Abstract
Mutations in the MTR gene, which encodes methionine synthase on human chromosome 1p43, result in the methylcobalamin deficiency G (cblG) disorder, which is characterized by homocystinuria, hyperhomocysteinemia, and hypomethioninemia. To investigate the molecular basis of the disorder, we have characterized the structure of the MTR gene, thereby identifying exon-intron boundaries. This enabled amplification of each of the 33 exons of the gene, from genomic DNA from a panel of 21 patients with cblG. Thirteen novel mutations were identified. These included five deletions (c.12-13delGC, c.381delA, c.2101delT, c.2669-2670delTG, and c.2796-2800delAAGTC) and two nonsense mutations (R585X and E1204X) that would result in synthesis of truncated proteins that lack portions critical for enzyme function. One mutation was identified that resulted in conversion of A to C of the invariant A of the 3′ splice site of intron 9. Five missense mutations (A410P, S437Y, S450H, H595P, and I804T) were identified. The latter mutations, as well as the splice-site mutation, were not detected in a panel of 50 anonymous DNA samples, suggesting that these sequence changes are not polymorphisms present in the general population. In addition, a previously described missense mutation, P1173L, was detected in 16 patients in an expanded panel of 24 patients with cblG. Analysis of haplotypes constructed using sequence polymorphisms identified within the MTR gene demonstrated that this mutation, a C→T transition in a CpG island, has occurred on at least two separate genetic backgrounds.
Introduction
The methylcobalamin deficiency G (cblG) disorder (MIM 250940) is the result of mutations affecting the MTR gene, which encodes the cytoplasmic enzyme methionine synthase (EC 2.1.1.13). This enzyme catalyzes the methylation of homocysteine to generate methionine, using 5-methyltetrahydrofolate as a methyl-group donor, and requires the presence of an enzyme-bound cobalamin prosthetic group for activity. The reaction proceeds by transfer of the methyl group from 5-methyltetrahydrofolate to the enzyme-bound cobalamin to form methylcobalamin and by subsequent transfer of the methyl group from methylcobalamin to homocysteine to form methionine (Matthews 1999; Matthews and Ludwig 2001). Activity of methionine synthase depends on the presence of a second protein, methionine synthase reductase, which maintains the methionine synthase–bound cobalamin in its fully reduced active state. When the cobalamin becomes oxidized, methionine synthase reductase catalyzes its reductive methylation, using S-adenosylmethionine as a methyl donor to regenerate methylcobalamin (Olteanu and Banerjee 2001).
Methionine synthase activity is important for maintaining adequate levels of methionine and for preventing accumulation of homocysteine. Increased blood levels of homocysteine have been associated with increased likelihood of developing cardiovascular disease, birth defects, and Down syndrome, and they may affect the development of some types of cancer (Carmel and Jacobsen 2001). Methionine synthase is also critical in the methylation cycle, which maintains the cellular level of the methionine derivative S-adenosylmethionine. This molecule acts as methyl group donor in a wide variety of cellular processes, including DNA and RNA methylation and neurotransmitter synthesis. Because methionine synthase is the only enzyme in mammalian cells that uses 5-methyltetrahydrofolate, deficient activity also results in the trapping of cellular folate as 5-methyltetrahydrofolate, which becomes unavailable for other folate-dependent reactions involved in purine and pyrimidine biosynthesis and other single-carbon–transfer reactions.
Inborn errors of metabolism resulting in functional deficiency of methionine synthase are characterized by homocystinuria, hyperhomocysteinemia, and hypomethioninemia. Patients have typically presented during the first 2 years of life with megaloblastic anemia and developmental delay, but patients who presented as adults have been reported (Rosenblatt and Fenton 2001). Complementation analysis demonstrated the existence of two forms of the disorder—cblG and a second form that is designated “methylcobalamin deficiency cblE type” (cblE)—that presumably result from mutations at separate loci (Watkins and Rosenblatt 1989). Cloning of the MTR gene (GDB accession number 119440) on chromosome 1q43 (Leclerc et al. 1996; Li et al. 1996; Chen et al. 1997) led to the demonstration that mutations in this gene are the cause of the cblG disorder (Gulati et al. 1996; Leclerc et al. 1996). It was subsequently shown that the cblE disorder is the result of mutations affecting the MTRR gene, on chromosome 5p15.2-15.3, that encodes methionine synthase reductase (Leclerc et al. 1998).
The cDNA for the human gene encoding methionine synthase was cloned on the basis of homology with the Escherichia coli gene encoding cobalamin-dependent methionine synthase, which had been previously identified. The human gene encodes a protein that is 1,265 amino acids in length. Several mutations in the gene have been described in patients with cblG (Gulati et al. 1996; Leclerc et al. 1996; Wilson et al. 1998). We report further characterization of this gene, defining its intronic and exonic structure. This has permitted sequencing of the entire coding sequence of methionine synthase from genomic DNA of a panel of patients with the cblG disorder. A panel of 18 patients with cblG was investigated by sequence analysis, and an expanded panel of 24 patients was investigated using restriction-endonuclease analysis. To date, 28 patients with the cblG disorder have been identified (Suormala et al. 2001; D.S.R., unpublished data). Thus, this panel of patients with cblG represents a majority of all recognized patients with this disorder.
Patients and Methods
Patients
DNA was extracted and sequenced from a panel of 18 fibroblast lines derived from patients with cblG. DNA from an additional six patients with cblG was tested for identified mutations, using restriction endonuclease–based assays, as described below. All cell lines were characterized by decreased functional activity of methionine synthase and decreased synthesis of methylcobalamin in intact cells, in the presence of normal functional activity of a second cobalamin-dependent enzyme, methylmalonyl-CoA mutase, and normal synthesis of its adenosylcobalamin coenzyme. The diagnosis of cblG was in all cases confirmed by somatic cell complementation analysis. These studies were approved by the research ethics board of the Royal Victoria Hospital.
Genomic Organization of the Coding Region of the Human Methionine Synthase Gene
Some of the cDNA clones used for the original sequencing of the human methionine synthase cDNA (Chen et al. 1997) contained intronic sequences. Additional exon-intron junctions were determined by PCR, using primers based on the cDNA sequence. PCR products that were larger than the expected size of the cDNA sequence were assumed to contain intronic sequences and were sequenced across the junctions. Additional intronic junctions were identified by genomic sequencing of putative exons. Intron sizes were determined by sequencing through the region or by PCR using flanking primers.
Sequencing of the MTR Gene
Sequencing was performed using genomic DNA from 18 cblG cell lines. Six additional cell lines (WG1655, WG1670, WG1671, WG2867, WG2918, and WG2989) were tested for presence of mutations detected by sequencing of the other cell lines, using the restriction endonuclease–based tests described below, but were not sequenced themselves.
