loci are not required for autism to occur. Thus, these families may have a high genetic load of autism high-risk alleles, and, even in an individual family, the disorder in sibs could be due to alleles at different subsets of susceptibility loci. For example, an affected subject without a deletion (e.g., subject AU09803) (fig. 5) could have a different complement of autism-susceptibility alleles than his or her sibling with the deletion. Likewise, subjects with the deletion but without ASD (e.g., all parents with the deletions and some sibs, as in the case of family AU044) (fig. 5) do not have a sufficient load of susceptibility alleles to have the disorder. The one deletion that is not consistent with this model is in family AU025, in which the shortest D8S264 deletion was only observed in the unaffected parent and child but not in the affected sibs. The argument against these deletions being susceptibility alleles is the way in which these sites were detected. The prior probability that an autism allele would be detected by sampling such an extremely small portion of the genome as was done here is extremely low. Because, in families with autism, the deletions at D7S630, D7S517, and D8S264 are rare, if these deletions are true susceptibility alleles, they would account for only a small fraction of the genetic factors contributing to autism in our families.
A second hypothesis is that alleles at autism-susceptibility loci cause deletions and other chromosomal rearrangements, possibly by increasing errors in meiosis. Under this model, the deletions observed here might not be the cause of autism, but rather the consequence of autism-susceptibility loci located elsewhere. Also, some of the events caused by these autism-susceptibility loci leading to the deletions occurred in previous generations. Autism would be the result of multiple specific deletions or mutations of genes critical to neurodevelopment early in life, and these critical deletions are not necessarily those described above. Thus the genes segregating with autism in the families studied here could be those that cause the deletions, the critical deletions themselves, or both. In either case, the genetics of autism would appear to be complex. The deletions presumably are related to meiosis rather than mitosis, because there is no evidence of mosaicism in genotype data generated from white blood cell DNA (data not shown). However, low levels of mosaicism might not be detected, so a mitotic mechanism cannot be excluded entirely. The deletions do not appear to be the result of unequal crossing over, because the breakpoint regions for a given deletion are not consistently homologous as is seen in genomic disorders in which deletion or duplication breakpoints occur in chromosome-specific low-copy repeats (duplicons) (Lupski 1998). Also, the breakpoints for each deletion do not share common repeat sequences (e.g., Alu repeats). That three different deletions were observed for both D7S517 (fig. 4) and D8S264 (fig. 8) indicates that these are independent events, possibly caused by sequences that predisposed these regions to meiotic errors. Because the different deletion junctions at a given site do not involve common repeated sequences, the predisposing factor(s) may involve higher-order chromatin structure. A similar model has recently been proposed to explain the complex genetics of panic disorder (Gratacos et al. 2001). A group of related duplications of a segment on chromosome 15 was observed in affected subjects with panic disorder and related phenotypes. The deletions are unstable and the de novo appearance and disappearance of deletions occur in the same pedigree. The model proposed for panic disorder is that this duplication region is predisposed to duplication/deletion events, because of the presence of duplicons, and that a mutation or variant in a gene(s) elsewhere causes an increased frequency of these duplication events. However, in contrast to autism, the duplications appear to involve mitosis errors, because mosaicism for the duplications is observed, and duplicons flank the duplicated sequences.
Evidence supporting an error-prone meiosis model for autism includes the broad range of deletions, duplications, and translocations reported in subjects with autism. These include a de novo ∼1-Mb deletion at 15q22-q23 in a subject with autism, developmental delay, and mild dysmorphism (Smith et al. 2000), an ∼6-Mb Xp22 deletion in a female subject with autism (Thomas et al. 1999), and a deletion of 17p11.2 in another subject with autism (Vostanis et al. 1994). Also, a 5–6-cM deletion of chromosome 20 was described in a subject with autism and Hirschsprung disease (HSD [MIM 235730]) (Michaelis et al. 1997). A 4,937-bp deletion at D15S822 was found in families with autism at a slightly higher frequency than in healthy control subjects (Nurmi et al. 2001). A number of subjects with deletions of 2q37.3 have been reported with autism and facial dysmophism (Burd et al. 1988; Ghazziuddin and Burnmeister 1999; Smith et al. 2001). Duplications of 15q11-q13 are observed in a subset of subjects with autism, either as an interstitial duplication of this region or as supernumerary pseudodicentric marker chromosome containing one or two extra copies of this region (Gillberg et al. 1991). In subjects with autism, numerous balanced and complex translocations have been reported, including several involving 7q. One is a balanced t(1;7)p22; q21 translocation within 1.5 Mb of D7S630 in a subject diagnosed with both autism and childhood-onsetschizophrenia (SCZD [MIM 181500]) (Yan et al. 2000). The second is an interstitial inversion of a large segment of 7q and the breakpoint nearest the centromere is in the proximity of D7S630 (Ashley-Koch et al. 1999). Several translocations involving 7q have been reported, and the chromosome 7 genes that are disrupted by two of these translocations have been identified (Vincent et al. 2000; Sultana et al., in press). Numerous translocations involving other chromosomes have been reported in subjects with autism and include unbalanced translocations with partial chromosomal monosomy (Vostanis et al. 1994) and balanced translocations with no obvious deleted segments. However, there is no obvious cluster of breakpoints at any specific chromosomal location. There is one report of a subject with a xeroderma pigmentosum group C (XPC [MIM 278720]) mutation who has autism (Khan et al. 1998). Whether both disorders occurred in this subject by chance or because of the DNA repair defect caused by the mutation in the XPC gene is unknown. It is difficult to determine whether the spectrum of genomic abnormalities seen in autism is unique, in part because ascertainment of these abnormalities is not systematic. Also, deletions the size detected here are not observable cytogenetically and have not been routinely sought in other disorders. However, autism does appear to be unique, in that there is no other disorder associated with cytogenetic abnormalities and deletions scattered throughout the genome.
More direct evidence that meiosis is altered in subjects with autism comes from the increased rate of recombination for a portion of chromosome 7 (Ashley-Koch et al. 1999) and for 15q11-q13 (Bass et al. 1999) seen in autism relative to control families. Additional work is needed to confirm the increased recombination rate in families with autism and to determine whether regions other than 7q and 15q11-q13 are involved. Increased recombination rates have not been observed as part of the phenotype of other inherited disorders, nor has there been a gene defect identified in a human disease leading to errors in meiosis.
