Abstract
There currently exists a significant gap in our understanding of how the detailed chemical characteristics of polycation gene carriers influence their delivery performances in overcoming an important cellular-level transport barrier, i.e., intranuclear gene transcription. In this study, a UV-degradable gene carrier material (ENE4-1) was synthesized by crosslinking low molecular weight branched polyethylenimine (bPEI-2k) molecules using UV-cleavable o-nitrobenzyl urethane (NBU) as the linker molecule. NBU degrades upon exposure to mild UV irradiation. Therefore, this UV-degradable carrier allows us to control the chemical characteristics of the polymer/DNA complex (polyplex) particles at desired locations within the intracellular environment. By using this photolytic DNA carrier, we found that the exact timing of the UV degradation significantly influences the gene transfection efficiencies of ENE4-1/DNA(pGL2) polyplexes in HeLa cells. Interestingly, even if the polyplexes were UV-degraded at different intracellular locations/times, their nuclear entry efficiency was not influenced by the location/timing of UV degradation. The UV treatment did not influence the size or binding strength of the polyplexes. However, we confirmed that the degradation of the carrier molecules impacts the chemical characteristics of the polyplexes (it produces carbamic acid and nitrosobenzyl aldehyde groups on ENE4-1). We believe that these anionic acid groups enhance the interaction of the polyplexes with nuclear transcription proteins and thus the final gene expression levels; this effect was found to occur, even though UV irradiation itself has a general effect of reducing transfection efficiencies. Excess (uncomplexed) ENE4-1 polymers appear to not play any role in the UV-enhanced gene transcription phenomenon.
1. Introduction
Polymer-based gene carriers, though promising as safer alternatives to viral gene carriers, have yet been limited in large part due to their poor transfection performance. Future design and development of better polycation gene carriers will be greatly facilitated by an improved understanding of the relationship between the polycation chemistry and performance mechanism. Prior studies over the years have revealed many useful information along these lines. However, the current state of knowledge in this area is still tentative and insufficient to serve the purpose [1-2].
In particular, an important gap exists in the lack of a discussion and explanation of seemingly inconsistent data regarding the exact optimal timing and location of DNA release from polycation/DNA complexes (polyplexes) during the post-internalization (i.e., post-endocytosis) intracellular trafficking pathway [2]. It was originally the objective of the present study to establish the correlations between various molecular parameters of polyplexes (such as polyplex size and compactness) and their performances in the above-mentioned delivery aspects (post-endosomal trafficking, and timely release of DNA) and ultimately in the expression of the delivered gene. Specifically, we sought to answer the following question: What is exactly the role that a polycation plays in promoting the nuclear import of DNA? In order to address this problem, we developed a UV-degradable DNA carrier. We thought that by using this photolytic DNA carrier it would be possible to control the precise location of the (partial or complete) disintegration of the polycation/DNA polyplex particles within the intracellular environment; thus, the effects of the location of the photo dissociation (i.e., cytosol, nucleus, or no degradation) on the nuclear localization and gene expression of the polyplexes could be studied.
Previously, several strategies for controlling the timing and location of the intracellular DNA release by external stimuli have been demonstrated by other researchers. Examples of these approaches include the use of such mechanisms as changes in pH [3], temperature [4] or redox potential [5-7], and UV [8] or IR [9] irradiation. In particular, photolytic DNA carriers allow to control the location/timing of the disintegration of the polyplexes within the intracellular environment without relying on inherent biochemical characteristics of intracellular compartments; using these photo-degradable gene delivery systems, it has been demonstrated that the intracellular degradation of the carrier material greatly enhances the delivery performance of DNA [8] (or siRNA [10]).
In many of these chemically degradable gene delivery systems, the chemical degradation not only impacts the physical strength of the polycation/DNA binding, but the chemical characteristics of the polyplexes too. For instance, the degradation of the UV-cleavable [8, 11] polymers typically produces anionic (acid) groups on polyplexes that can alter the interaction of the polyplexes with intracellular proteins (e.g., transcription-related proteins present in the nucleus) and thus the final gene expressions levels. However, this aspect of the intracellular carrier degradation has not been investigated. In this regard, one remarkable recent finding is that when an anionic component (such as hyaluronic acid [12], heparin [13], poly(aspartic acid) [14], poly(γ-glutamic acid) [15], carboxylic acid pendant groups [16] or oligonucleotides [17]) is incorporated into the DNA/polycation complex, the transfection efficiency is increased by multiple orders of magnitude. A common explanation for this phenomenon has been that the anionic additive facilitates efficient release of DNA in the cell’s nucleus and thus the gene transcription process, by loosening the binding between the polycation and DNA. However, the isolated effects of carrier’s anionic groups on the individual steps of the intracellular trafficking processes (i.e., endosome escape, nuclear entry, and interaction/binding with transcription-related proteins) have not been determined.
In the present study, we developed a polyethylenimine(PEI)-based DNA carrier that is degradable upon exposure to mild UV irradiation (Figure 1). This polycation carrier contains UV-cleavable o-nitrobenzyl urethane (NBU) linkages; upon exposure to UV, the NBU groups degrade and produce carbamic acid groups. This photolytic DNA carrier allows us to control the location of the generation of negative acid groups on the DNA polyplex nanoparticles. We studied the effects of the UV degradation on the strength/state of the binding between DNA and the polymer, and the nuclear entry efficiency, the charge characteristics and the gene expression levels of the polyplexes. We also examined how the exact timing of the UV degradation of the carrier influences its gene transfection efficiency.
Figure 1.
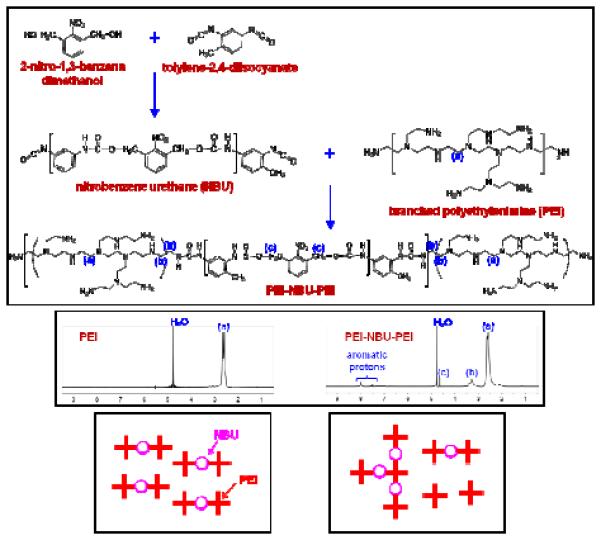
(Upper) Synthesis route for the preparation of a UV-degradable crosslinked polyethylenimine (PEI) material (named “ENE4-1”). Low molecular weight branched PEI (bPEI-2k) polymers were crosslinked with a UV-cleavable o-nitrobenzyl urethane (NBU) spacer. (Middle) 1H NMR spectra of the bPEI-2k precursor and the PEI-NBU-PEI product in D2O. In the synthesis of the ENE4-1 material, bPEI-2k was reacted with NBU in a 2:1 stoichiometric molar ratio. As the bottom right cartoon describes, the random nature of the coupling reaction results in a polydisperse product (rather than a monodisperse product such as shown in bottom left of the figure).