Each of the 33 exons of the MTR gene was amplified separately, using PCR primers within the flanking intronic sequences, such that the entire exon (as well as exon-intron boundaries) could be sequenced. Primers were designed using the Primer 3.0 software available on the Whitehead Institute/MIT Center for Genome Research server. The sequences of the PCR primers used are shown in table 1. The annealing temperature for all primer pairs was 60°C. PCR was performed in a total volume of 50 μl containing 1.25 U AmpliTaq Gold polymerase (Perkin-Elmer Cetus) in the buffer provided by the manufacturer, 2.5 mM MgCl2, 0.2 mM dNTPs, primers at a final concentration of 0.5 μM, and 100 ng of template genomic DNA. PCR products were purified with BioMag DNA Sep magnetic beads (PerSeptive Biosystems). Sequencing reactions were performed using the BigDye Primer Cycle Sequencing Ready Reactions-21M13 kit (PE Applied Biosystems), and products were analyzed on ABI 377 automated DNA sequencers (PE Applied Biosystems). Gel files were processed using Sequence Analysis software (PE Applied Biosystems) and then were assembled and analyzed using Autoassembler 2.0 (PE Applied Biosystems).
Table 1.
PCR Primers Used in Sequencing[Note]
| Exon | Sense | Antisense | Product Size(bp) |
| 1 | 5′-TGTAAAACGACGGCCAGTGTACCCGGGAAGAAAGCAC-3′ |
5′-GTGGAGCAAAACACGTAGCC-3′ | 457 |
| 2 | 5′-AAGCTCCTCCCTCACACATC-3′ | 5′-TGTAAAACGACGGCCAGTTTTATTCTCCGGAGCACTCG-3′ |
384 |
| 3 | 5′-TGTAAAACGACGGCCAGTATGTTTATTTTGGGGGCACA-3′ |
5′-GCAAACGCTCGTAAGCATCT-3′ | 202 |
| 4 | 5′-TGTAAAACGACGGCCAGTCTCATGGTTTGGGAGGAGAA-3′ |
5′-AGGACCACAGAGTTGGCATC-3′ | 513 |
| 5 | 5′-TGTAAAACGACGGCCAGTATTGGCTATTCATTCACGAC-3′ |
5′-GAGTACTTCCCCAAGATCAA-3′ | 209 |
| 6 | 5′-TGTAAAACGACGGCCAGTATCCCTAGGCCATTCAGAGG-3′ |
5′-TACCCCAGCCTTGTCTGTTT-3′ | 207 |
| 7 | 5′-TGTAAAACGACGGCCAGTGTGCTGGGGTTGTGCCTTAT-3′ |
5′-AACAGTTGCCTTCCATCAGC-3′ | 452 |
| 8 | 5′-TGTAAAACGACGGCCAGTTCTGTTTCTGTGGCTTCCTG-3′ |
5′-GCAGCGATCTTCTGAAACCT-3′ | 483 |
| 9 | 5′-TTAAGGAGTCCACCCACAGG-3′ | 5′-TGTAAAACGACGGCCAGTGAAACACTGCACCCAGACAC-3′ |
490 |
| 10 | 5′-GGGGGTGTGGTATCTCTTTG-3′ | 5′-TGTAAAACGACGGCCAGTTGGGATTTCTTCAGGGAAAA-3′ |
432 |
| 11 | 5′-TCCTATGTCTAGGGCATGTGAT-3′ | 5′-TGTAAAACGACGGCCAGTCAGCCCACATTCTGAAACAG-3′ |
388 |
| 12 | 5′-TGTAAAACGACGGCCAGTAAAGGCATGATTGTTGCTG-3′ |
5′-TACCTGGGCAAATCACCTG-3′ | 479 |
| 13 | 5′-TTGTAACAAATAGCCTTTACTGCAT-3′ | 5′-TGTAAAACGACGGCCAGTCTGCAAACACACATTACCA-3′ |
451 |
| 14 | 5′-AGGGAGTCTGGCGAGGTAGT-3′ | 5′-TGTAAAACGACGGCCAGTCAACTGGCACGGTTAAATGA-3′ |
338 |
| 15 | 5′-TGTAAAACGACGGCCAGTTCCAAGTATTTTGGGCTTAAAGA-3′ |
5′-ACGTTGGCACAATGACAAAA-3′ | 597 |
| 16 | 5′-TGTAAAACGACGGCCAGTGGAAGCTGCTTGGCATTTAG-3′ |
5′-GCGAGTTCATCAGGCTCTGT-3′ | 601 |
| 17 | 5′-TGTAAAACGACGGCCAGTTGTAATACGGCAAGCTTTGG-3′ |
5′-TAGGTGTGGCTCTATCCCAAT-3′ | 425 |
| 18 | 5′-TGTAAAACGACGGCCAGTCTGTGGCTGCTTTTATGCTG-3′ |
5′-CACCACCATCTCTGCAACTC-3′ | 453 |
| 19 | 5′-TGTAAAACGACGGCCAGTCAGTCTACTAGGGGCTGGAAAA-3′ |
5′-CTCCCCAGGTTGAGAATCAG-3′ | 344 |
| 20 | 5′-TGAGGAAACTGAGGCTTATTGA-3′ | 5′-TGTAAAACGACGGCCAGTACTCTGAACGCGGATTCTTG-3′ |
520 |
| 21 | 5′-TGTAAAACGACGGCCAGTGCTGTCCTGAAGTCAAAGCA-3′ |
5′-AAGGGAGAGATGTGGGATCA-3′ | 363 |
| 22 | 5′-TGTAAAACGACGGCCAGTACTGTTTGGGCTTCGTTTTC-3′ |
5′-TCAAAAACACAGCAAGCAAAA-3′ | 400 |
| 23 | 5′-TTAGCATAGTGCCTGGCGTA-3′ | 5′-TGTAAAACGACGGCCAGTAAATGGCAATGAAAGCAACC-3′ |
432 |
| 24 | 5′-TGTAAAACGACGGCCAGTGGGCAACAGAATGAGACTCC-3′ |
5′-GCAAGCCAAAGAAATCCAAG-3′ | 542 |
| 25 | 5′-TGTAAAACGACGGCCAGTAATGGCGGTGAACAAGAGTT-3′ |
5′-TGCAAGCTTTCCAACACATC-3′ | 207 |
| 26 | 5′-TGTAAAACGACGGCCAGTCCAAGCCCACTGAGTTTACC-3′ |
5′-GACACTGAAGACCTCTGATTTGAA-3′ | 433 |
| 27 | 5′-GGCCATGGGAAGACCAGTTA-3′ | 5′-TGTAAAACGACGGCCAGTCACACAACACCTGCTCCATAA-3′ |
596 |
| 28 | 5′-ACTCCTACGGGTGACAGGAG-3′ | 5′-TGTAAAACGACGGCCAGTATCCAAATCCACTGCTCCTC-3′ |
376 |
| 29 | 5′-AAGAAGCATTGTTTGATGGT-3′ | 5′-TGTAAAACGACGGCCAGTTGAAGCAGACCCTCATTTAT-3′ |
355 |
| 30 | 5′-TGTAAAACGACGGCCAGTTCCAAGTACTGGGGATGTAACAG-3′ |
5′-TCAAGGTTTTTCTTTGCCTCA-3′ | 5,675 |
| 31 | 5′-TGTAAAACGACGGCCAGTGTGTCCTCATGGTTCTAATTCA-3′ |
5′-TACTGAAGAAGGAAGACAGGA-3′ | 332 |
| 32 | 5′-TGTAAAACGACGGCCAGTAGACACTGAGTCCATAAGCA-3′ |
5′-GCAAGCCACATATAATGAA-3′ | 189 |
| 33 | 5′-GAAACTTCTATTCCAAAAGTC-3′ | 5′-TGTAAAACGACGGCCAGTTTGTTGTTGTTATTTTTAAGG-3′ |
230 |
Note.— Primers are those used for sequencing of the methionine synthase gene from genomic DNA. The M13 tail included in one of the PCR primers is indicated by underlining.