The D8S272 deletion is a polymorphism allele present in control subjects at a frequency of 0.026–0.104 (table 4). Although other polymorphic in/del alleles have been reported, the D8S272 site is unique in terms of the size of the segment deleted. Other large in/del polymorphisms include D15S63, where a 28-kb deletion allele was characterized (Silverstein et al. 2001). This site is in the 15q11-q13 PWS/AS region, which is known to be highly unstable. Another large in/del is on chromosome 15, 25 kb downstream of the PML gene (MIM 102578) (Goy et al. 1995; Gilles et al. 2000). This polymorphic system has five common alleles that comprise different numbers of 30-kb tandem repeat sequences. The difference between the smallest and largest allele is ∼140 kb. Recently, a 33,048-bp insertion allele polymorphism was detected in the process of determining the DNA sequence of chromosome 20 (Deloukas et al. 2001). Numerous large deletions and insertions have been characterized as causing different genetic diseases, affecting either single or multiple genes. However, these disease-related duplications and deletions are not polymorphisms. Likewise, large deletions are common in tumors, although, again, these are acquired as somatic cell events and are not polymorphisms. At present, it is not clear how frequently large deletions comparable to the D8S272 site occur as non–disease-related polymorphism alleles. The detection of the D8S272 by sampling only a small fraction of the genome suggests that large-deletion alleles may be relatively common. Methods designed specifically to detect polymorphic sites—such as RFLP analysis, screening short and long tandem repeats for polymorphisms, and detection of SNPs and in/del sites either by direct DNA sequencing or by database sequence comparisons—will potentially miss the type of deletion allele observed here. Further work is needed to determine how common these sites are, by use of methods designed specifically to detect large-deletion alleles.
The frequency of the D8S272 deletion allele varied significantly between the young and elderly control groups and between the control groups and the autism sample, suggesting that the two alleles could have different phenotypic effects. However, because the sampling schemes were different for each of the control groups and the subjects with autism, a more likely explanation of these results is that the deletion allele frequency varies substantially in different populations.
The physiologic consequences, if any, of the deletion alleles detected here are difficult to predict. No gene of known function was affected by the deletions. A number of GenScan-predicted genes are in the deletions. One of these, in the D8S264 deletion region, matches a known EST that appears to be the product of a spliced message and thus is almost certainly a real expressed gene. The predicted gene in the center of the D8S272 region also matches a known EST. This EST was expressed from a promoter vector inserted randomly throughout the genome to generate a RAGE cDNA library for identification of novel undetected genes (Harrington et al. 2001) and thus might not be a real expressed gene. However, the EST sequence, when compared with the genomic sequence, is consistent with intron-exon splicing. Analysis of novel sequences from RAGE libraries suggests that >55% are actually expressed genes. Although, at the present time, it is unknown whether the other predicted genes are actually expressed, a recent evaluation of novel GenScan-predicted genes from chromosome 22 suggests that at least 27% of the predicted genes are expressed (Das et al. 2001).
The deletions encountered in the autism linkage study were unexpected, since similar findings have not been reported for other large linkage studies. Thus, it is not possible to evaluate the statistical significance of the deletion frequencies detected here. Since large-deletion alleles are not routinely detected by other methods for polymorphism discovery, large-deletion–allele polymorphisms may be an underappreciated class of genetic variability in humans. Deletions of the size reported here potentially could contribute to human phenotype variability, either by deleting one or more genes or by influencing the expression of neighboring genes. Thus, identification and characterization of large-deletion–allele polymorphisms may be important for understanding the influence of genetic variability on normal and disease-related phenotypes.
Acknowledgments
This research was supported by program project grant P01HD34565, from the National Institute of Child Health and Human Development (NICHD) and the National Institute on Deafness and Communication Disability (NIDCD), which is part of the NICHD/NIDCD Collaborative Program of Excellence in Autism. We gratefully acknowledge the contributions of the Diagnostic and Statistical Cores of the University of Washington Autism Program Project; Cathy Brock, who assisted in recruitment of participants; and the parents and children who participated in this study. This work was also supported in part by the Veterans Affairs Administration (support to G.D.S.), the Mental Illness Research Education and Clinical Center (support to W.H.R.), and the National Institutes of Health Learning Disabilities Center (grant P50 HD33812, to W.H.R. and E.M.W.). Work at the University of Utah was supported by grant HD35476 from NICHD, grant M01-RR00064 from the National Center for Research Resources, the Devonshire Foundation, and the Utah Autism Research Foundation. W.M.M. also thanks Janet Lainhart, Sally Ozonoff, Pamula Bennett, and Hilary Coon. We thank Thomas D. Bird, Stanley Gartler, Raymond Monnet, and Arno Motulsky for reading the manuscript.
Online-Only Appendix: Primer Sequences for Amplification of Polymorphic Sites Not Provided in the Text
Primer pairs have the same initial designation, followed by either an “F” (forward) or an “R” (reverse). For example, in table A1, 2127-ATCT-F and 2127ATCT-R are a primer pair for site 2127-ATCT. SNPs and STRPs are labeled as such. Unless otherwise noted, SNP sites are designated with a number followed by “-F” or “-R,” and the number is from the annotation of the sequence contig in GenBank and dbSNP. The “Comments” column annotates each primer, indicating if it was a site listed in one of the figures of the text. The size of the PCR product is given in the “Size” column; for SNPs, the exact size is given, and, for STRP sites, the size of a single allele is given.
Initial primer pairs used for D7S630, D7S517, D8S272, and D8S264 were from ABI, and the sequences are not provided by the manufacturer. Primers designed for these four loci to test for primer-site–polymorphism artifacts are listed at the end of each table as “alternate primers.”
Table A1.