2. Experimental Procedures
2.1 Synthesis and characterization of a UV-degradable gene carrier
A UV-degradable DNA carrier material (named “ENE4-1”) was prepared by crosslinking low molecular weight branched polyethylenimine (bPEI-2k, molecular weight ≈ 1.8 kDa, purchased from Polysciences, Inc.) with a UV-degradable o-nitrobenzyl urethane (NBU) linker (Figure 1). Following the literature procedure [18] (with necessary modifications), the NBU compound was first obtained by reacting 2-nitro-1,3-benzenedimethanol (NBDM, synthesized in our laboratory using the reported procedure [19]) with two molar excess of tolylene 2,4-diisocyanate (TDI, purity > 95%, Sigma-Aldrich) in anhydrous dimethylformamide (DMF, purity 99.8%, Sigma-Aldrich). Briefly, NBDM (110 mg, 0.60 mmol) was charged into a flask, and dissolved in anhydrous DMF (10 ml) under slight heating at 60 °C for 1 h. The flask was then cooled down to 0 °C in ice bath, and kept under protection from light using aluminum foil. In a separate flask, TDI (209 mg, 1.20 mmol) was diluted with anhydrous DMF (10 ml). This TDI solution was then cooled down to 0 °C, and quickly added to the flask containing the NBDM solution under vigorous stirring under argon atmosphere. The reaction mixture was slowly heated to 60 °C over 1 h, and the reaction was allowed to continue at that temperature for 24 h. Afterwards, bPEI-2k (1.08 g, 0.6 mmol) dissolved in anhydrous DMF (50 ml) was added in one portion to the reaction mixture under vigorous stirring to initiate the crosslinking reaction. The crosslinking reaction was allowed to proceed at 80 °C for 24 h. After the reaction, the product was concentrated using a rotary evaporator, and the resulting dense solution (~ 1 ml) of the crossliked polymer product was added dropwise into 100 ml diethyl ether for precipitation. The precipitate was collected by filtration through filter paper, and the collected crosslinked PEI product (yellowish powder) was further dried under vacuum at 100 °C overnight (1.20 g, yield 86%). 1H NMR characterization of this polymer product (ENE4-1) in D2O was performed using a Bruker ARX 300 spectrometer (300 MHz).
2.2 UV degradation of ENE4-1
1H NMR and UV-visible (UV-VIS) light absorption spectra measurements were performed to confirm the degradation of ENE4-1 upon exposure to UV radiation. The NMR sample was prepared by dissolving ENE4-1 in D2O at 10 mg/ml, and the measurement was conducted using a Bruker ARX 300 spectrometer. For the UV-VIS measurement, ENE4-1 was dissolved in water at 1 mg/ml, and a Cary 100 Bio UV-VIS spectrometer (Varian) was used. The ENE4-1 solution was exposed to UV light for 20 minutes. The UV exposure was performed using a UVP’s B-100AP lamp (wavelength = 365 nm, power density = 20 mW/cm2) mounted on an illumination environmental chamber designed in our laboratory for homogeneous illumination and better heat dissipation via the vent at the bottom (Figure S1 of the Supplementary Information (SI)); during the irradiation, air was continuously blown through the vent to present the heating of the illuminated sample. Unless described otherwise, all UV exposure experiments were performed using the same procedure. The NMR and UV-VIS measurements were conducted at 30 minutes after the UV exposure.
2.3 Preparation, and size and ζ-potential characterizations of polyplexes
To prepare a polyplex sample at a desired N/P ratio (N/P ratio is defined as the molar ratio of the polycation’s amine (N) groups relative to the DNA’s phosphate (P) groups), 800 μl of an appropriately diluted polycation solution (in 10 mM Tris-HCl buffer, pH 7.5) was added to 400 μl of a pGL2 (Invitrogen) stock solution (50 μg/ml pGL2 in 10 mM Tris-HCl buffer, pH 7.5); thus, in the final polyplex solution, the pGL2 concentration was always 16.7 μg/ml. Immediately after the mixing of the polycation and pGL2 solutions, the mixture was homogenized by gentle pipetting for 15 seconds. The resulting polyplex solution was allowed to sit quiescently for 30 minutes at room temperature before characterization. The size and surface charge characteristics of polyplexes prepared at various N/P ratios were characterized by dynamic light scattering (DLS) and ζ-potential measurements using a Malvern Zetasizer Nano ZS instrument.
2.4 Gel electrophoresis (retardation) assay
For gel retardation assays, polyplex samples were prepared in the same way as above except that the final volume was 30 μl. 30 μl of the polyplex solution (pGL2 16.7 μg/ml) was mixed with 6 μl of the gel loading buffer (2.5 mg/ml bromophenol blue in a 30:70 (by volume) mixture of glycerol and water), and immediately loaded into an agarose gel (1.0% (w/v) agarose in Tris-acetate-EDTA (TAE) buffer) containing 0.5 μg/ml ethidium bromide (EtBr). The gel was run for 90 minutes at 70 volts. The gel was digitally imaged with a PhotoDoc-it UV transillumination system (UVP).
2.5 Polyanion competition assay
10 μl of an appropriately diluted heparin (purified grade, Fisher Scientific) solution (in 10 mM Tris-HCl buffer, pH 7.5) was added to 30 μl of a polyplex solution containing pGL2 at a concentration 16.7 μg/ml in 10 mM Tris-HCl buffer (pH 7.5); the final concentration of heparin was in the range of 2 – 30 IU heparin per μ g of pGL2. The mixture was then homogenized by gentle pipetting for 15 seconds, and incubated at 25 °C for 30 minutes prior to the gel electrophoresis analysis. Heparin was measured international units (IU); one IU of heparin is defined to be the amount needed to suppress clotting of 1 ml of whole blood for 3 minutes. Referring to the data shown in Figure 10, for both the “no UV” and “UV for 20 min” samples, the heparin assay was performed at the same time (i.e., at one hour) after formation of the polyplexes. ENE4-1 and pGL2 stock solutions were mixed by gentle pipetting for 15 seconds and then allowed to sit quiescently for 30 minutes. A portion of this ENE4-1/pGL2 polyplex solution (the “UV for 20 min” sample) was taken and exposed to UV radiation for 20 minutes and then kept statically for 10 minutes prior to the heparin treatment. The other portion of the polyplex solution (the “no UV” sample) was allowed to sit for the entire additional 30 minute period before the addition of heparin.
Figure 10.
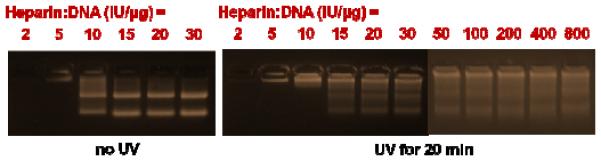
Release of pGL2 from UV-treated (365 nm, 20 mW/cm2) vs. non-UV-treated ENE4-1/pGL2 polyplexes (N/P = 60) in response to the addition of varying amounts of a polyanion dissociation agent (heparin).
2.6 SYBR Green assay
100 μl of an appropriately diluted SYBR Green (SYBR Safe, Invitrogen) solution (in 10 mM Tris-HCl buffer, pH 7.5) was added to 100 μl of a polyplex solutions containing pGL2 at a pGL2 concentration of 16.7 μg/ml in 10 mM Tris-HCl buffer (pH 7.5); the final concentration of SYBR Green was in the range of 0.1 – 100× SYBR Green concentration units (no information available in conventional units). After 10 minutes of incubation at 25 °C, the fluorescence intensity was measured at 536 nm (excitation at 495 nm) using a microplate reader (SpectraMax M2, Molecular Devices).
2.7 Cell culture
HeLa cells (purchased from ATCC) were incubated in a humidified atmosphere of 5% CO2 in air at 37 °C in Dulbecco's modified Eagle’s medium (DMEM, Life Technologies) containing 25 mM D-glucose supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
2.8 Cell viability after UV exposure
The cell viability was measured by the trypan blue assay; dead cells are stained with trypan blue due to ruptured membranes, and viable cells remain unstained. HeLa cells were seeded in 6-well plates at an initial density of 2 × 105 cells/well with 2 ml complete DMEM medium, and allowed to grow for 24 h before UV exposure. Prior to the UV treatment, the medium was replaced with fresh complete medium. The cells were then exposed to UV irradiation for designated durations, and incubated for additional 24 h. The cells were washed with CMF saline, trypsinized, and suspended in fresh complete medium. 10 μl of the cell suspension was mixed with 10 μl of 0.4% trypan blue solution (HyClone, Fisher Scientific). Immediately afterwards, the entire 20 μl of cell suspension was loaded into a counting chamber of a hemocytometer, and allowed to sit for 1 minute. The numbers of total and stained cells were counted. The cell viability was calculated as the percentage of unstained cells.