Restriction Endonuclease Analysis
MaeIII was purchased from Boehringer Mannheim; the remaining restriction endonucleases (Bsp1286I, BsrI, BstNI, BstUI, DdeI, HinfI, MspI, and NspI) were purchased from New England Biolabs. When a sequence change detected on sequence analysis resulted in the creation or destruction of a restriction site, DNA was amplified by the same primers used for sequencing, and the PCR product was digested with the appropriate restriction endonuclease, using the buffer and reaction conditions specified by the manufacturer. Digestion products were separated by electrophoresis on 2% agarose gels or on 10% polyacrylamide (29:1 acrylamide:bis-acrylamide) gels, depending on the sizes of the products to be separated. Two sequence changes that did not result in the creation or destruction of a restriction site were confirmed by use of primers that resulted in the creation of artificial restriction sites in PCR products. For the c.1310C→A sequence change in exon 14, alternative PCR primers were designed (sense: 5′-CCAGATTTTGCAACTTAATTGATT-3′; antisense: 5′-CCCTGTTTTCGAT GTTGAGA-3′) that resulted in the creation of a HinfI restriction site in wild-type DNA that was absent in DNA containing the sequence change. For the c.2669-2670delTG sequence change in exon 25, alternative PCR primers were designed (sense: 5′-CTGGACGCGTCCAAGACT-3′; antisense: 5′-GACATGGTCACCTGGACCTC-3′) that resulted in the creation of a BsrI restriction site when the mutant sequence was present, a sequence that was absent when wild-type DNA was amplified. For a third deletion that did not result in the creation or destruction of a restriction site (c.2796-2800delAAGTC), the amplicon was incubated with NspI to generate smaller DNA fragments, and mutations were recognized on the basis of heteroduplex formation on polyacrylamide gel electrophoresis.
Results
Organization of the Human MTR Gene
The coding region of the human methionine synthase gene is composed of 33 exons and 32 introns. The nucleotide sequences at the splice junctions and the approximate size of each intron are summarized in table 2. All the introns, except for intron 21, follow the GT-AG rule. The start ATG (+1) is in exon 1.
Table 2.
Intron-Exon Junctions in the Coding Region of the Human MTR Gene[Note]
| Exon | Position | 5′ Splice Donor | Intron | Size(kb) | 3′ Splice Acceptor |
| 1 | −394 to 34 | TCGCAACCCGgtaacgctgc | 1 | 4.0 | ttctttaaagAAGGTCTGAA |
| 2 | 35–249 | AATCCATAAGgtaaagtatt | 2 | 2.4 | cttgttgcagGAATACTTGC |
| 3 | 250–339 | TGAACACTTGgtaagaattc | 3 | 2.4 | gatacgttagGCCTACCGGA |
| 4 | 340–409 | TGAACACTTGgtaagaattc | 4 | 1.7 | tttcccaaagGAATTAAGAG |
| 5 | 410–502 | AGGAACATCAgtgagtattt | 5 | 2.0 | ttttttgcagCATTTGATGA |
| 6 | 503–609 | CAATGCCAAGgtgagttaag | 6 | 2.7 | tccctgacagGTAGCCTTGT |
| 7 | 610–669 | GCCTATCTTTgtaagttcta | 7 | .7 | gtcaattcagATTTCAGGGA |
| 8 | 670–764 | AACCACTCTGgtgagtgatc | 8 | ∼3.5 | gtttttctagCATTGGATTA |
| 9 | 765–865 | AACCACTCTGgtgagtgatc | 9 | 1.1 | ctttactcagGTCTTCCCAA |
| 10 | 866–927 | GCACCTAAAGgtcaggggtc | 10 | 1.5 | ctaaatgcagGATTTTGCTA |
| 11 | 928–995 | ATCATATCAGgtcaggggtc | 11 | 2.2 | ttcaactcagGGAAATTGCT |
| 12 | 996–1075 | TTACTGTCTGgtgagtcata | 12 | 2.5 | tccttttaagGTCTAGAGCC |
| 13 | 1076–1188 | AACTATGAAGgtgagtggtt | 13 | 3.0 | cttccttcagGAAGCCTTGT |
| 14 | 1189–1329 | CATCGCAAAGgttatacaaa | 14 | 2.5 | ctgatctcagGTACCTTTGT |
| 15 | 1330–1515 | AGAAGGACAGgtgagtggtt | 15 | ND | ttttccctagGCAACAGAAA |
| 16 | 1516–1695 | AGTCATTAAAgtaagtgtag | 16 | 2.0 | tttttgccagGAAACATTAC |
| 17 | 1696–1812 | TGCAATCAAGgtatggtaga | 17 | .3 | ttgcccttagTCTGGCATGG |
| 18 | 1813–1953 | TTATGCCCAGgtagagagac | 18 | 4.5 | ttcaattcagACTCAAGGCA |
| 19 | 1954–2043 | CCTTGTGAAGgtaagttaca | 19 | 1.3 | tcttttttagGGCATTGAAA |
| 20 | 2044–2196 | TCTACCTCAGgttagcaaaa | 20 | .9 | tttttctcagGTTATAAAGT |
| 21 | 2197–2304 | AGAAGAAGAGgcaagtcatt | 21 | 1.0 | tcctttccagGACCCTTACC |
| 22 | 2305–2405 | ATAATTTCCGgtaagttagg | 22 | ∼3.0 | tgtgcctcagAGTTATTGAT |
| 23 | 2406–2473 | CACAAAGCAGgtactgtgca | 23 | .9 | aaaaaaatagATATAATTGG |
| 24 | 2474–2594 | CCACTTCAAAgtaagttata | 24 | 6.0 | ttctttttagAACCCACACA |
| 25 | 2595–2676 | TGTGGTGGTGgtaagtgggt | 25 | ND | gcattttcagTGTTCCCAGC |
| 26 | 2677–2775 | GTCTCTCAAGgtaagtggta | 26 | 2.5 | tttttaacagGAGAGGAGAT |
| 27 | 2776–2851 | CCTCACCCAGgtctgtttgg | 27 | 1.0 | tctcctgtagTGAAGCCCAC |
| 28 | 2852–3007 | AAAACAGTAGgttagtgcag | 28 | 1.8 | tttctaatagGTGGAGAGGC |
| 29 | 3008–3204 | AAGGCAACAGgtatggaagg | 29 | 2.5 | tggttttaagGCTGAGAAGG |
| 30 | 3205–3405 | GCTGGCAGAGgtaaggcaga | 30 | .8 | gggtcccaagGCCTTTGCAG |
| 31 | 3406–3598 | CAGTCTACAGgtaggaagcc | 31 | 1.4 | ctccctctagGCATTAGGTT |
| 32 | 3599–3711 | CAAGGATCAGgtaagctagc | 32 | .5 | tcctttgcagGTTGAGGATT |
| 33 | 3712– |
Note.— Nucleotides are numbered from the translation start site. Uppercase letters refer to coding sequence and lowercase letters indicate intronic sequences; ND = not determined.