Primers for D7S630[Note]
| Primer Name | Sequence(5′→3′) | Size(bp) | Comments |
| 2127-ATCT-F | GAATCTGCCTTACTAAGTAACAGG | STRP (fig. 2) | |
| 2127-ATCT-R | AGATTGGTAAGTAAGTAGGTAGG | 251 | STRP (fig. 2) |
| 2127-CA1-F | CAATTTGTAAGTAGTGACTATCCC | STRP (fig. 2) | |
| 2127-CA1-R | GTGAGATACAGATGTGTGACCC | 156 | STRP (fig. 2) |
| 2127-CT-F | GGGAAGTGTTTTTTGTTGTCCC | STRP (fig. 2) | |
| 2127-CT-R | GCGCCCAGCTAGTTATTAGTC | 123 | STRP (fig. 2) |
| 2127-GT-F | GAAGAATGAGCATTTAAGCTGGG | STRP (fig. 2) | |
| 2127-GT-R | CTTTAACATATGCCCTCTGCTG | 121 | STRP (fig. 2) |
| 2127-sn13-F | TTATCAGCAAAGCTAGAGGTCC | SNP (data not shown) | |
| 2127-sn13-R | TGATTGGTATGTCTGGTTCTCC | 204 | SNP (data not shown) |
| 16645-F | GCAGTAGTGAGGGTTAACAGG | SNP (data not shown) | |
| 16645-R | ACATGAGGTCAATAGACTTAAGG | 312 | SNP (data not shown) |
| 2127-sn8-F | TAAAATGTTCTCAGTAGTGACGG | SNP (data not shown) | |
| 2127-sn8-R | TCTTGAATTAAAGCTGTACCAGG | 257 | SNP (data not shown) |
| 2127-sn9-F | TGGCCTACTAGTACATTTTTACC | SNP (data not shown) | |
| 2127-sn9-R | CCCCAGGCTTCCAGTAAACC | 293 | SNP (data not shown) |
| 4746-AT-F | CGAAGTGACCTTGTGGGATGG | SNP (data not shown) | |
| 4746-AT-R | CTAAGCTAATGTTTGAATGAAATGC | 188 | SNP (data not shown) |
| 4746-CA-F | ATAATGTCTGTTAGCATAATCACC | STRP (fig. 2) | |
| 4746-CA-R | CGACATTGAAATATAGAGGACC | 198 | STRP (fig. 2) |
| 4746-GT-F | GAAATAGAGATTGAAGTAGAGTGG | STRP (fig. 2) | |
| 4746-GT-R | CAAGCACCTAGAATATGCCAGG | 193 | STRP (fig. 2) |
| 1016807-F | GGATATTGATTACTTCTGGCAGG | SNP (fig. 2) | |
| 1016807-R | ATTTCTGTATCACGTATTCTTTAGG | 206 | SNP (fig. 2) |
| 4746-tet-F | TTCACACACACAGAAAATAAGGG | STRP (fig. 2) | |
| 4746-tet-R | AAAAAAAGCGAAGAAAGGGAAGG | 328 | STRP (fig. 2) |
| D7S630n-F | TCACTGTCTCTTACCTCCCTT | Alternate primer for D7S630 | |
| D7S630n-R | TTGCCTCAACTGAGTCTCAT | 138 | Alternate primer for D7S630 |
Note.— All sequences are in contig NT_017168.5. SNPs 2127-sn8, 2127-sn9, and 2127-sn13 were in contig NT_000409 in GenBank, which was withdrawn, along with associated SNP annotations. The locations of these SNPs in NT_017168.5 of the polymorphic nucleotides, with the allelic nucleotides in parentheses, are as follows: SNP 2127-sn8, 3490356 (A/G); SNP 2127-sn9, 3474959 (C/T); and SNP 2127-sn13, 3441445 (C/T).
Table A2.
Primers for D7S517[Note]
| Primer Name | Sequence(5′→3′) | Size(bp) | Comments |
| 1006344-F | CAGCAAGTCCTGGTTTTACACC | SNP (data not shown) | |
| 1006344-R | GAGGATTCCTGGTGCAAAACC | 415 | SNP (data not shown) |
| 1009527-F | AGCCTACGTGCAGGGTAAAGG | SNP (data not shown) | |
| 1009527-R | CAATAGAGGTGAATGTGTGTTGG | 214 | SNP (data not shown) |
| 1075415-F | CGAGTCAGATGAAGTAATTAGG | SNP (data not shown) | |
| 1075415-R | TACGTCACTAATGTACTTGTCTTG | 600 | SNP (data not shown) |
| 1447398-F | CCTTTCCTCTATGCCTGGACC | SNP (data not shown) | |
| 1447398-R | AACTAACTGCTTTGTGACTGACC | 392 | SNP (data not shown) |
| 1447401-F | CCAGGAAAGTTGATTCTGCAGTG | SNP (data not shown) | |
| 1447401-R | TCTGTGATTGAAGCTCTGTTTGG | 416 | SNP (data not shown) |
| 1467933-F | ATCTCTGATTGATTGAAAGGTACC | SNP (data not shown) | |
| 1467933-R | CCTCCAGTCATAGCAAGAAGCC | 715 | SNP (data not shown) |
| 1557961-F | GGTTAGAAGAGTTGGTGTGACC | SNP (data not shown) | |
| 1557961-R | TCTCCCCAAATTTATTTCAAAGCC | 298 | SNP (data not shown) |
| 16898-AT-F | AGAATGAGCCTGGCTTTAATTAC | SNP (fig. 4) | |
| 16898-AT-R | AAGGTTTTGACGTGTTACTCAGG | 420 | SNP (fig. 4) |
| 2005745-F | GCGCGTCTGCGGTTTGCTCC | SNP (fig. 4) | |
| 2005745-R | GACCTGGAGAAACTTTGTGTCC | 474 | SNP (fig. 4) |
| 314591-F | CAAAAGGCAAGCGTTCTTCTGG | SNP (data not shown) | |
| 314591-R | GAGTCACCCAACTCTTCCAGG | 455 | SNP (data not shown) |
| 314605-F | CTTTCTCTGGTACATTTCAGTGG | SNP (data not shown) | |
| 314605-R | CTACTGCATGTTGAAGCCTTGG | 683 | SNP (data not shown) |