The cell viability was also measured by the methylthiazol tetrazolium (MTT) assay; this test measures the reduction of MTT into formazan by mitochondria in living cells. HeLa cells were seeded and grown using the same procedure as above. Twenty eight (or sixteen) hours prior to the MTT assay, the cell’s medium was replaced with serum-free medium, and the cells were exposed to UV irradiation for 20 minutes at that point. After 4 hours of serum deprivation period, the medium was changed back to complete medium, i.e., at 24 (or 12) hours prior to the MTT assay. Immediately before the MTT assay was to be performed, the medium was once again replaced with fresh complete medium. 50 μl of the 12 mM MTT stock solution (Invitrogen) was then added to the cells, and the cells were incubated at 37 °C for 4 h. Afterwards, half of the medium was removed from each well, and 250 μl of dimethyl sulfoxide (DMSO) was added. The plate was then further incubated at 37 °C for 10 minutes for dissolution of the formazan crystals formed by MTT reduction. The absorbance of the sample was read at a wavelength of 540 nm on a microplate reader. The cell viability was calculated as the absorbance of the UV-treated cells normalized by that of non-UV-treated cells.
2.9 In vitro gene transfection assay
HeLa cells were seeded in 24-well plates at an initial density of 5 × 104 cells/well in 0.5 ml complete DMEM medium, and allowed to grow for 24 h. Prior to the transfection with polyplexes, the medium was replaced with serum-free medium. A polyplex solution prepared at a designated N/P ratio was diluted using the same medium, i.e., 10 mM Tris-HCl buffer (pH 7.5), to a final pGL2 concentration of 10 μg/ml. To each cell well, 50 μl of the diluted polyplex solution was added; the final pGL2 concentration per well was 0.91 μg/ml. After 4 hours of polyplex transfection, the medium was replaced with complete medium. The cells were incubated for additional 24 h to allow time for pGL2 expression. Afterwards, the cells were washed with CMF saline, and lysed with 1× passive lysis buffer (Promega) at room temperature for 30 minutes under constant shaking. 10 μl of the resulting cell lysate was mixed with 50 μl of luciferase substrate solution (Promega), and then the luminescence from this mixture was measured using a luminometer (Lumat LB 9501, Berthold Technologies) and recorded in relative light units (RLU). This measured luciferase activity was normalized by the total protein concentration estimated by the BCA (Pierce Biotechnology). Experiments were performed in four replicates. Data presented represent the mean and standard deviation of these four independent experiments.
2.10 In vitro gene transfection efficiency after exposure to UV light
The procedure was the same as above except that the cells were exposed to UV light for 20 minutes at various different times during the 28-hour assay period.
2.11 Confocal (laser scanning) microscopy
HeLa cells were seeded on a glass cover slip (Micro Cover Glasses Square No. 1, 22 × 22 mm, Fisher Scientific) at a density of 4 × 105 cells per slip, and were grown for 24 h. The cells were then transfected with YOYO-1-labeled polyplexes using the same procedure as describe above. The procedure for the YOYO-1-labeling of pGL2 is presented in Section S4 of the SI. At various different times after the transfection with the polyplexes, the cells were stained and immediately fixed (as described below) for confocal imaging.
The cells were first washed with 1× phosphate buffered saline (PBS) three times, and incubated with 5 mg/ml of Hoechst 33342 (Molecular Probes) and 5 mM of LysoTracker Red (Molecular Probes) in complete medium at 37 °C in 5% CO2-air for 20 minutes; Hoechst 33342 and LysoTracker Red stain the nuclei and late endosomes/lysosomes of the cells, respectively. After the staining, the cells were washed three times with 1× PBS, and then fixed with freshly prepared 4% paraformaldehyde in 1× PBS for 15 minutes at room temperature. Afterwards, the fixed cells were mounted on a glass slide (76 × 25.4 mm, Fisher Scientific) with a drop (~ 50 μl) of antifade fluorescent mounting medium (VectaShield H-1000, Vector Laboratories). Imaging was conducted using a Nikon A1R confocal microscope.
Confocal measurements on the UV-treated cells were conducted using the same general procedure as above. The HeLa cells transfected with YOYO-1-labeled polyplexes were exposed to UV light for 20 minutes at several different times after the transfection. The UV-exposed cells were further incubated, and stained and fixed at 24 hours after the onset of transfection for the imaging measurements at that point.
3. Results and Discussion
3.1 Synthesis and characterization of a UV-degradable branched polyethylenimine (PEI) gene carrier
A UV-degradable gene carrier material (designated as “ENE4-1”) was synthesized by crosslinking low molecular weight branched polyethylenimine (bPEI-2k) molecules using UV-cleavable o-nitrobenzyl urethane (NBU, molar mass ≈ 0.5 kDa) as the linker moiety (Figure 1). The proton NMR spectra (Figure 1) confirm that NBU (the presence of which is confirmed by the o-nitrobenzyl carbamate peak at δ ≈ 4.6 ppm) successfully crosslinks bPEI-2k (as indicated by the appearance of the peak at δ ≈ 3.1 – 3.5 ppm).
The water-dispersed ENE4-1 molecules have a size appropriate for DNA condensation and delivery. The hydrodynamic diameter of ENE4-1 in water was measured by dynamic light scattering (DLS) to be about 3 nm (Figure S8(A) of the Supplementary Information (SI)), which is comparable to that of a ~10-kDa crosslinked PEI material prepared by crosslinking bPEI-2k with dithiobis(succinimidyl propionate) (DSP, molar mass ≈ 0.4 kDa) [20]; therefore, it is inferred that the average molecular weight of ENE4-1 is also close to that value, ~ 10 kDa. It is known that crosslinked PEI polymers in the molecular weight range between 5 and 10 kDa are effective in mediating gene transfer [5, 20-22].
A 1H NMR measurement was performed to confirm the UV degradability of ENE4-1. Figure 2 displays the NMR spectra obtained from ENE4-1 in D2O after a 20-minute exposure to UV light (wavelength = 365 nm, power density = 20 mW/cm2) using a commercial UV lamp and the UV environmental chamber described in Figure S1. As can be seen from the NMR data, the o-nitrobenzyl peak at δ = 4.6 ppm decreases in height after the UV exposure. Analysis of the peak area suggests that about 68 percent of the NBU bonds have been cleaved after the 20-minute UV exposure, which is consistent with the results obtained from a similar experiment performed by other researchers [23]. The UV degradation was also evidenced by a change in the UV-visible light absorption spectra for ENE4-1 (Figure S2).
Figure 2.
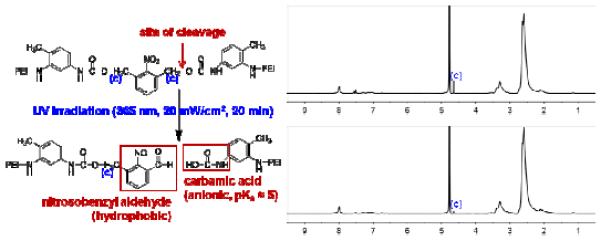
1H NMR spectra of ENE4-1 in D2O before and after 20-minute exposure to UV light (wavelength: 365 nm, power density: 20 mW/cm2).
3.2 DNA (pGL2) condensation behavior of the UV-degradable polymer (ENE4-1)
The DNA complexation behavior of ENE4-1 was examined at various N/P ratios by using the gel electrophoresis technique. As shown in Figure 3(A), at all N/P ratios greater than or equal to 5 the gel electrophoretic migration of the pGL2 molecules was seen to be completely retarded because of the complete complexation of pGL2 with ENE4-1. The ethidium bromide(EtBr)-staining intensity at the loading position gradually decreased with increasing N/P ratio up to an N/P value of 10, and this EtBr signal completely disappeared at higher N/P ratios, which suggests that ENE4-1 is capable of completely shielding pGL2 from access by EtBr at high N/P conditions. It is suggested in the literature that a crosslinked PEI material exhibits a poor DNA condensing capability, when the material is too tightly crosslinked, i.e., when the material is crosslinked to the extent that the crosslinked structure has an insufficient cationic charge density, or it becomes too stiff, to accommodate the formation of dense DNA condensates [24]. ENE4-1 appears to possess appropriate charge/stiffness characteristics for DNA condensation and delivery. The N/P-dependent gel migration patterns of ENE4-1/pGL2 polyplexes are quite comparable to the gel patterns observed for bPEI-2k/pGL2 polyplexes (Figure 3(B)).
Figure 3.