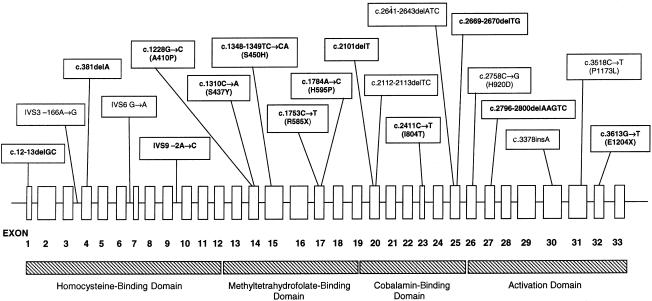
All 33 exons of the methionine synthase gene were sequenced in a panel of 18 cblG fibroblast lines. Genomic DNA from six additional patients with cblG was analyzed for the presence of mutations detected in the original panel using restriction endonuclease–based techniques described below. This panel included cell lines that had been investigated previously (WG1655, WG1670, WG1671, WG1765, WG1892, and WG2290) (Gulati et al. 1996; Leclerc et al. 1996; Wilson et al. 1998). Thirteen novel mutations were identified. Furthermore, one previously identified mutation (c.3518C→T [P1173L]) was identified in several additional cell lines. All mutations in methionine synthase that have been identified to date are shown in figure 1. All detected mutations were present in the heterozygous state (table 3).
Figure 1.
Identified mutations in human methionine synthase. The exon structure of the methionine synthase gene and the locations of the mutations identified in the present study and in previous studies are shown. Mutations identified for the first time in the present publication are shown in bold. The regions of the gene encoding the four domains of the methionine synthase enzyme are also shown.
Table 3.
Summary of Mutations Found in Patients with cblG[Note]
|
Patient Characteristics |
||||
| CellLine | Sex/Racea | Age at Onset | First Mutation | Second Mutation |
| WG2292 | F/W | 2 wk | c.3518C→T (P1173L) | c.3613G→T (E1204X) |
| WG1975 | M/W | 1 mo | c.3518C→T (P1173L) | c.1753C→T (R585X) |
| WG1308 | M/W | 1.5 mo | c.3518C→T (P1173L) | IVS9−2A→C |
| WG1505 | M/W | 2 mo | c.3518C→T (P1173L) | c.2411T→C (I804T) |
| WG1386 | F/W | 3 mo | c.3518C→T (P1173L) | c.1784A→C (H595P) |
| WG2507 | F/W | 6 mo | c.3518C→T (P1173L) | c.2796-2800delAAGTC |
| WG2306 | F/W | 7 mo | c.3518C→T (P1173L) | c.1310C→A (S437Y) |
| WG1352 | F/W | 1 year | c.3518C→T (P1173L) | c.1348-1349TC→CA (S450H) |
| WG1321 | M/W | 1.7 years | c.3518C→T (P1173L) | c.2669-2670delTG |
| WG1892 | M/W | 3.5 years | c.3518C→T (P1173L)b | c.2641-2643delATCb,c |
| WG1408 | F/W | 21 years | c.3518C→T (P1173L) | c.2101delT |
| WG2009 | M/W | 38 years | c.3518C→T (P1173L) | c.1228G→C (A410P) |
| WG1671 | M/B | Neonatal | IVS3 –166A→Gd | c.2112-2113delTCd |
| WG1670 | F/B | 2 days | IVS3 –166A→Gd | c.2112-2113delTCd |
| WG1655 | M/W | 1.7 mo | IVS6 G→Ad | c.3378insAd |
| WG1223 | F/W | 5.5 mo | c.3518C→T (P1173L) | |
| WG2989 | F/W | 1 year | c.3518C→T (P1173L) | |
| WG2867 | M/W | 13 years | c.3518C→T (P1173L) | |
| WG2724 | M/W | 31 years | c.3518C→T (P1173L) | |
| WG1765 | M/W | 2.5 mo | c.12-13delGC | |
| WG2725 | M/W | 2.5 mo | c.1348-1349TC→CA (S450H) | |
| WG2290 | M/W | 2.7 mo | c.2758C→G (H920D)c | |
| WG2829 | F/W | 5 mo | c.381delA | |
| WG2918 | F/B | 3 wk | ||
Note.— Mutations detected in the methionine synthase gene in patients with cblG are summarized. Amino acid changes are given in brackets for missense mutations. The recurrent c3518C→T mutation is underlined. Cell lines WG2867, WG2918, and WG2989 were not sequenced but were tested, by use of restriction endonuclease–based methods, for the presence of mutations detected in other cell lines. Note that at least one mutation was detected from 23 of 24 cell lines and that two mutations were identified in 15 cell lines; the single cell line in which no mutation was identified (WG2918) was not sequenced, but investigated using restriction endonuclease–based tests only. References are given for mutations already published elsewhere. Note that the precise location of the G→A change within intron 6 of WG1655 has not been determined.
F = female; M = male; W = white; B = black.
Gulati et al. (1996).
Leclerc et al. (1996).
Wilson et al. (1998).