| 314654-F | TGTGTGCGATATATACCTCTTGG | SNP (data not shown) | |
| 314654-R | TGAATAGCAGCTCCATTAGTTGG | 1032 | SNP (data not shown) |
| 730813-F | CCGTAGCAAAGGACCACAGG | SNP (fig. 4) | |
| 730813-R | CCAGATTCCAGATTTGGTTAGG | 514 | SNP (fig. 4) |
| 740128-F | CGAGTTGAGGGAACATAGATCC | SNP (data not shown) | |
| 740128-R | AATCAATAATTGTTGCTAAGTTGCC | 510 | SNP (data not shown) |
| 879946-F | CAGGGTGGCTTTGGTTGTTCC | SNP (data not shown) | |
| 879946-R | AGACTAGATGTTTCCTTGAGGC | 550 | SNP (data not shown) |
| 895705-F | GATTCAGGACAGCTTGGTAAGG | SNP (data not shown) | |
| 895705-R | CACATCACACGTCTTAGGAATTGCTGG | 854 | SNP (data not shown) |
| 895706-F | GGAATAGGGTATGCATGCTGG | SNP (data not shown) | |
| 895706-R | AAAAACTTTTAGGCTGGTCATGG | 418 | SNP (data not shown) |
| 917118-F | AGCAATCTCAGACACAGAATCC | SNP (data not shown) | |
| 917118-R | ATATTCAGACAAGTAGTCAATCAC | 247 | SNP (data not shown) |
| 929347-F | CTCTGACACAGCTCTGGTGG | SNP (data not shown) | |
| 929347-R | TTGGATTACGAATTGTCTCTTGG | 248 | SNP (data not shown) |
| 929372-F | GGAAGCTTCTGTCCTAGCTCC | SNP (data not shown) | |
| 929372-R | TCAGGACCCAAGGGGCAACC | 434 | SNP (data not shown) |
| 975926-F | GCCACAGCAGATAGTTTATGCC | SNP (fig. 4) | |
| 975926-R | TCCTCTGCCTCAATATGTGACC | 945 | SNP (fig. 4) |
| 979365-F | ACTCAATATTATGACAACAGCAGG | SNP (data not shown) | |
| 979365-R | TGAACCACCGCACACAGACTG | 470 | SNP (data not shown) |
| CE1R-AGAT-F | ATATACAGATTAGATAGAAAGATAGG | STRP (fig. 4) | |
| CE1R-AGAT-R | GTCAAGTAACACATGCTCTCTATG | 204 | STRP (fig. 4) |
| CE1R-AGGG-F | TGGAGCAGAGAGGCACAAGCC | STRP (fig. 4) | |
| CE1R-AGGG-R | CTCCCTTTTCCCGTTTTCCTCC | 276 | STRP (fig. 4) |
| CE1R-CA-F | TACACGTGACAAATTTGCACAGAG | STRP (fig. 4) | |
| CE1R-CA-R | GTTTTATGTGCACTCTTGCATGTG | 93 | STRP (fig. 4) |
| CE1R-GT-F | GAGGCATGAACTTGAAGATGTGG | STRP (fig. 4) | |
| CE1R-GT-R | TCTGCATGAAAACCTTCAGGTGG | 178 | STRP (fig. 4) |
| D7S3056-F | CTCAATAGCCCTGACCTTATGC | STRP | |
| D7S3056-R | TACCTACCTACCTACCTCTATGGC | 179 | STRP |
| D7S472-F | GGTGTCAAAAAGAGTTATGCTTG | STRP (fig. 4) | |
| D7S472-R | GAGTGTTTAATTCCATAAGCCCA | 120 | STRP (fig. 4) |
| D7S517-F1 | AGACTTCGAGTTTAAAAGGGAGG | Alternate primer for D7S517 | |
| D7S517-R1 | GAAAGCTATTTTAAGTAATTACGTGG | 257 | Alternate primer for D7S517 |
| D7S517-F2 | CTCAGATTTGGGGCTGTTACC | Alternate primer for D7S517 | |
| D7S517-R2 | CCCTGGCCCCATAGGTAACC | 226 | Alternate primer for D7S517 |
Note.— All sequences are in contig NT_007784.5. Sites with designations beginning with “CE” were originally from Celera contig CE1R and are all STRP sites.
Table A3.
Primers for D8S272[Note]
| Primers | Sequence(5′→3′) | Size(bp) | Comments |
| 1020849-F | CAGCTTGGATGTGCTGCATGG | SNP (data not shown) | |
| 1020849-R | ACTGCATATGTACCTGATCCTG | 280 | SNP (data not shown) |
| 1020850-F | CATTTGCTCTAGCCACAGATGG | SNP (data not shown) | |
| 1020850-R | CTGTCAAACTTTGATGACTCGG | 145 | SNP (data not shown) |
| 1157761-F | CCCACACCACCTCATGTCACC | SNP (data not shown) | |
| 1157761-R | GTTTGGTCAGATGTTCTCCAAACC | 215 | SNP (data not shown) |
| 12410-CA-F | TCAGTTAAGACTTTTATAGCTGTTGG | STRP (fig. 6) | |
| 12410-CA-R | GCTTTGCTGACTACAGATCTTAG | 136 | STRP (fig. 6) |
| 12410-GT-F | TATGGTAAGTGAGATTTCTAAAAACC | STRP (fig. 6) | |
| 12410-GT-R | CCATTTTCCTATCCACCAGCTCC | 303 | STRP (fig. 6) |
| 13646-ATTT-F | TGGGAAAATGCCCATGAGATTCC | STRP (fig. 6) | |
| 13646-ATTT-R | GTTATGATAGAATCTACAGTCAACTC | 158 | STRP (fig. 6) |
| 13646-CTTT-F | ACTGTCAATATCTGTTCACTTAATGG | STRP (fig. 6) | |
| 13646-CTTT-R | AATAAATACTTTTGAACTCAAATACAGG | 223 | STRP (fig. 6) |
| 13646-GTT-F | GAGTACTTGGGTTATGTGTGTCGG | STRP (fig. 6) | |
| 13646-GTT-R | ATATAACTTATAATTATATGTAATCATTATG | 158 | STRP (fig. 6) |
| 13646-TA-F | TGGCTTGGGGTCTGTAGATCTTC | STRP (fig. 6) | |