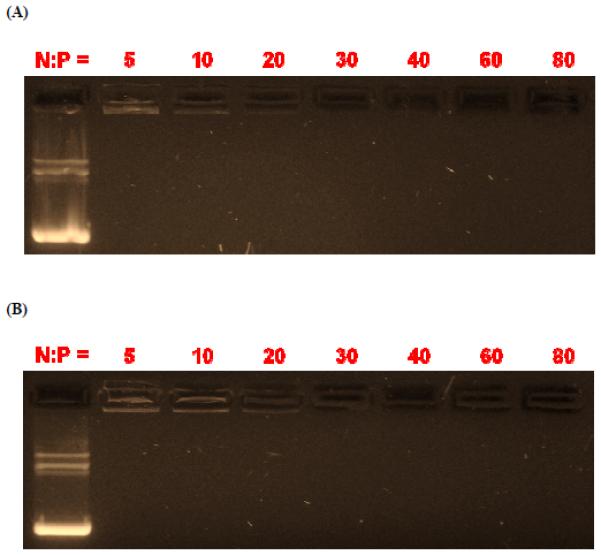
Electrophoretic gel migration patterns of (A) ENE4-1/pGL2 and (B) bPEI-2k/pGL2 polyplexes at various N/P ratios at a fixed concentration of pGL2 (16.7 μg/ml) in 10 mM Tris-HCl buffer (pH 7.5). In each gel, the uncomplexed pGL2 DNA is shown in the first lane.
The actual size and surface charge characteristics of ENE4-1/pGL2 polyplexes prepared at various N/P ratios were characterized by dynamic light scattering (DLS) and ζ-potential measurements. As shown in Figure 4, the mean hydrodynamic diameter of the ENE4-1/DNA polyplexes decreased with increasing N/P ratio, and then plateaued at about 75 nm at N/P ratios above 15. At N/P > 15, the polyplex particles were highly positively charged with their ζ-potential value exceeding +20 mV. We also confirmed that when compared at a fixed N/P ratio (i.e., at N/P = 60), ENE4-1 produces smaller-sized, more compact polyplex particles than bPEI-2k (Figures S8(C) and S8(D)). This result is consistent with the previous report that bPEI-2k is less efficient at condensing DNA to form small complexes than higher molecular weight PEI [25].
Figure 4.
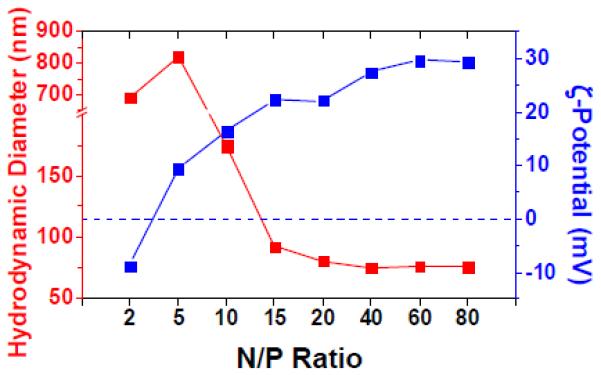
Hydrodynamic diameter (red) and ζ-potential (blue) values of ENE4-1/pGL2 polyplexes in 10 mM Tris-HCl buffer (pH 7.5) at various N/P ratios at a fixed pGL2 concentration of 16.7 μg/ml.
3.3 ENE4-1-mediated pGL2 transfection in HeLa cells
The gene transfection efficiencies of ENE4-1/pGL2 polyplexes in HeLa cells were measured at various N/P ratios. The cells were exposed (i.e., “transfected”) to a serum-free medium containing ENE4-1/pGL2 polyplexes at a pGL2 concentration of 0.91 μg/ml for four hours (t = 0 – 4 h). After this process, the cells were fed with the “complete” medium, and were further incubated for additional 24 hours. At t = 28 h, the gene expression levels were measured by the luciferase assay. For comparison, the same measurements were also performed with bPEI-2k/pGL2 and lPEI-25k/pGL2 polyplexes. The results are summarized in Figure 5.
Figure 5.
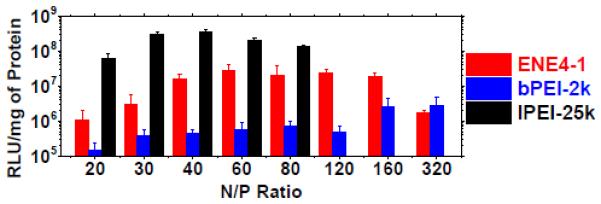
In vitro gene transfection efficiencies of ENE4-1/pGL2, bPEI-2k/pGL2 and lPEI-25k/pGL2 polyplexes in HeLa cells at a pGL2 concentration of 0.91 μg/ml at various N/P ratios. The luciferase expression in relative light units (RLU) was normalized to the total protein in each well. The data represent the mean and standard deviation of four independent experiments.
As shown in the figure, the transfection efficiency of ENE4-1/pGL2 polyplexes exhibits a maximum at an N/P ratio of around 60. It appears that free/uncomplexed ENE4-1 molecules coexisting with ENE4-1/pGL2 polyplexes play a significant role in enhancing the pGL2 expression at high N/P ratio (e.g., N/P = 60), as has been suggested in the literature [18]. The transfection efficiency of ENE4-1/pGL2 polyplexes stayed almost constant up to an N/P ratio of about 160, and then was seen to be reduced at N/P = 320, likely because of the cytotoxicity of ENE4-1. In the bPEI-2k/pGL2 and lPEI-25k/pGL2 cases, maximum transfection efficiencies were observed at N/P ratios of about 80 and 40, respectively. The N/P value that gives a maximum transfection efficiency appears to inversely correlate with the molecular weight of the PEI carrier; the molecular weight of ENE4-1 is estimated to be about 10 kDa (Section 3.1). This result also supports the explanation that the decrease in transfection efficiency at high N/P ratio is due to the PEI’s toxicity to the cells. We note that typically (for instance, at N/P = 60) ENE4-1 is about 100-fold more efficient at transfecting pGL2 into HeLa cells than bPEI-2k, but it is about a factor of ten less efficient than lPEI-25k. The viability of the HeLa cells after transfection with ENE4-1/pGL2 polyplexes at N/P = 60 (ENE4-1 = 8.87 μg/ml, pGL2 = 0.91 μg/ml) was measured to be 83.4 ± 0.5% (based on the methylthiazol tetrazolium (MTT) assay).
Based on these data, we chose to proceed with further study of the effect of the UV degradation of ENE4-1 on its gene transfer efficiency (as will be discussed in the next section). We would like to note that the ENE4-1 material was selected for this study out of more than ten candidate PEI-(P)NBU-PEI (ENE) materials having different molecular weights, architectures and compositions; the chemical structures of these other candidate polymers examined are explained in Figure S3 of the SI. The detailed molecular and micellization characteristics, DNA complexation properties, and in vitro gene transfection efficiencies of these other ENE materials are also presented in Tables S1 through S3 and Figure S4. The preliminary results suggested that the efficiency of crosslinked/micellized PEI materials as gene carrier depends on such parameters as PEI molecular weight and architecture, crosslink density, and linker type and size, as has been reported by other researchers [5, 20-22].
3.4 Effect of the UV degradation of ENE4-1 on the pGL2 transfection
We examined the effect of the UV degradation of ENE4-1/pGL2 polyplexes (N/P = 60) on the pGL2 transfection efficiency in HeLa cells. HeLa cells were first transfected with ENE4-1/pGL2 polyplexes in a serum-free medium (pGL2 0.91 μg/ml) for four hours (t = 0 – 4 h). Afterwards, the medium was replaced with the complete medium, and the cells were further incubated for additional 24 hours. At t = 28 h, the gene expression levels were measured by the luciferase assay. At various times during the t = 0 – 28 h period, the samples were exposed to UV light (365 nm, 20 mW/cm2) for 20 minutes. The same experiments were also performed on lPEI-25k/pGL2 polyplexes (N/P = 60) for comparison. The results are presented in Figure 6.
Figure 6.