The Recurrent c.3518C→T Mutation (P1173L)
The c3518C→T mutation, which results in replacement of proline by leucine at position 1173 of the amino acid sequence, had been previously identified by Gulati et al. (1996). It was identified by sequence analysis in 14 of the 18 cblG lines in the panel of the present study (table 3), including WG1892, the cell line in which it was originally identified. The c3518C→T mutation results in loss of an MspI site (Gulati et al. 1996). Restriction-endonuclease analysis confirmed the presence of the sequence change in DNA from the 14 cell lines; in addition, the sequence change was detected in DNA from two additional cblG cell lines that had not undergone sequencing. The c3518C→T mutation was thus detected in 16 of 24 patient cell lines in total. The sequence change was not detected in a panel of 50 anonymous control DNA samples tested by restriction-endonuclease digestion.
Novel Methionine Synthase Mutations
c.12-13delGC
Sequence analysis of DNA from WG1765 demonstrated the presence of a 2-bp deletion at position 12 in exon 1, resulting in a frame shift, with the loss of normal amino acid sequence starting at codon 5 and creation of a stop codon at position 20. This sequence change resulted in loss of a BstUI site. Restriction analysis with BstUI confirmed the presence of c.12-13delGC in WG1765; this change was not detected in DNA from any other patient with cblG.
c.381delA
Sequence analysis indicated a single–base-pair deletion in exon 4 of DNA from WG2829. This resulted in a frame shift, with altered amino acid sequence, beginning at codon 128, and the creation of a stop signal at codon 132.
IVS9−2A→C
Sequence analysis of exon 10 from WG1308 showed a sequence change resulting in alteration of the invariant A at the splice site at the 3′ end of intron 9. This mutation is expected to result in deletion of exon 10 from the methionine synthase transcript. This sequence change resulted in the creation of an MspI restriction site. Restriction-endonuclease analysis confirmed the sequence change in WG1308. The sequence change was not detected in DNA from any other patient with cblG, nor was it detected in a panel of 50 anonymous control DNA samples.
c.1228G→C (A410P)
This sequence change, detected in DNA from WG2009, resulted in conversion of alanine to proline at amino acid 410 of the methionine synthase protein. The sequence change resulted in the creation of a Bsp1286I restriction site. Restriction-endonuclease analysis confirmed that the sequence change was present in DNA from WG2009 but not in DNA from any other patients. The sequence change was not detected in a panel of 50 anonymous control DNA samples.
c.1310C→A (S437Y)
This sequence change, detected in WG2306, resulted in replacement of serine by tyrosine at amino acid 437 of the methionine synthase protein. No restriction site was created or destroyed by this sequence change. Alternative PCR primers were designed that resulted in the creation of a HinfI restriction site in wild-type DNA that was absent in DNA containing the sequence change. Restriction-endonuclease analysis using these primers demonstrated that the sequence change was present in DNA from WG2306 but not from any other patient DNA. The sequence change was not detected in a panel of 50 anonymous control DNA samples.
c.1348-1349TC→CA (S450H)
This sequence change was observed in DNA from two unrelated patients with cblG, WG1352 and WG2725, and caused conversion of serine to histidine at amino acid 450 of the methionine synthase protein. This sequence change resulted in loss of a HinfI site in exon 15. Restriction analysis confirmed that the sequence change was present in DNA from WG1352 and WG2725 but not in DNA from other patients. The sequence change was not detected in a panel of 50 anonymous control DNA samples.
c.1753C→T (R585X)
This sequence change in exon 17 of the MTR gene was detected in DNA from WG1975 and resulted in the creation of a stop codon at position 585 of the cDNA sequence. This sequence change resulted in the creation of a DdeI site. Restriction-endonuclease analysis confirmed the presence of c1753C→T in DNA from WG1975; the sequence change was not detected in DNA from other patients with cblG, nor was it detected in a panel of 50 anonymous control DNA samples.
c1784A→C (H595P)
This sequence change, detected in exon 17 of DNA from WG1386, resulted in replacement of a histidine residue by proline at amino acid position 595 of the methionine synthase protein. This sequence change resulted in the creation of a BstNI restriction site in the patient DNA. Restriction-endonuclease analysis confirmed the existence of the sequence change in WG1386 DNA; the sequence change was not present in DNA from other patients with cblG and was not detected in a panel of 50 anonymous control DNA samples.
c.2101delT
This single–base-pair deletion, detected in DNA from WG1408, resulted in a frame shift that involved the loss of normal amino acid sequence (starting at position 701 of the methionine synthase protein) and the creation of a stop codon at position 707.
c.2411T→C (I804T)
This sequence change resulted in conversion of isoleucine to threonine at amino acid 804 of the methionine synthase protein in WG1505. This sequence change resulted in the creation of a MaeIII restriction site in the patient DNA. Restriction-endonuclease analysis confirmed the existence of the sequence change in WG1505. The sequence change was not present in DNA samples from other patients with cblG and was not detected in a panel of 50 control DNA samples.
c.2669-2670delTG
This 2-bp deletion was detected by sequence analysis in DNA from WG1321. It resulted in a frame shift, starting at codon 890, and in the creation of a stop codon at position 898. Alternate PCR primers were designed that resulted in the creation of an artificial BsrI site in DNA containing the deletion; this site was absent in wild-type DNA. Analysis confirmed that the mutation was present in WG1321 but was not present in any other patient in the panel.
c.2796-2800delAAGTC
A 5-bp deletion was detected on sequence analysis of DNA from WG2507. This resulted in a frame shift, beginning at codon 935, and in the creation of a stop codon at position 945. This mutation did not result in the creation or destruction of a restriction site. The amplicon (613 bp) was digested with NspI, yielding fragments of 268, 205, and 140 bp for wild-type DNA. The 5-bp deletion affected the 205-bp fragment, resulting in heteroduplex formation in the heterozygous individual. Heteroduplex formation occurred in digested DNA from WG2507 and not in DNA from any other patient with cblG.
c.3613G→T (E1204X)
This sequence change was detected on sequence analysis of exon 32 of DNA from WG2292 and resulted in the creation of a stop codon at position 1204 of the methionine synthase protein. This sequence change resulted in the loss of a HinfI restriction site. Restriction-endonuclease analysis confirmed the presence of the sequence change in WG2292. The sequence change was not detected in DNA from any other patient with cblG.
Polymorphisms in the MTR Gene
Sequencing of the MTR gene also demonstrated the existence of polymorphisms not associated with cblG disease. Several of these polymorphisms were detected in multiple cell lines, including both patients and control individuals. Two of these, c.3492A→C (R1164R) and c.3576C→T (L1192L), both in exon 31, were seen as being different from sequences of the MTR gene published elsewhere (Leclerc et al. 1996; Chen et al. 1997). Additional polymorphisms that were observed include c.2756A→G (D919G) (Leclerc et al. 1996), c.3144A→G (A1048A) (Tang et al. 2001), and polymorphisms within introns 9 (IVS9−47insA) and 26 (IVS26+43G→A). In addition, each of two sequence changes (IVS3 –6C→T and c.858C→T [P286P]) that would not be expected to affect enzyme function was observed in a single cell line.