| 13646-TA-R | GAAGACATAGTGTTTATCTTAGTTCC | 198 | STRP (fig. 6) |
| 13707-AAAT-F | GGAAGTGTAATATGAGCCTTTGGG | STRP (fig. 6) | |
| 13707-AAAT-R | CAGTGGTTATTCTGTTGTTTGTTTGG | 121 | STRP (fig. 6) |
| 13707-ATCT-F | TGGCTCTCAAAGCTGGCCACGG | STRP (fig. 6) | |
| 13707-ATCT-R | CACATGGACGCACATGAAAGATAG | 177 | STRP (fig. 6) |
| 1381019-F | TCAAGGTATTGCTAAAAGTCCTGG | SNP (data not shown) | |
| 1381019-R | AAATAGGGTCAGTCCAACACAGG | 190 | SNP (data not shown) |
| 1381020-F | ATAAATCTCAGTGGTTTTTGAAAAGG | SNP (data not shown) | |
| 1381020-R | TTTAAGGGATTATAATGCTGGATGG | 275 | SNP (data not shown) |
| 1381022-F | CTAGCACAACGCCTACAAGTGG | SNP (data not shown) | |
| 1381022-R | TGGCTAATAATGTGTCCTCTATGG | 240 | SNP (data not shown) |
| 1461001-F | TTTGTCCAAAGACTTGTGCCAGG | SNP (data not shown) | |
| 1461001-R | AGATACAACCAATAATATGGCTGTG | 210 | SNP (data not shown) |
| 1461002-F | GAATAAAGGCAAGAATTGCTTCACG | SNP (data not shown) | |
| 1461002-R | GGCTGCAACTGCCCTGTCCG | 250 | SNP (data not shown) |
| 1461003-F | AGCTCTGTGTCATTCATTAGTAGC | SNP (data not shown) | |
| 1461003-R | CAAACTACACTTGTACCCTGACC | 180 | SNP (data not shown) |
| 1499456-F | AAAGTGAATAGCAAAACCACAGCC | SNP (data not shown) | |
| 1499456-R | AAGGCAGTCTAACGTTACATTTCC | 240 | SNP (data not shown) |
| 1499457-F | TACATGTCATACATGACATACACC | SNP (data not shown) | |
| 1499457-R | GGCAGTCTAACGTTACATTTCC | 200 | SNP (data not shown) |
| 1550705-F | CAAAGGACTGTGGTGCATTTCC | SNP (data not shown) | |
| 1550705-R | TTTTCCTTCAGCACACTTAAGCC | 179 | SNP (data not shown) |
| 16080-GT-F | TGTCCTCTAGTTCCATCTAAGTGG | STRP (data not shown) | |
| 16080-GT-R | GTCCCCGTCAATGGATGAGTGG | 147 | STRP (data not shown) |
| 16080-TA-F | GTGTTCTCCCATTGAATTGAAAAGG | STRP (data not shown) | |
| 16080-TA-R | TGCTCCAAACACTTCTCTGCTGG | 246 | STRP (data not shown) |
| 17045-F | AGCTAACTCCCTCAATTGGCCA | SNP (data not shown) | |
| 17045-R | TGCTCTAATAACTAGCAGGCTGG | 147 | SNP (data not shown) |
| 26261-CT-F | CGTCTTATCGTAACCTAAGGTGCC | STRP (fig. 6) | |
| 26261-CT-R | CTAATTTGGTATCTGTTTAATATGTTCC | 218 | STRP (fig. 6) |
| 305301-F | GTTATTTGCTTAGCCCTGGATCC | SNP (data not shown) | |
| 305301-R | GGAGGAACAGACATATGTATACC | 294 | SNP (data not shown) |
| 305347-F | AAAAAGAATATCATACTTCTACTCTC | SNP (data not shown) | |
| 305347-R | AACTTGGGTCAACACTCTCTCC | 237 | SNP (data not shown) |
| 305350-F | ATTGAGACTACCCCATATTCAGG | SNP (data not shown) | |
| 305350-R | GTCATGGAAACACTATAAGTCCTG | 185 | SNP (data not shown) |
| 305361-F | GTTCACTATAGTCAACTGTGACC | SNP (data not shown) | |
| 305361-R | GGTCATGAGAATTGATCTGGCTC | 192 | SNP (data not shown) |
| 305365-F | GGTGATACACACTGGCAGTTCC | SNP (data not shown) | |
| 305365-R | TTCAGAGTCTCTCAGCAGGACC | 298 | SNP (data not shown) |
| 305369-F | TGAGGTGTTATAGAAAGAGTATGAG | SNP (data not shown) | |
| 305369-R | TTCCTGTGTCCATGATAGAATGG | 228 | SNP (data not shown) |
| 8104-CCCT-F | TTACAATTCAGCAATAGTACTTGTTAG | STRP (fig. 6) | |
| 8104-CCCT-R | CACGGCACGTAACTTGGCAAATG | 162 | STRP (fig. 6) |
| 8104-TG-F | AGGACAGCAGTGGTGTCCTGG | STRP (fig. 6) | |
| 8104-TG-R | AGATATTAAGAGAGACAAAATGCTAAG | 125 | STRP (fig. 6) |
| 990663-F | GAAAGAGCATCGGTTCTTGATGG | SNP (data not shown) | |
| 990663-R | GTTCTGAATGCCTTTACGTGTGG | 175 | SNP (data not shown) |
| AFM175xb4a | GAGAACTAATCCCTTCTGGC | STRP (data not shown) | |
| AFM175xb4m | AGCTTCATAAAGAGTCTGGAAAAT | 192 | STRP (data not shown) |
| D8S272-2F | CTTATGACATAATGAAACTCAACC | Alternate primer for D7S272 | |
| D8S272-2R | ATAACAAAATGCTATTGAGTTCTTCC | 224 | Alternate primer for D7S272 |
| D8S272-3F | AGCTTCATAAAGAGTCTGGAAAAT | Alternate primer for D7S272 | |
| D8S272-3R | CACACACTCTTACACACAACGG | 221 | Alternate primer for D7S272 |
Note.— All sequences are in contig NT_008104.5.
Table A4.