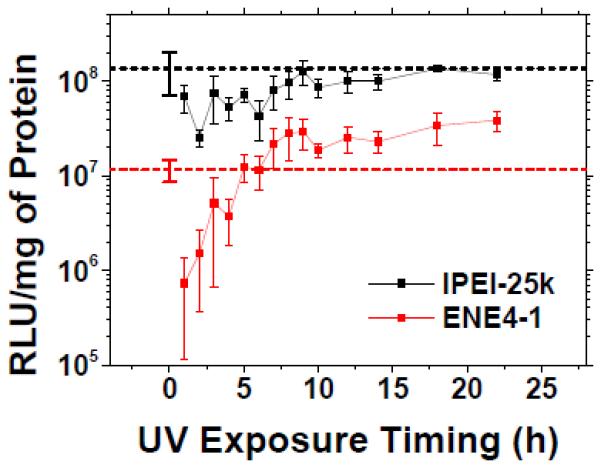
In vitro gene transfection efficiencies of ENE4-1/pGL2 and lPEI-25k/pGL2 polyplexes (pGL2 0.91 μg/ml, N/P = 60) in HeLa cells after 20-minute exposure to UV light (365 nm, 20 mW/cm2) at various time points after treatment with polyplexes. After the UV exposure at a designated time, the cells were further incubated until the pGL2 expression was measured at t = 28 h after the transfection (t = 0 – 4 h). The data represent the mean and standard deviation of four independent experiments. The dashed horizontal lines represent the transfection efficiencies of the respective polyplexes (N/P = 60) obtained without UV exposure.
As shown in the figure, the gene transfection efficiency of ENE4-1/pGL2 polyplexes is significantly influenced by the exact timing of the UV degradation. An exposure of ENE4-1/pGL2 polyplexes to UV light at a time earlier than t ≈ 5 h caused a dramatic reduction in the transfection efficiency below that of the non-UV-treated situation (shown by the dotted red horizontal line); the premature degradation of ENE4-1 during the “transfection” period (t = 0 – 4h) appears to significantly compromise the cellular internalization of the polyplexes. However, a UV exposure at t > ~ 6 h resulted in a significant enhancement in the transfection efficiency; the enhancement for the samples UV-exposed at t ≈ 22 h amounted to a factor of 3 to 4. To the contrary, the UV exposure of (non-UV degradable) lPEI-25k/pGL2 polyplexes did not produce any enhancement in the gene transfection efficiency relative to what was observed without UV exposure (marked by the dotted black horizontal line). With the aid of UV light, ENE4-1 performs almost comparably to lPEI-25k; the difference becomes within a factor of 2 – 3.
In order to confirm that in these “transfection under UV” experiments, the UV exposure at different times indeed caused the degradation of ENE4-1 at different locations within the cell, HeLa cells transfected with ENE4-1/pGL2 polyplexes were imaged by confocal microscopy at various times after the transfection. For this study, pGL2 was labeled with YOYO-1 (green); as demonstrated in Figure S5, this YOYO-1 labeling did not alter the conformational or complexation characteristics of pGL2. The nuclei and late endosomes/lysosomes of the HeLa cells were stained with Hoechst 33342 (blue) and LysoTracker Red (red), respectively. Figure 7 presents representative images taken at t = 4, 8, 18 and 22 h after transfection. At t = 4 h, the pGL2 signals appeared in isolated clusters, indicating that ENE4-1/pGL2 polyplexes were mostly confined within endosomal vesicles. As time passed, these green clusters were transported to the nuclear periphery. Also, the green fluorescence gradually became more diffuse with time. By 22 hours, most pGL2 molecules were seen to be diffusely localized within the nuclei of the cells. These results confirm that in the UV transfection measurements, the ENE4-1 carrier was UV-degraded indeed at various different intracellular locations.
Figure 7.
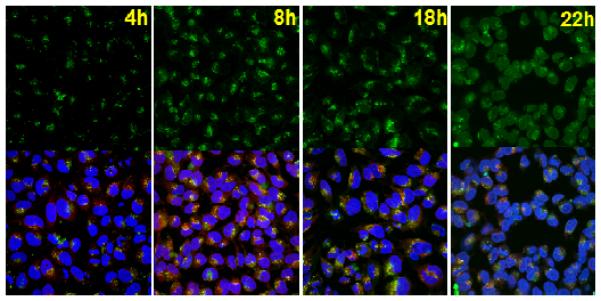
Confocal microscopy images of HeLa cells transfected with ENE4-1/pGL2 polyplexes (pGL2 0.91 μg/ml, N/P = 60). The images were taken at various times (t = 4, 8, 18 and 22 h) after the transfection (t = 0 – 4 h). The pGL2 DNA, nuclei, and late endosomes/lysosomes were stained with YOYO-1 (green), Hoechst 33342 (blue), and LysoTracker (red), respectively. The upper images were obtained using a green channel. Blue and red channel images are presented in Figure S6 of the Supplementary Information. The lower images represent merged data from the green, blue and red channels.
We also measured the nuclear entry efficiencies of ENE4-1/pGL2 polyplexes after exposure to UV light at different times during the intracellular trafficking process, again by using the confocal imaging technique. As shown in Figure 8, HeLa cells exposed to UV light at different times (t = 4, 8 and 22 h) after transfection with ENE4-1/pGL2 polyplexes (t = 0 – 4 h) exhibit indistinguishable amounts of pGL2 delivered to the nuclei of the cells at t = 24 h. It is clear that (surprisingly) the nuclear entry is not influenced by the UV exposure timing.
Figure 8.
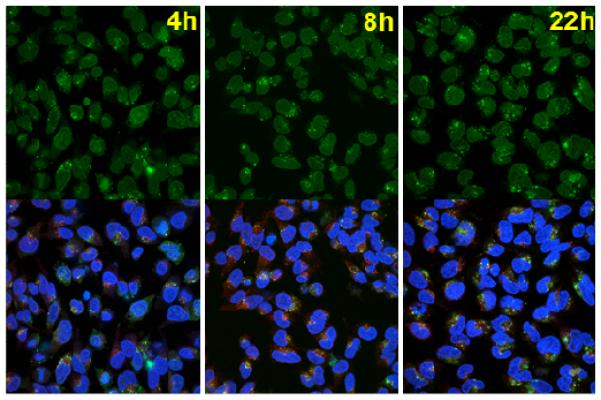
Confocal microscopy images of HeLa cells transfected with ENE4-1/pGL2 polyplexes (pGL2 0.91 μg/ml, N/P = 60). The transfected cells were exposed to UV irradiation (365 nm, 20 mW/cm2) at various times (t = 4, 8 and 22 h) after the transfection (t = 0 – 4 h). The images were taken at t = 24 h after the onset of transfection (t = 0 h). The pGL2 DNA, nuclei, and late endosomes/lysosomes were stained with YOYO-1 (green), Hoechst 33342 (blue), and LysoTracker (red), respectively. The upper images were obtained using a green channel. Blue and red channel images are presented in Figure S7 of the Supplementary Information. The lower images represent merged data from the green, blue and red channels.
3.5 Effects of UV irradiation on the binding and charge characteristics of the ENE4-1/pGL2 polyplexes
To determine exactly how/why the UV degradation of ENE4-1/pGL2 polyplexes causes an increase in the pGL2 expression, we examined the effects of UV radiation on various properties of the polyplexes. First, we examined how the UV degradation of ENE4-1/pGL2 polyplexes influences their gel migration patterns. Figure 9 displays the results obtained at various UV exposure durations and N/P ratios. At all conditions examined the UV irradiation did not cause release of pGL2 from ENE4-1. It did not even cause the weakening of the binding between pGL2 and ENE4-1 (i.e., no decrease in EtBr fluorescence at the loading position). By DLS, we also confirmed that the UV degradation does not influence the polyplex size (Figure S8(C)).
Figure 9.