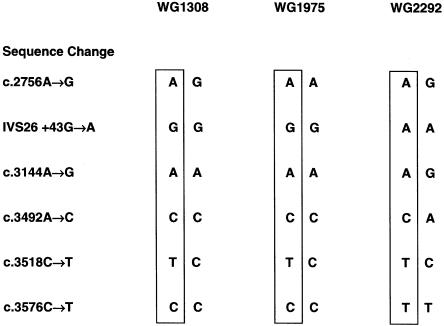
Haplotype Analysis of Patients with the Recurrent c.3518C→T Mutation
Using the results of typing for polymorphisms within the MTR gene, we were able to determine the haplotypes of the alleles carrying the c.3518C→T mutation in some patients (fig. 2). These results demonstrated that the mutation had arisen on at least two different backgrounds. Thus, in WG1308 and WG1975, c.3518C→T was associated with the G allele at the IVS26+43G→A polymorphism and with the C allele at the c.3576C→T polymorphism in exon 31. In WG2292, c.3518C→T was associated with the A and the T alleles, respectively, of these polymorphisms. It was not possible to identify haplotypes associated with the mutation in the remaining patients.
Figure 2.
Haplotypes of patients with the c.3518C→T mutation. Haplotypes were constructed using the genotypes of patients for the polymorphisms c.2756A→G (exon 26), IVS26+43G→A, c.3144A→G (exon 29), c.3492A→ C (exon 31) and c.3576C→T (exon 31) within the methionine synthase gene. Results are shown for patients that were informative. Note that WG1308 and WG1975 are homozygous for the G form of the IVS26+43G→A polymorphism 5′ to the mutation; WG2292 is homozygous for the A form. Similarly, WG1308 and WG1975 are homozygous for the C form of the c.3576C→T polymorphism 3′ to the mutation, whereas WG2292 is homozygous for the T form.
Genotype-phenotype correlations
Table 3 summarizes the mutations detected in the MTR gene in the panel of patients with cblG, together with clinical information on the patients. In 15 of the 24 patients, two causal mutations were identified; in 8 patients, a single mutation was identified; and in 1 patient (WG2918, whose DNA was analyzed by restriction-endonuclease digestion for known mutations but was not sequenced), no causal mutation was identified. No patient was homozygous for a single mutation. Four mutations were identified in more than one patient. The shared mutations in siblings WG1671 and WG1672 (IVS3 –166A→G and c.2112-2113delTC) have been described elsewhere (Wilson et al. 1998). S450H was found in two unrelated patients (WG1352 and WG2725). The P1173L mutation was present in 16 patients. The identification of several compound heterozygotes with P1173L plus a second mutation suggested that it might be possible to estimate the effects of the second mutations by comparing clinical presentations. Mutations that would be expected to result in production of truncated, inactive proteins (E1204X, R585X, and c.2796-2800delAAGTC) were present together with P1173L in three patients with presentation during the first months of life. However, patient WG1408—who had P1173L plus c.2101delT, which would be expected to produce a protein lacking enzyme activity—did not come to medical attention until adulthood.
Discussion
In the present study, we report the organization of the MTR gene encoding methionine synthase. It is a highly compressed gene of 33 exons and 32 introns that spans at least 60 kb, with two intron sizes undefined (table 2). The gene has also been identified in E. coli (Banerjee et al. 1989) and Caenorhabditis elegans (GenBank accession number Z46828), and the cDNA has been identified in the rat (Yamada et al. 1998). The amino acid sequence is 55% identical with that of the cobalamin-dependent methionine synthase from E. coli (Li et al. 1996) and is 92% identical to that of the rat (Yamada et al. 1998). A survey of sequence similarity allows identification of conserved residues that may be important for enzyme function. This allows meaningful comparisons to be made between the enzyme from humans and that from E. coli, in which the function of the cobalamin-dependent methionine synthase has been extensively studied. Methionine synthase from E. coli is a modular enzyme. Domains have been identified by tryptic digestion of native E. coli methionine synthase and by expression of truncated metH genes. In E. coli, residues 2–353 constitute a domain involved in the binding and activation of homocysteine (Goulding et al. 1997). Residues 354–649 are believed to constitute the 5-methyltetrahydrofolate binding (Goulding et al. 1997). Residues 650–896 constitute the cobalamin-binding domain of methionine synthase (Banerjee et al. 1989). Residues 897–1227 make up the activation domain and contain the binding site for S-adenosylmethionine, which is required for reductive methylation of enzyme-bound cobalamin (Dixon et al. 1996). The crystal structures of the domains involved in binding of cobalamin (Drennan et al. 1994) and of S-adenosylmethionine (Dixon et al. 1996) have been determined, and amino acids directly involved in binding have been identified. The structure of mammalian methionine synthase appears similar to that of the bacterial enzyme, and domains and critical residues can be identified when the bacterial and mammalian forma have been aligned.
Of the 13 novel sequence changes identified in the present report, 7 are predicted to result in premature termination of the amino acid chain. These range from c.12-13delGC, which would result in total absence of functional protein, to c.3613G→T, which would result in loss of the terminal 62 amino acids of the methionine synthase protein. On the basis of the modular nature of methionine synthase, the effects of the various mutations on protein function can be predicted. Thus, c.12-13delGC would result in a protein lacking all four functional domains. c.381delA would result in a protein lacking most of the homocysteine-binding domain, as well as all of the 5-methyltetrahydrofolate-binding, cobalamin-binding, and activation domains. c.1753C→T would result in loss of part of the 5-methyltetrahydrofolate-binding domain, as well as all of the cobalamin-binding domain and the activating domain. c.2101delT would result in loss of most of the cobalamin-binding domain and loss of the entire activation domain. The three remaining truncating mutations would result in loss of parts of the activating domain, including residues known to be critical for binding of S-adenosylmethionine. It has been shown that an intact activation domain is critical to the function of methionine synthase. Thus, a fragment of E. coli methionine synthase lacking the activation domain is initially able to support methylation of homocysteine, but activity decays with time and is ultimately lost (Drummond et al. 1993). Although these are predictions of resulting structure, it is often the case that mutations that introduce premature stop codons are associated with instability of the mRNA. This was found to be the outcome for c.2112delTC and c.3378insA, which, coupled with mutations involving intronic inserts, gave fibroblast mRNA nearly undetectable by northern blot analysis (Wilson et al. 1998).