Primers for D8S264[Note]
| Primers | Sequence(5′-3′) | Size(bp) | Comments |
| 1190556-F | AACACTCCTTTATCTAAAGAGTCG | SNP (data not shown) | |
| 1190556-R | TAATTAGCAACGGACCTTACACC | 510 | SNP (data not shown) |
| 1382608-F | GTCTACCATGACTGTCCGAGG | SNP (fig. 8) | |
| 1382608-R | CCATGCTTCAGGCCAATACAGG | 199 | SNP (fig. 8) |
| 1440290-F | AAAGATGCCAAGTCTTCCATTCC | SNP (data not shown) | |
| 1440290-R | CCACATTTGAACGGCATCACATC | 912 | SNP (data not shown) |
| 1455635-F | GAGGATACAAGGGACTCTGTAC | SNP (data not shown) | |
| 1455635-R | AGGAGAATGGGTCTTTTTCACC | 430 | SNP (data not shown) |
| 1455638-F | CTATCTTCAGTTTTTTTGAGGAACC | SNP (fig. 8) | |
| 1455638-R | GCAAAAGCAGGGCACTACATCC | 249 | SNP (fig. 8) |
| 1455641-F | TCCTTTATGCTCCTCCAAACAGG | SNP (data not shown) | |
| 1455641-R | CATTGTTATCAATGCACTCCTAGG | 280 | SNP (data not shown) |
| 1455642-F | TTCCACCTCATGGCTCGTACC | SNP (data not shown) | |
| 1455642-R | ATTACAAATTTGGACTGGAACCTC | 292 | SNP (data not shown) |
| 1457445-F | TTGGTTATTGAGATTTGTGAAATTC | SNP (data not shown) | |
| 1457445-R | TTTTACTGCTTCAGGTATCACAC | 300 | SNP (data not shown) |
| 1457450-F | CTCTATCCAACGCTGTCTTTGG | SNP (data not shown) | |
| 1457450-R | ATCATAAATAACATTAGATTGACATG | 310 | SNP (data not shown) |
| 153108-F | TAGCAACAAAAGCTGTACTTTTGG | SNP (data not shown) | |
| 153108-R | GTGAGAAAACCATAATCATTCCATG | 312 | SNP (data not shown) |
| 1625100-F | AAGCCTGCTGAGATTACTACACC | SNP (data not shown) | |
| 1625100-R | ACCAGCCCCAATGAGACGAACC | 240 | SNP (data not shown) |
| 1677799-F | AAGACACGGGAGACCTCATTGG | SNP (data not shown) | |
| 1677799-R | GACTGTGTGACGGAACCACAGG | 185 | SNP (data not shown) |
| 1824863-F | GTTAATGAGCATGTGTATGTCAC | SNP (fig. 8) | |
| 1824863-R | GCGCTTCGGGATATGTTGTCC | 320 | SNP (fig. 8) |
| 1870118-F | GAGGTACGGGCTTCTGAGAGG | SNP (data not shown) | |
| 1870118-R | GAACAGAGACCTGAACCCTGG | 427 | SNP (data not shown) |
| 1871320-F | CTGAGAAACATGACCTTTCATCC | SNP (data not shown) | |
| 1871320-R | CAGACAGCAGACAGGGTATCC | 282 | SNP (data not shown) |
| 1947488-F | CAGAGTGATGGAGCATGCAACC | SNP (data not shown) | |
| 1947488-R | TTGGACTTCTGTTGCCTCATCC | 451 | SNP (data not shown) |
| 2119237-F | AAATCACTTGGTGAAGATATCCAC | SNP (fig. 8) | |
| 2119237-R | CCAGTGGCAGGTCCTGACTC | 214 | SNP (fig. 8) |
| 22257-ATCT-F | GAGTTTATGAACCATGCTCAAGG | STRP (fig. 8) | |
| 22257-ATCT-R | TATATACAGAAAAATAGATAGTAGAGG | 201 | STRP (fig. 8) |
| 22257-comp-F | GGCTCTTACATTCTCTAATACTATGGG | STRP (data not shown) | |
| 22257-comp-R | GAAGCTATTCTGTGAGTTTATGTGTGG | 976 | STRP (data not shown) |
| 767271-F | TGTCCATGGTTTGCTGAGTTCC | SNP (data not shown) | |
| 767271-R | ACCTCCAGGAATTCGCAGTCC | 332 | SNP (data not shown) |
| 8087-GT-F | TGGTCAATGTAATCAGTGTAACTGG | STRP (data not shown) | |
| 8087-GT-R | AATGGGTATATGTGAGTACATAGAC | 210 | STRP (data not shown) |
| 8152018-CA-F | CTGATCCTAATTAATAGAAAGCAACC | STRP (fig. 8) | |
| 8152018-CA-R | AATCACTGTACCTTGTTACTTGACC | 201 | STRP (fig. 8) |
| 897445-F | CCTTACCTGTTAGCATCTGTTGG | SNP (fig. 8) | |
| 897445-R | GATTAGAAACCATGATAGCTCTTGG | 188 | SNP (fig. 8) |
| 899397-F2 | CCGCTGCCTTTATTCAACTTCTC | SNP (data not shown) | |
| 899397-R | ATATGACCTAATAGTAATCATAACTC | 280 | SNP (data not shown) |
| 899410-F | TGCATTCTTACCATCAAAAGTCC | SNP (fig. 8) | |
| 899410-R | TACGGTCTGCGGTTTACTGCC | 488 | SNP (fig. 8) |
| 899411/3-F | GAAGAGTCCTAAGTTTAGAGTCC | SNP (data not shown) | |
| 899411/3-R | AGCCTGGGACACCGTGTCAC | 294 | SNP (data not shown) |
| 922501-F | GCATTCTGTTCAAACTAATATCAGG | SNP (fig. 8) | |
| 922501-R | GTCCATTATAAAGGCATTTCAAAGAG | 198 | SNP (fig. 8) |
| 930843-F | TCTTCTAAGCTGAGCTCATCGC | SNP (data not shown) | |
| 930843-R | GGTTCACCGTTATTTCCTAGTCC | 794 | SNP (data not shown) |
| 936553-F | AGTGCCCAATACACGTAAATGTG | SNP (fig. 8) | |
| 936553-R | TGTACTGTCAAAATGAAGGCTCG | 780 | SNP (fig. 8) |
| 958660-F | GACTGACTTGTGTCTCCTCTACC | SNP (fig. 8) | |
| 958660-R | CTCCCCTTTGTGTAAGGTCACC | 180 | SNP (fig. 8) |
| AFM143xd8a | ACATCTGCGTCGTCTTCATA | STRP (data not shown) | |
| AFM143xd8m | CCAACACCTGAGTCAGCATA | 121 | STRP (data not shown) |
| D8S264-2F | TTGGGGAGGCAAAGTGCTCC | Alternate primer for D7S264 | |
| D8S264-2R | GAGAAGATGTGAATGGGAAAGCC | 200 | Alternate primer for D7S264 |
| D8S264-3F | GGCCTCCTGGGAGATTCGG | Alternate primer for D7S264 | |
| D8S264-3R | CCGGCAGGACTGAGCGTGG | 224 | Alternate primer for D7S264 |
Note.— All sequences are in contig NT_008087.5.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Database, http://www.ncbi.nlm.nih.gov/SNP/ (for data on individual SNPs)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequence and SNP information for genomic contigs NT_017168.5 [accession number GI:15298273], NT_007784.5 [accession number GI:15299808], NT_008104.5 [accession number GI:15300207], and NT_008087.5 [accession number GI:15300158])
- Genetic Epidemiology of Complex Traits, http://www.stat.washington.edu/thompson/Genepi/ (for the computer program Loki)
- Seqhelp, http://biocommons.bcc.washington.edu/services/psoftware.seqhelp/seqhelpinput.html (for identification of repeat sequences using RepeatMaster, ProcessRepeats, and Repbase)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism [MIM 209850], FMR1 [MIM 309550], TS [MIM 191100], PWS [MIM 176270], AS [MIM 105830], HOXA1 [MIM 142955], HOXB1 [MIM 142968], EN-2 [MIM 131310], SLC6A4 [MIM 182138], RELN [MIM 600514], WNT2 [MIM 147870], VIPR2 [MIM 601970], RAY1 [MIM 600833], GABRB3 [MIM 137192], VCFS [MIM 192430], MYOM2 [MIM 603509], LOAD [MIM 104300], HSD [MIM 235730], SCZD [MIM 181500], XPC [MIM 278720], and PML [MIM 102578])