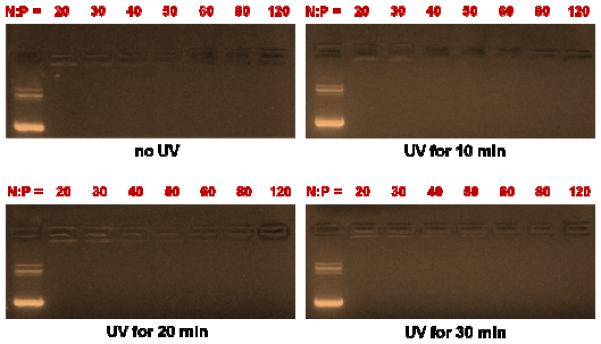
Electrophoretic gel migration patterns of ENE4-1/pGL2 polyplexes at a pGL2 concentration of 16.7 μg/ml after exposure to UV radiation (365 nm, 20 mW/cm2) for varying periods of time (0 – 30 minutes). In each gel, the leftmost lane represents the migration of the uncomplexed pGL2 control.
In order to determine whether the UV treatment decreases (or increases) the ENE4-1/pGL2 binding, we performed a polyanion competition assay, in which a polyanion dissociation agent, heparin, was added in different amounts to ENE4-1/pGL2 polyplexes, and the amount of heparin needed to dissociate the polyplexes was determined. As shown in Figure 10, at heparin:pGL2 ratios greater than 5 IU heparin per μg of pGL2, non-UV-treated ENE4-1/pGL2 polyplexes released pGL2 in its pristine form, as indicated by the appearance of the original pGL2 bands. However, for UV-degraded ENE4-1/pGL2 polyplexes, a similar treatment with heparin failed to cause complete release of pGL2 (only smeared bands are seen on the gel), which supports that the strength of the ENE4-1/pGL2 binding is increased after the UV degradation; this trend was also confirmed by a separate, SYBR Green binding analysis (Figure S9). Interestingly, even when a large surplus amount of heparin (e.g., 800 IU heparin per μg of pGL2) was added, pGL2 was not completely released from the UV-degraded ENE4-1/pGL2 polyplexes. This result suggests that the nature of the ENE4-1/pGL2 binding has been altered after the UV exposure, perhaps from predominantly electrostatic to partially hydrophobic; it is possible that upon the cleavage of the NBU linkers the resulting (hydrophobic) nitrosobenzyl urethane groups (Figure 2) become exposed to and associate with the hydrophobic bases of pGL2. In theory, it is also possible that the aldehyde groups produced by the UV degradation react with excess amine groups on pGL2 and (reversibly) form Schiff bases, resulting in crosslinking between pGL2 and the degraded segments of ENE4-1. However, this latter explanation is less likely because the covalent crosslinking between pGL2 and the ENE4-1 fragments would not eventually be able to cause the observed enhancement in the pGL2 expression. At this point, it remains unanswered exactly why the ENE4-1/pGL2 binding is increased upon UV treatment.
Therefore, the only plausible explanation for the increased pGL2 transfection after the UV degradation of ENE4-1 is that the negatively charged carbamic acid groups on ENE4-1 produced by UV irradiation (Figure 2) facilitate the transfection of pGL2. There is growing recognition in the literature that “amphoteric” (or “zwitterionic”) carriers (such as ternary DNA/polycation/polyanion complexes) are better gene transfection agents than binary DNA/polycation complexes; that is, the incorporation of anionic groups into carrier architecture enhances the transfection efficiency [12, 26]. In order to confirm the generation of (carbamic) acid groups upon UV exposure, ζ-potential measurements were performed on UV-degraded ENE4-1/pGL2 polyplexes. As summarized in Figure S11(A), we confirmed that the ζ-potential decreases (becomes less positive) by about 6 mV after the UV treatment.
To further demonstrate the effect of the UV degradation of ENE4-1 on its physicochemical properties, we prepared a “pre-degraded” sample of ENE4-1 by exposing it to UV light (365 nm, 20 mW/cm2, 20 minutes), and characterized the DNA complexation and binding characteristics of this pre-degraded ENE4-1 polymer. As shown in Figure S11(A), the pre-degraded polymer is not able to condense pGL2 into compact complexes. This result, and also the fact that the structure of the pre-formed ENE4-1/pGL2 polyplexes is not normally disrupted upon UV degradation, appear to be in analogy to the previous observation that compact, small-sized, ternary DNA/polycation/polyanion polyplex particles can only be achieved by adding the polyanion component to pre-formed binary DNA/polycation polyplexes at the final stage of preparation; mixing these three components at the same time does not produce small compact complex particles [12]. A common explanation for the enhanced gene transfection observed with ternary DNA/polycation/polyanion polyplexes has been that the polyanion causes loosening of the DNA/polycation binding, thereby enhancing the accessibility of transcription proteins to DNA in the nucleus [12]. However, our results suggest that the binding strength does not play a critical role. Therefore, the most reasonable explanation for the increased transfection efficiency of ENE4-1/pGL2 polyplexes upon UV exposure is that the anionic groups produced after the UV irradiation enhance the interactions of the polyplexes with transcription proteins in the nuclei of the cells. Currently, a study is in progress to directly test this hypothesis.
At the high N/P ratio used in the transfection experiments (N/P = 60), the majority of the ENE4-1 molecules exist in the free uncomplexed state [18, 27-28]. To examine the role of excess (uncomplexed) ENE4-1 polymers in the UV-enhanced gene transcription, we carried out the following experiment. We prepared a “hybrid” polyplex sample (named “lPEI-25k/pGL2 + ENE4-1” polyplexes) by first mixing pGL2 and lPEI-25k at an N/P ratio of 5 and then subsequently adding an excess amount of ENE4-1 to the pre-formed lPEI-25k/pGL2 polyplexes to make the overall N/P ratio equal to 60; as shown in Figure S12 (and also as reported previously [20]), at N/P = 5, lPEI-25k already fully condenses pGL2 into compact structures, so ENE4-1 (added after the formation of the lPEI-25k/pGL2 polyplexes) is not expected to be incorporated into the polyplexes. As summarized in Figure S13, without UV exposure the “lPEI-25k/pGL2 + ENE4-1” polyplexes mediated pGL2 transfection at intermediate levels, i.e., higher than the ENE4-1/pGL2 polyplexes but lower than the lPEI-25k/pGL2 polyplexes. When exposed to UV radiation, the “lPEI-25k/pGL2 + ENE4-1” polyplexes exhibited the lowest pGL2 transfection efficiency. Regardless of the UV exposure timing, the transfection efficiencies of the “lPEI-25k/pGL2 + ENE4-1” polyplexes obtained under the influence of UV radiation were always below the level of pGL2 transfection obtained without UV exposure, which suggests that the uncomplexed ENE4-1 molecules do not contribute to the UV-enhanced pGL2 expression observed with the ENE4-1/pGL2 polyplexes.
3.6 Effect of UV treatment on inherent gene transcription processes
Looking carefully at the graphs in Figure 6 (in particular, from the transfection data for the non-degradable lPEI-25k/pGL2 polyplexes), it was obvious that UV irradiation, in general, significantly compromises the pGL2 transfection (see also the transfection data for the semi-UV-degradable “lPEI-25k/pGL2 + ENE4-1” polyplexes in Figure S13). In an attempt to understand the mechanism responsible for this effect, we performed further investigations as discussed below.
First, we measured the viability of HeLa cells after 20-minute exposure to UV light (365 nm, 20 mW/cm2) by the trypan blue and MTT methods. As shown in Figures S14 (trypan blue data) and S15 (MTT data) of the SI, the results from these experiments indicate that the viability of HeLa cells is not influenced by the treatment with UV light. Therefore, the decrease in the pGL2 transfection level is not due to the immediate cytotoxic effect of UV radiation.
This suggests two other possibilities; UV radiation causes damage on pGL2 or the DNA of HeLa cells. To test these possibilities, two control experiments were run. In one, HeLa cells were transfected with lPEI-25k/pGL2 polyplexes (N/P = 60) prepared from pGL2 that had been exposed to UV light (365 nm, 20 mW/cm2, 20 minutes) prior to the complexation with lPEI-25k; in the other, HeLa cells that had been exposed to UV light (365 nm, 20 mW/cm2, 20 minutes) were transfected with the normal lPEI-25k/pGL2 polyplexes (N/P = 60). As shown in Figure 11, the former test revealed that pre-exposure of pGL2 to UV irradiation decreases the transfection efficiency to about a half of the level obtained without UV exposure. It is well-documented that UV light in the wavelength range 315 to 400 nm can cause indirect (oxidative) damage on DNA due to the generation of hydroxyl and oxygen radicals [29-31]. It appears that the oxidative effect of UV radiation, though it does not alter the gel migration behavior of pGL2 (Figure S16), is capable of adversely affecting the transcriptional activity of the pGL2 sequence presumably due to the arrest of RNA polymerases at sites of damage on pGL2 [32].