Five missense mutations within the coding sequence of the MTR gene were identified in the present study. Although these mutations have not been expressed to confirm that they disrupt methionine synthase activity, none were detected in a survey of DNA samples from the control population. Three of the mutations (A410P, S450H, and H595P) affect residues that are conserved in methionine synthase from E. coli, C. elegans, and the rat. S437Y affects a residue conserved in C. elegans and the rat; E. coli contains glycine at this position. I804T affects a residue conserved in methionine synthase from the rat; the equivalent residue in E. coli and C. elegans is valine. Thus, in four of the five missense mutations, the affected residue is conserved over considerable phylogenetic distance, suggesting that these sequence changes have a significant effect on methionine synthase function. The final mutation, IVS9−2C→A, was not observed in the general population and would be expected to result in deletion of part of the homocysteine-binding domain of methionine synthase.
One previously identified mutation, P1173L, was highly prevalent in the patient samples of the present study. It was detected in 16 of 24 mutant fibroblast lines in the panel, including WG1892, in which the mutation had been previously identified (Gulati et al. 1996). This is clearly a disease-causing mutation, given its prevalence in patients and its absence in control subjects (consisting of a total of 210 control individuals analyzed in the present study and the study by Gulati et al. [1996]). In addition, it results in replacement of a highly conserved proline residue at position 1173 by leucine; P1173 lies between two residues, R1172 and A1174, that interact directly with S-adenosylmethionine (Dixon et al. 1996).
The high frequency of the P1173L mutation in patients with cblG raises the question of whether the mutation has arisen more than once during history. The majority of the members of the patient panel were of European descent, including all of the patients who carried the P1173L mutation. Analysis of haplotypes in patients who carried the P1173L mutation (fig. 2) demonstrated that it has arisen on at least two different backgrounds. The mutation represents a C→T transition at a CpG island; such sites are known to be prone to mutation (Templeton et al. 2000). These data suggest that P1173L is truly a recurrent mutation that has arisen more than once during human history.
Clinical findings in patients with the cblG disorder vary widely. The identification—by the present study and others—of causal mutations in the cblG disorder means that it is now possible to attempt to correlate phenotype with genotype in this disorder, although such a correlation is limited by the fact that both causal mutations have not been found in all patients. Variation in clinical presentation in patients carrying one copy of P1173L in combination with a second mutation may reflect the effect of the second mutation. Patients with P1173L in combination with a nonsense mutation that would be expected to result in production of a truncated, inactive protein showed a wide range of age at onset (table 3). Some patients (WG2292, WG1975, and WG2507) showed onset of symptoms within the first months of life. However, WG1408, who had P1173L plus a deletion resulting in loss of the cobalamin-binding and activation domains of methionine synthase, came to medical attention in adulthood. These results suggest that genetic modifiers and environmental factors may significantly affect clinical presentation in patients with cblG.
Elevated blood levels of homocysteine have been associated with increased risk of cardiovascular disease and adverse pregnancy outcomes (Ueland et al. 2001). Homocysteine has also been associated with increased likelihood of occurrence of birth defects, such as neural tube defects and orofacial clefting (Christensen and Rosenblatt 1995; Wong et al. 1999) and Down syndrome (James et al. 1999), and it may affect development of some types of cancer (Kim 1999). Polymorphisms in genes encoding methylenetetrahydrofolate reductase and methionine synthase reductase (enzymes involved in homocysteine remethylation) have been shown to affect blood levels of homocysteine and to be associated with altered risk of cardiovascular disease and birth defects (Rozen 2001). To date, studies of the c.2576A→G (D919G) MTR polymorphism have been inconclusive. The G form appears to be associated with lowered plasma total homocysteine (Wang et al. 1999; Chen et al. 2001; Dekau et al. 2001). However, no consistent effect on risk of developing cardiovascular disease (Morita et al. 1999; Hyndman et al. 2000; Chen et al. 2001) or birth defects (Christensen et al. 1999; Shaw et al. 1999) has been documented. Further study of MTR mutations and polymorphisms such as those described in the present publication may shed light on the function of methionine synthase that will be important beyond development of the cblG phenotype.
Acknowledgments
The authors offer thanks to the clinicians who provided patient samples and clinical information, to Nora Matiaszuk for complementation analysis, and to Gail Dunbar for growth of patient fibroblasts. This work was supported by an operating grant from the Canadian Institutes for Health Research (CIHR) and by the CIHR Group Grant in Medical Genetics. The work of B.S. and H.-Y.H. was supported by Public Health Service grants HL58991 and DK42033 from the United States Department of Health and Human Services. This is a publication of the Hess B. and Diane Finestone Laboratory in the Memory of Jacob and Jenny Finestone.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for C. elegans methionine synthase gene sequence [accession number Z46828])
- Genome Database, http://gdbwww.gdb.org (for MTR gene [GDB accession number 119440])
- Online Mendelian Inheritance in Man (OMIM),http://www.ncbi.nlm.nih.gov/Omim/ (for cblG disorder [MIM 250940])
- Whitehead Institute/MIT Center for Genome Research,http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for Primer 3.0 software)
References
- Banerjee RV, Johnston NL, Sobeski JK, Datta P, Matthews RG (1989) Cloning and sequence analysis of the Escherichia coli metH gene encoding cobalamin-dependent methionine synthase and isolation of a tryptic fragment containing the cobalamin-binding domain. J Biol Chem 264:13888–13895 [PubMed] [Google Scholar]
- Carmel R, Jacobsen DW (eds) (2001) Homocysteine in health and disease. Cambridge University Press, Cambridge [Google Scholar]
- Chen J, Stampfer MJ, Ma J, Selhub J, Malinow MR, Hennekens CH, Hunter DJ (2001) Influence of a methionine synthase (D919G) polymorphism on plasma homocysteine and folate levels and relation to risk of myocardial infarction. Atherosclerosis 154:667–672 [DOI] [PubMed] [Google Scholar]
- Chen LH, Liu M, Hwang H, Chen L, Korenberg J, Shane B (1997) Human methionine synthase: cDNA cloning, gene localization and expression. J Biol Chem 272:3628–3634 [PubMed] [Google Scholar]