References
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders (4th ed). American Psychiatric Association, Washington, DC, pp 65–78 [Google Scholar]
- Asano E, Kuivaniemi H, Huq M, Tromp G, Behen M, Rothermel R, Herron J, Chugani DC (2001) A study of novel polymorphisms in the upstream region of vasoactive intestinal peptide receptor type 2 gene in autism. J Child Neurol 16:357–363 [DOI] [PubMed] [Google Scholar]
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, Delong GR, Cuccaro ML, Pericak-Vance MAJ (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Baker P, Piven J, Schwartz S, Patil S (1994) Brief report: duplication of chromosome 15q11-13 in two individuals with autistic disorder. J Autism Dev Disord 24:529–535 [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Bass MP, Menold MM, Wolpert CM, Donnelly SL, Ravan SA, Hauser ER, Maddox LO, Vance JM, Abramson RK, Wright HH, Gilbert JR, Cuccaro ML, DeLong GR, Pericak-Vance MA (1999) Genetic studies in autistic disorder and chromosome 15. Neurogenetics 2:219–226 [DOI] [PubMed] [Google Scholar]
- Bolton P, MacDonald H, Pickles A, Rios P, Goode S, Crowson M, Bailey A, Rutter M (1994) A case-control family history study of autism. J Child Psychol Psychiatry 35:877–900 [DOI] [PubMed] [Google Scholar]
- Burd L, Martsolf JT, Kerbeshian J, Jalal SM (1988) Partial 6p trisomy associated with infantile autism. Clin Genet 33:356–359 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E (1998) Linkage disequilibrium mapping of autistic disorder, with 15q11-12 markers. Am J Hum Genet 62:1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Courchesne R, Lord C, Cox NJ, Yan S, Lincoln A, Haas R, Couchesne E, Leventhal BL (1997a) Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry 2:247–250 [DOI] [PubMed] [Google Scholar]
- Cook EH, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E (1997b) Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet 60:928–934 [PMC free article] [PubMed] [Google Scholar]
- Das M, Burge CB, Park E, Colinas J, Pelletier J (2001) Assessment of the total number of human transcription units. Genomics 77:71–78 [DOI] [PubMed] [Google Scholar]
- Deloukas P, Matthews LH, Ashurst J, Burton J, Gilbert JGR, Jones M, Starvrides Geal (2001) The DNA sequence and comparative analysis of human chromosome 20. Nature 414:865–871 [DOI] [PubMed] [Google Scholar]
- Ewen KR, Bahlo M, Treloar SA, Levinson DF, Mowry B, Barlow JW, Foote SJ (2000) Identification and analysis of error types in high-throughput genotyping. Am J Hum Genet 67:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977a) Genetic influences and infantile autism. Nature 265:726–728 [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977b) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18:297–321 [DOI] [PubMed] [Google Scholar]
- Ghazziuddin M, Burnmeister M (1999) Deletion of chromosome 2 q37 and autism: a distinct subtype? J Autism Dev Disord 29:259–263 [DOI] [PubMed] [Google Scholar]
- Gillberg C, Steffenburg S, Wahlstrom J, Gillberg IC, Sjostedt A, Martinsson T, Liedgren S, Eeg-Olofsson O (1991) Autism associated with marker chromosome. J Am Acad Child Adolesc Psychiatry 30:489–494 [DOI] [PubMed] [Google Scholar]
- Gilles F, Goy A, Remache Y, Manova K, Zelenetz AD (2000) Cloning and characterization of a Golgin-related gene from the large-scale polymorphism linked to the PML gene. Genomics 70:364–374 [DOI] [PubMed] [Google Scholar]
- Goy A, Passalaris T, Xiao YH, Miller WH, Siegel DS, Zelenetz AD (1995) The PML gene is linked to a megabase-scale insertion/deletion restriction fragment length polymorphism. Genomics 26:327–333 [DOI] [PubMed] [Google Scholar]
- Gratacos M, Nadal M, Martin-Santos R, Pujana MA, Gago J, Peral B, Armengol L, Ponsa I, Miro R, Bulbena A, Estivill X (2001) A polymorphic genomic duplication on human chromosome 15 is a susceptibility factor for panic and phobic disorder. Cell 106:367–379 [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372 [PubMed] [Google Scholar]
- Harrington JJ, Sherf B, Rundlett S, Jackson PD, Perry R, Cain S, Leventhal C, Thorntonm M, Ramachandran R, Whittington J, Lerner L, Costanzo D, McElligott K, Boozer S, Mays R, Smith E, Veloso N, Klika A, Hess J, Cothren K, Lo K, Offenbacher J, Danzig J, Ducar M (2001) Creation of genome-wide protein expression libraries using random activation of gene expression. Nat Biotechnol 19:440–445 [DOI] [PubMed] [Google Scholar]
- Ingram JL, Stodgell CJ, Hyman SL, Figlewicz DA, Weitkamp LR, Rodier PM (2000) Discovery of allelic variants of HOXA1 and HOXB1: genetic susceptibility to autism spectrum disorders. Teratology 62:393–405 [DOI] [PubMed] [Google Scholar]
- IMGSAC (2001a) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMGSAC (2001b) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 10:973–982 [DOI] [PubMed] [Google Scholar]