Figure 11.
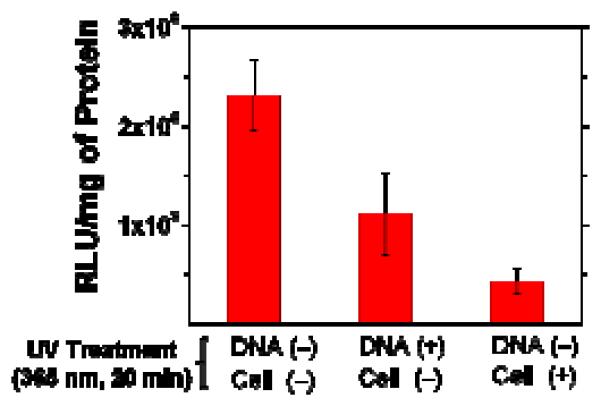
In vitro gene transfection efficiencies of lPEI-25k/pGL2 polyplexes (pGL2 0.91 μg/ml, N/P = 60) in HeLa cells; the measurements were performed by using either pre-UV-treated (365 nm, 20 mW/cm2) pGL2 (2nd bar), or pre-UV-treated HeLa cells (3rd bar), for comparison with the non-UV-treated situation (1st bar). The data represent the mean and deviation between two independent experiments.
The data in Figure 11 indicate that pre-exposure of HeLa cells to UV light prior to the transfection with lPEI-25k/pGL2 polyplexes has a more severe effect; the transfection efficiency reduces to about 19% of the non-UV-treated level. Again, we suspect that this reduced pGL2 expression in pre-UV-treated cells is associated with UV damage on the genomic DNA of the cells. It is known that genotoxic DNA damage can cause a delay in DNA replication due to DNA repair to prevent genomic instability and mutagenesis [33]. Therefore, it is reasonable to speculate that under such situation the expression of the exogenous pGL2 DNA is also delayed accordingly.
There still remains a question: Then why does the transfection depend on the UV exposure timing (Figures 6 and S13)? We believe that the answer to this question lies in the fact that in the cell populations used in the transfection assay, the mitotic cell division cycles are not synchronized; given that the nuclear entry of polyplexes is facilitated during the cell division (“mitosis”) period when the nuclear envelope becomes disintegrated [34], it is expected that at different times during the transfection interval a different number of cells have expressed the pGL2 gene in accordance with the mitotic stages of individual cells. Therefore, the later the cells are exposed to UV irradiation, the less number of cells will remain in the unexpressed cell population, resulting in an increased level of overall gene expression (as evidenced in the data presented in Figures 6 and S13).
This concept now allows us to estimate the isolated effect of the UV degradation of ENE4-1/pGL2 polyplexes on the pGL2 transfection in HeLa cells. Based on the data for lPEI-25k/pGL2 polyplexes shown in Figure 6, we estimate that at t = 22 h, about 95% of the cells have already expressed the pGL2 gene. The data obtained from ENE4-1/pGL2 polyplexes (Figure 6) show that when exposed to UV at 22 h, the ENE4-1/pGL2 transfected cells show a ~4-fold higher transfection efficiency, which gives the following equation: 95%·x + 5%·y ≈ 4, where x and y denote the respective factors by which the pGL2 transfection (measured at the end of the 28-hour period) will be reduced in the 95% already pGL2-expressing cells, or increased in the 5% unexpressed cell population, after UV exposure at t = 22 h relative to the non-UV-treated condition. Here, it is reasonable to assume that x ≈ 1, and thus the value of y is calculated to be about 60. Therefore, we conclude that the isolated effect of the UV degradation of ENE4-1 at t = 22 h is a 60-fold enhancement in the pGL2 transfection (measured as of t = 28 h). These x and y factors are expected to be increasing functions of the timing of UV exposure (otherwise the plot of the pGL2 transfection efficiency of ENE4-1/pGL2 polyplexes versus the UV exposure timing in Figure 6 would show a negative slope) because of the fact that the luciferase protein has a relatively short half-life in cells of around 2 – 3 hours [35-36]; note that, as has been shown using the RNA interference method, however, the effect of pre-existing luciferase proteins may last longer (up to about a day) even in rapidly dividing cells having a cell doubling time of about one day [37], and this uncertainty associated with the stability of luciferase makes the quantitative interpretation of the UV-transfection data rather challenging at this point.
4. Conclusions
The most important conclusions of the present study can be summarized as follows. (1) We developed a UV-degradable polymer DNA carrier. (2) The UV degradation of the carrier molecules modifies the chemical characteristics of the polyplexes (i.e., produces acid groups on the polyplexes) in such a way that it enhances the interaction of the polyplexes with nuclear transcription proteins and thus the final gene expression levels. (UV irradiation, however, does not cause release of DNA from the polyplexes or weaken the polymer/DNA binding.) (3) The exact timing of the UV degradation significantly influences the gene transfection efficiencies of the polymer/DNA complexes; this result is suspected to be due to a combined effect of the following three factors: (i) the negative influence of UV irradiation on transgene expression, (ii) a heterogeneity in the mitotic cell cycle progression, and (iii) a short half-life of the reporter protein in cells. Our results suggest that the in situ intracellular surface modification of polyplexes to incorporate anionic groups is a useful strategy for improving the transfection efficiency of polyplexes.
Supplementary Material
Acknowledgement
This work was supported by the US National Science Foundation (CBET-0828574 and DMR-0906567) and the Purdue University School of Chemical Engineering (YYW) and by the National Institutes of Health (DK55489 and CA124586) (SFK). YYW gratefully acknowledges the Research Fellowship from the Bindley Bioscience Center at Purdue University and also the Global RNAi Carrier Initiative Program Fellowship from the Biomedical Research Institute of the Korea Institute of Science and Technology (KIST). We are also grateful to Professor Julie C. Liu (School of Chemical Engineering, Purdue University), Professor Chongli Yuan (School of Chemical Engineering, Purdue University), and Professor Jue Chen (Department of Biological Sciences, Purdue University) for allowing us to use their instruments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Information. Procedures for the ENE polymer synthesis (Section S1), the preparation of ENE solutions in aqueous media (Section S2), the DNase I assay (Section S3), and the labeling of pGL2 with YOYO-1 (Section S4); molecular characteristics of ENE polymers (Table S1); size and charge characteristics of ENE/pGL2 polyplexes (Table S2); gene transfection abilities of ENE/pGL2 polyplexes (Table 3); UV illumination chamber (Figure S1); UV-Vis spectra of ENE4-1 before and after UV exposure (Figure S2); chemical structures of the ENE polymers examined (Figure S3); gene transfection efficiencies of ENE/pGL2 polyplexes (Figure S4); gel migration patterns of YOYO-1-labeled pGL2 before and after complexation with ENE4-1 (Figure S5); confocal images of HeLa cells transfected with ENE4-1/pGL2 polyplexes at various times after the transfection (Figure S6); confocal images of HeLa cells transfected with ENE4-1/pGL2 polyplexes after UV exposure at various time points (Figure S7); DLS data for various polymers and polyplexes (Figure S8); SYBR Green binding assay results for ENE4-1/pGL2 polyplexes before and after UV exposure (Figure S9); gel migration patterns of pGL2 dissociated from various polyplexes after treatment with DNase I (Figure S10); DLS, ζ-potential, SYBR Green binding and gel electrophoresis data for polyplexes prepared using pre-UV-treated ENE4-1 (Figure S11); gel migration patterns of lPEI-25k/pGL2 polyplexes at various N/P ratios (Figure S12); gene transfection efficiency of “hybrid” lPEI-25k/pGL2+ENE4-1 polyplexes as a function of UV exposure timing (Figure S13); viability of HeLa cells after exposure to UV radiation by the trypan blue (Figure S14) and MTT (Figure S15) methods; gel migration patterns of pGL2 after exposure to UV radiation for varying periods of time (Figure S16).