- Christensen B, Arbour L, Tran P, Leclerc D, Sabbaghian N, Platt R, Gilfix BM, Rosenblatt DS, Gravel RA, Forbes P, Rozen R (1999) Genetic polymorphisms in methylenetetrahydrofolate reductase and methionine synthase, folate levels in red blood cells, and risk of neural tube defects. Am J Med Genet 84:151–157 [DOI] [PubMed] [Google Scholar]
- Christensen B, Rosenblatt DS (1995) Effects of folate deficiency on embryonic development. In: Wickramasinghe SN (ed) Baillière's clinical haematology: megaloblastic anemia. Baillière Tindall, London, pp 617–638 [DOI] [PubMed] [Google Scholar]
- Dekau V, Gudnason V, Hawe E, Miller GJ, Stansbie D, Humphries SE (2001) Gene-environment and gene-gene interaction in the determination of plasma homocysteine levels in healthy middle-aged men. Thromb Haemost 85:67–74 [PubMed] [Google Scholar]
- Dixon MM, Huang S, Matthews RG, Ludwig M (1996) The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure 4:1263–1275 [DOI] [PubMed] [Google Scholar]
- Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML (1994) How a protein binds B12: a 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science 266:1669–1674 [DOI] [PubMed] [Google Scholar]
- Drummond JT, Huang S, Blumenthal RM, Matthews RG (1993) Assignment of enzymatic function to specific protein regions of cobalamin-dependent methionine synthase from Escherichia coli. Biochemistry 32:9290–9295 [DOI] [PubMed] [Google Scholar]
- Goulding CW, Postigo D, Matthews RG (1997) Cobalamin-dependent methionine synthase is a modular protein with distinct regions for binding homocysteine, methyltetrahydrofolate, and adenosylmethionine. Biochemistry 36:8082–8091 [DOI] [PubMed] [Google Scholar]
- Gulati S, Baker P, Li YN, Fowler B, Kruger WD, Brody LC, Banerjee R (1996) Defects in human methionine synthase in cblG patients. Hum Mol Genet Suppl 5:1859–1865 [DOI] [PubMed] [Google Scholar]
- Hyndman ME, Bridge PJ, Warnica JW, Fick G, Parsons HG (2000) Effect of heterozygosity for the methionine synthase 2756 A→G mutation on the risk for recurrent cardiovascular events. Am J Cardiol 86:1144–1146 [DOI] [PubMed] [Google Scholar]
- James SJ, Pogribna M, Pogribny II, Melnyk S, Hine RJ, Gibson JB, Yi P, Tafoya DL, Swenson DH, Wilson VL, Gaylor DW (1999) Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase gene may be maternal risk factors for Down syndrome. Am J Clin Nutr 70:495–501 [DOI] [PubMed] [Google Scholar]
- Kim Y-I (1999) Folate and cancer prevention: a new medical application of folate beyond hyperhomocysteinemia and neural tube defects. Nutr Rev 57:314–321 [DOI] [PubMed] [Google Scholar]
- Leclerc D, Campeau E, Goyette P, Adjalla CE, Christensen B, Ross M, Eydoux P, Rosenblatt DS, Rozen R, Gravel RA (1996) Human methionine synthase: cDNA cloning and identification of mutations in patients of the cblG complementation group of folate/cobalamin disorders. Hum Mol Genet 5:1867–1874 [DOI] [PubMed] [Google Scholar]
- Leclerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D, Heng HHQ, Rommens JM, Scherer SW, Rosenblatt DS, Gravel RA (1998) Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc Natl Acad Sci USA 95:3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YN, Gulati S, Baker PJ, Brody LC, Banerjee R, Kruger WD (1996) Cloning, mapping and RNA analysis of the human methionine synthase gene. Hum Mol Genet 5:1851–1858 [DOI] [PubMed] [Google Scholar]
- Matthews RG (1999) Cobalamin-dependent methionine synthase. In: Banerjee R (ed) Chemistry and biochemistry of B12. John Wiley & Sons, New York, pp 681–706 [Google Scholar]
- Matthews RG, Ludwig ML (2001) Microbial modeling of human disease: homocysteine metabolism. In: Carmel R, Jacobsen DW (eds) Homocysteine in health and disease. Cambridge University Press, Cambridge, pp 100–112 [Google Scholar]
- Morita H, Kurihara H, Sugiyama T, Hamada C, Kurihara Y, Shindo T, Oh-hashi Y, Yazaki Y (1999) Polymorphism of the methionine synthase gene: association with homocysteine metabolism and late-onset vascular diseases in the Japanese population. Arterioscler Thromb Vasc Biol 19:298–302 [DOI] [PubMed] [Google Scholar]
- Olteanu H, Banerjee R (2001) Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J Biol Chem 276:35558–35563 [DOI] [PubMed] [Google Scholar]
- Rosenblatt D, Fenton WA (2001) Inherited disorders of folate and cobalamin transport and metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B (eds) The metabolic and molecular bases of inherited disease, 8th ed. McGraw-Hill, New York, pp 3897–3933 [Google Scholar]
- Rozen R (2001) Polymorphisms of folate and cobalamin metabolism. In: Carmel R, Jacobsen DW (eds) Homocysteine in health and disease. University of Cambridge Press, Cambridge, pp 259–269 [Google Scholar]
- Shaw GM, Todoroff K, Finnell RH, Lammer EJ, Leclerc D, Gravel RA, Rozen R (1999) Infant methionine synthase variants and risk for spina bifida. J Med Genet 36:86–87 [PMC free article] [PubMed] [Google Scholar]
- Suormala ST, Baumgartner ER, Fowler B (2001) Characterization of methylmalonic aciduria and/or hyperhomocysteinemia: studies in cultured fibroblasts. J Inher Metab Dis 24:55 [Google Scholar]
- Tang B, Li YN, Kruger WD (2000) Defects in methylthioadenosine phosphorylase are associated with but not responsible for methionine-dependent tumor cell growth. Cancer Res 60:5543–5547 [PubMed] [Google Scholar]
- Templeton AR, Clark AG, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF (2000) Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am J Hum Genet 66:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueland PM, Nygard O, Vollset SE, Refsum H (2001) The Hordaland homocysteine studies. Lipids 36:S33–S39 [DOI] [PubMed] [Google Scholar]
- Wang XL, Duarte N, Cai H, Adachi T, Sim AS, Cranney G, Wilcken DEL (1999) Relationship between total plasma homocysteine, polymorphisms of homocysteine metabolism related enzymes, risk factors and coronary artery disease in the Australian hospital-based population. Atherosclerosis 146:133–140 [DOI] [PubMed] [Google Scholar]
- Watkins D, Rosenblatt DS (1989) Functional methionine synthase deficiency cblE and cblG): clinical and biochemical heterogeneity. Am J Med Genet 34:427–434 [DOI] [PubMed] [Google Scholar]
- Wilson A, Leclerc D, Saberi F, Phillips JA III, Rosenblatt DS, Gravel RA (1998) Functionally null mutations in patients with the cblG variant form of methionine synthase deficiency. Am J Hum Genet 63:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WY, Eskes TKAB, Kuijpers-Jagtman A-M, Sprauwen PHM, Steegers EAP, Thomas CMG, Hamel BCJ, Blom HJ, Steegers-Theunissen RPM (1999) Non-syndromic orofacial clefts: association with maternal hyperhomocysteinemia. Teratology 60:253–257 [DOI] [PubMed] [Google Scholar]
- Yamada K, Tobimatsu T, Toraya T (1998) Cloning, sequencing, and heterologous expression of rat methionine synthase cDNA. Biosci Biotechnol Biochem 62:2155–2160 [DOI] [PubMed] [Google Scholar]