- Khan SG, Levy HL, Legerski R, Quackenbush E, Reardon JT, Emmert S, Sancar A, Li L, Schneider TD, Cleaver JE, Kraemer KH (1998) Xeroderma pigmentosum group C splice mutation associated with autism and hypoglycinemia. J Invest Dermatol 111:791–796 [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M (1996) A broader phenotype of autism: the clinical spectrum in twins. J Child Psychol Psychiatry 37:785–801 [DOI] [PubMed] [Google Scholar]
- Liu JJ, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, Gilliam TC (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed] [Google Scholar]
- Michaelis RC, Skinner SA, Deason R, Skinner C, Moore CL, Phelan MC (1997 ) Intersitial deletion of 20p: new candidate region for Hirschsprung disease and autism? Am J Med Genet 71:298–304 [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Bradford Y, Chen YH, Hall J, Arnone B, Gardiner MB, Hutcheson HB, Gilbert JR, Pericak-Vance MA, Copelandyates SA, Michaelis RC, Wassink TH, Santangelo SL, Sheffield VC, Piven J, Folstein SE, Haines JL, Sutcliffe JS (2001) Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics 77:105–113 [DOI] [PubMed] [Google Scholar]
- Persico AM, Dagruma L, Maiorano N, Totaro A, Militerni R, Bravaccio C, Wassink TH, Schneider C, Melmed R, Trillo S, Montecchi F, Palermo M, Pascucci T, Puglisiallegra S, Reichelt KL, Conciatori M, Marino R, Quattrocchi CC, Baldi A, Zelante L, Gasparini P, Keller F (2001) Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatry 6:150–159 [DOI] [PubMed] [Google Scholar]
- Petit E, Herault J, Martineau J, Perrot A, Barthelemy C, Hameury L, Sauvage D, Lelord G, Muh JP (1995) Association study with two markers of a human homeogene in infantile autism. J Med Genet 32:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloudbataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Vanmalldergerme L, Penet C, Feingold J, Brice A, Leboyer M (1999) Genome-wide scan for autism susceptibility genes. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim C-H, Rutter M (1995) Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet 57:717–726 [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK (2001) Are rare variants responsible for susceptibility to complex disease? Am J Hum Genet 69:124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM (1985) Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry 142:74–77 [DOI] [PubMed] [Google Scholar]
- Robinson W, Binkert F, Gine R, Vazquez C, Muller W, Rosenkranz W, Schinzel A (1993) Clinical and molecular analysis of five inv dup(15) patients. Eur J Hum Genet 1:37–50 [DOI] [PubMed] [Google Scholar]
- Silverstein S, Lerer I, Buiting K, Abeliovich D (2001) The 28-kb deletion spanning D15S63 is a polymorphic variant in the Ashkenazi Jewish population. Am J Hum Genet 68:261–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Escamilla JR, Filipek P, Bocian ME, Modahl C, Flodman P, Spence MA (2001) Molecular genetic delineation of 2q37.3 deletion in autism and osteodystrophy: report of a case and of new markers for deletion screening by PCR. Cytogenet Cell Genet 94:15–22 [DOI] [PubMed] [Google Scholar]
- Smith M, Filipek PA, Wu C, Bocian M, Hakim S, Modahl C, Spence MA (2000) Analysis of a 1-megabase deletion in 15q22-q23 in an autistic patient: identification of candidate genes for autism and of homologous DNA segments in 15q22-q23 and 15q11-q13. Am J Med Genet 96:765–770 [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg I, Jakobsson G, Bohman M (1989) A twin study of autism in Denmark, Finland, Iceland, Norway, and Sweden. J Child Psychol Psychiatry 30:405–416 [DOI] [PubMed] [Google Scholar]
- Sultana R, Yu C-E, Yu J, Munson J, Chen D, Hua W, Estes A, Cortes F, de la Barra F, Yu D, Haider ST, Trask BJ, Green ED, Raskind WH, Disteche CM, Wijsman E, Dawson G, Storm DR, Schellenberg GD, Villacres EC. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics (in press) [DOI] [PubMed] [Google Scholar]
- Thomas NS, Sharp AJ, Browne CE, Skuse D, Hardie C, Dennis NR (1999) Xp deletions associated with autism in three females. Hum Genet 104:43–48 [DOI] [PubMed] [Google Scholar]
- Vincent JB, Herbrick J-A, Gurling HMD, Bolton PF, Roberts W, Scherer SW (2000) Identification of a novel gene on chromosome 7q31 that is interrupted by a translocation breakpoint in an autistic individual. Am J Hum Genet 67:510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostanis P, Harrington R, Prendergast MFP (1994) Case reports of autism with interstitial deletion of chromosome 17 (p11.2 p11.2) and monosomy of chromosome 5 (5pter-5p15.3). Psychiatr Genet 4:109–111 [DOI] [PubMed] [Google Scholar]
- Wassink TH, Piven J, Vieland VJ, Huang J, Swiderski RE, Pietila J, Braun T, Beck G, Folstein SE, Haines JL, Sheffield VC (2001) Evidence supporting WNT2 as an autism susceptibility gene. Am J Med Genet 105:406–413 [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128 [DOI] [PubMed] [Google Scholar]
- Yan WL, Guan X-Y, Green ED, Nicolson R, Yap TK, Zhang JH, Jacobsen LK, Krasnewich DM, Kumra S, Lenane MC, Gochman P, Damschroder-Williams PJ, Esterling LE, Long RT, Martin BM, Sidransky E, Rapoport JL, Ginns EI (2000) Childhood-onset schizophrenia/autistic disorder and t(1;7) reciprocal translocation: identification of a BAC contig spanning the translocation breakpoint at 7q21. Am J Med Genet 96:749–753 [DOI] [PubMed] [Google Scholar]