References
- 1.Nguyen J, Szoka FC. Nucleic acid delivery: the missing pieces of the puzzle? Acc Chem Res. 2012;45:1153–62. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Won Y-Y, Sharma R, Konieczny SF. Missing pieces in understanding the intracellular trafficking of polycation/DNA complexes. J Control Release. 2009;139:88–93. doi: 10.1016/j.jconrel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You J-O, Auguste DT. Nanocarrier cross-linking density and pH sensitivity regulate intracellular gene transfer. Nano Lett. 2009;9:4467–73. doi: 10.1021/nl902789s. [DOI] [PubMed] [Google Scholar]
- 4.Lavigne MD, Pennadam SS, Ellis J, Yates LL, Alexander C, Górecki DC. Enhanced gene expression through temperature profile-induced variations in molecular architecture of thermoresponsive polymer vectors. J Gene Med. 2007;9:44–54. doi: 10.1002/jgm.992. [DOI] [PubMed] [Google Scholar]
- 5.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12:989–94. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 6.Oupický D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2001;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 7.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, et al. Block catiomer polyplexes with regulated densities of charge and disulfide cross-linking directed to enhance gene expression. J Am Chem Soc. 2004;126:2355–61. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 8.Kim MS, Gruneich J, Jing H, Diamond SL. Photo-induced release of active plasmid from crosslinked nanoparticles: o-nitrobenzyl/methacrylate functionalized polyethyleneimine. J Mater Chem. 2010;20:3396–403. [Google Scholar]
- 9.Braun GB, Pallaoro A, Wu G, Missirlis D, Zasadzinski JA, Tirrell M, et al. Laser-activated gene silencing via gold nanoshell–siRNA conjugates. ACS Nano. 2009;3:2007–15. doi: 10.1021/nn900469q. [DOI] [PubMed] [Google Scholar]
- 10.Huschka R, Barhoumi A, Liu Q, Roth JA, Ji L, Halas NJ. Gene silencing by gold nanoshell- mediated delivery and laser-triggered release of antisense oligonucleotide and siRNA. ACS Nano. 2012;6:7681–91. doi: 10.1021/nn301135w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobolčiak P, Špírek M, Katrlík J, Gemeiner P, Lacík I, Kasák P. Light-switchable polymer from cationic to zwitterionic form: synthesis, characterization, and interactions with DNA and bacterial cells. Macromol Rapid Commun. 2013;34:635–9. doi: 10.1002/marc.201200823. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, Iida-Tanaka N, Niidome T, Kawano T, Kubo K, Yoshikawa K, et al. Hyaluronic acid and its derivative as a multi-functional gene expression enhancer: Protection from non-specific interactions, adhesion to targeted cells, and transcriptional activation. J Control Release. 2006;112:382–8. doi: 10.1016/j.jconrel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang CF, Luo X, Zhao YF, Han LN, Zeng X, Feng M, et al. Influence of the polyanion on the physico-chemical properties and biological activities of polyanion/DNA/polycation ternary polyplexes. Acta Biomater. 2012;8:3014–26. doi: 10.1016/j.actbio.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 14.Wang CF, Luo X, Zhao YF, Han LN, Zeng X, Feng M, et al. Influence of the polyanion on the physico-chemical properties and biological activities of polyanion/DNA/polycation ternary polyplexes. Acta Biomaterialia. 2012;8:3014–26. doi: 10.1016/j.actbio.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 15.Peng SF, Tseng MT, Ho YC, Wei MC, Liao ZX, Sung HW. Mechanisms of cellular uptake and intracellular trafficking with chitosan/DNA/poly(gamma-glutamic acid) complexes as a gene delivery vector. Biomaterials. 2011;32:239–48. doi: 10.1016/j.biomaterials.2010.08.081. [DOI] [PubMed] [Google Scholar]
- 16.Yoshihara C, Shew CY, Ito T, Koyama Y. Loosening of DNA/Polycation Complexes by Synthetic Polyampholyte to Improve the Transcription Efficiency: Effect of Charge Balance in the Polyampholyte. Biophysical Journal. 2010;98:1257–66. doi: 10.1016/j.bpj.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung YC, Hsieh WY, Young TH. Polycation/DNA complexes coated with oligonucleotides for gene delivery. Biomaterials. 2010;31:4194–203. doi: 10.1016/j.biomaterials.2010.01.116. [DOI] [PubMed] [Google Scholar]
- 18.Han D, Tong X, Zhao Y. Fast photodegradable block copolymer micelles for burst release. Macromolecules. 2011;44:437–9. [Google Scholar]
- 19.Pavia MR, Moos WH, Hershenson FM. Benzo-fused bicyclic imides. J Org Chem. 1990;55:560–4. [Google Scholar]
- 20.Deng R, Yue Y, Jin F, Chen Y, Kung H-F, Lin MCM, et al. Revisit the complexation of PEI and DNA — how to make low cytotoxic and highly efficient PEI gene transfection non-viral vectors with a controllable chain length and structure? J Control Release. 2009;140:40–6. doi: 10.1016/j.jconrel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Kloeckner J, Wagner E, Ogris M. Degradable gene carriers based on oligomerized polyamines. Eur J Pharm Sci. 2006;29:414–25. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Peng Q, Zhong Z, Zhuo R. Disulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectors. Bioconjug Chem. 2008;19:499–506. doi: 10.1021/bc7003236. [DOI] [PubMed] [Google Scholar]
- 23.Lv C, Wang Z, Wang P, Tang X. Photodegradable polyurethane self-assembled nanoparticles for photocontrollable release. Langmuir. 2012;28:9387–94. doi: 10.1021/la301534h. [DOI] [PubMed] [Google Scholar]
- 24.Neu M, Sitterberg J, Bakowsky U, Kissel T. Stabilized nanocarriers for plasmids based upon cross-linked poly(ethylene imine) Biomacromolecules. 2006;7:3428–38. doi: 10.1021/bm060788z. [DOI] [PubMed] [Google Scholar]
- 25.Petersen H, Kunath K, Martin AL, Stolnik S, Roberts CJ, Davies MC, et al. Star-shaped poly(ethylene glycol)-block-polyethylenimine copolymers enhance DNA condensation of low molecular weight polyethylenimines. Biomacromolecules. 2002;3:926–36. doi: 10.1021/bm025539z. [DOI] [PubMed] [Google Scholar]
- 26.Yao J, Fan Y, Du R, Zhou J, Lu Y, Wang W, et al. Amphoteric hyaluronic acid derivative for targeting gene delivery. Biomaterials. 2010;31:9357–65. doi: 10.1016/j.biomaterials.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 27.Bonner DK, Zhao X, Buss H, Langer R, Hammond PT. Crosslinked linear polyethylenimine enhances delivery of DNA to the cytoplasm. J Control Release. 2013;167:101–7. doi: 10.1016/j.jconrel.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gary DJ, Min J, Kim Y, Park K, Won Y-Y. The Effect of N/P Ratio on the In Vitro and In Vivo Interaction Properties of PEGylated Poly(2-(dimethylamino)ethyl methacrylate)-Based siRNA Complexes. Macromolecular Bioscience. 2013 doi: 10.1002/mabi.201300046. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peak MJ, Peak JG, Carnes BA. Induction of direct and indirect single-strand breaks in human cell DNA by far- and near-ultraviolet radiations: action spectrum and mechanisms. Photochem Photobiol. 1987;45:381–7. doi: 10.1111/j.1751-1097.1987.tb05390.x. [DOI] [PubMed] [Google Scholar]
- 30.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res-Fund Mol M. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Sinha RP, Hader D-P. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. 2002;1:225–36. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 32.Scicchitano DA, Mellon I. Transcription and DNA damage: a link to a kink. Environ Health Perspect. 1997;105(1):145–53. doi: 10.1289/ehp.97105s1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA- binding protein. EMBO J. 1994;13:2114–23. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol Ther. 2002;5:80–6. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 35.Ignowski JM, Schaffer DV. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol Bioeng. 2004;86:827–34. doi: 10.1002/bit.20059. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JF, Hayes LS, Lloyd DB. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–7. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 37.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–33. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


