Abstract
Rothmund-Thomson syndrome (RTS) is an autosomal recessive disorder caused by deleterious mutations in the RECQL4 gene on chromosome 8. The RECQL4 gene structure is unusual because it contains many small introns <100 bp. We describe a proband with RTS who has a novel 11-bp intronic deletion, and we show that this mutation results in a 66-bp intron too small for proper splicing. Constraint on intron size may represent a general mutational mechanism, since human-genome analysis reveals that ∼15% of genes have introns <100 bp and are therefore susceptible to size constraint. Thus, monitoring of intron size may allow detection of mutations missed by exon-by-exon approaches.
Rothmund-Thomson syndrome (RTS [MIM #268400]) is an autosomal recessive disorder characterized by a poikilodermatous rash, skeletal anomalies, small stature, alopecia, cataracts, and strong predisposition to malignancy, particularly osteosarcoma (Vennos et al. 1992; Wang et al. 2001). Truncating mutations in the RECQL4 gene (MIM #603780) have been previously reported in four kindreds with RTS (Kitao et al. 1999b; Lindor et al. 2000; Wang et al. 2000).
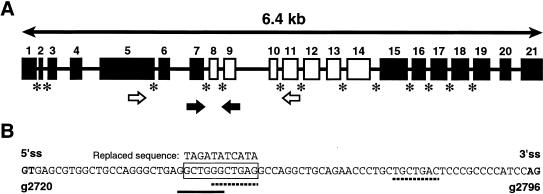
The RECQL4 gene structure is unusual because it contains 21 exons in only 6 kb of genomic DNA sequence (Kitao et al. 1999a). Unlike most human genes, which have large introns, RECQL4 has 13 introns that are <100 bp in length (fig. 1A). Introns less than ∼80 bp have been shown to splice inefficiently in model systems, despite the presence of consensus splice-donor and -acceptor sites and branch-point sites (van Santen and Spritz 1985; Berget 1995).
Figure 1.
Intronic deletion in a proband with RTS. A, Schematic diagram of RECQL4 gene (adapted from Kitao et al. 1999a). The helicase region is unshaded. Asterisks indicate introns <100 bp. B, Sequence of intron 8, showing 11-bp deletion in FCP-102 (boxed), potential branch points (dotted line), and potential intronic splicing enhancer (underline). The sequence used to create the “replaced construct” is shown above the deletion.
One of our probands with RTS (individual FCP-102), who had osteosarcoma and was from a consanguineous Mexican family, was found to be homozygous for an 11-bp deletion (g2746del11) in intron 8 (fig. 1B). A 10-bp direct repeat flanks the deleted bases. This intronic deletion was not found in 100 population control subjects, including 33 Hispanic control subjects, and sequencing of the RECQL4 coding region in FCP-102 revealed no other truncating mutations. The deletion (intronic base-pair position 25–35) reduced the 77-bp intron to 66 bp in length, leaving the 3′ and 5′ consensus splice sites intact. Within the deleted sequence there is a potential branch-point site (44 bp upstream of the 3′ splice site) with a partial match to the YNYURAY branch-point consensus sequence (Li and Pritchard 2000). However, 20 bp downstream, in a region of the intron not affected by the deletion, there is another sequence, with a perfect match to the branch-point consensus sequence. The deleted sequence (GCTGGGCTGAG) does contain a sequence that matches the consensus G2(X1-4)G3 for an intronic splicing enhancer (ISE) (McCarthy and Phillips 1998).
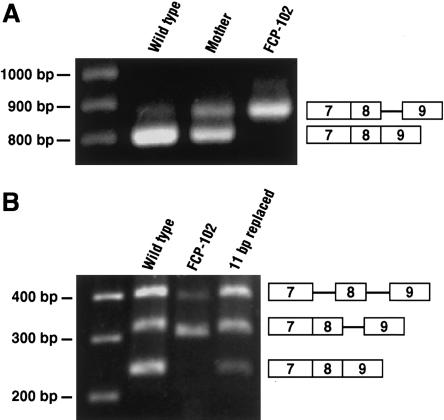
We hypothesized that this deletion represents the disease-causing mutation because it reduces the intron to a size incompatible with proper splicing. RT-PCR using primers within exons 5 and 11 would be expected to yield a product of 791 bp, as was seen in analysis of a wild-type control sample (fig. 2A). In contrast, RT-PCR of RNA from lymphoblasts and fibroblasts of FCP-102 demonstrated a predominant, larger aberrant product, of 857 bp. The proband’s mother is heterozygous for the deletion and has both the expected 791-bp RT-PCR product and the aberrant product. Cloning and sequencing of the major RT-PCR product in FCP-102 showed retention of intron 8 containing the 11-bp deletion. Analysis of other cloned RT-PCR products from FCP-102 revealed a 750-bp product that retained intron 8 but that utilized a cryptic 5′ splice site in exon 9. Both of these RNA species are predicted to generate a premature stop codon. The major RT-PCR product from wild-type RNA showed proper splicing of intron 8. Additional bands resulting from misspliced products were also seen in analysis of RNA from wild-type control subjects and were found to retain intron 8. However, these were not the predominant bands, and, conversely, multiple RT-PCRs performed with FCP-102 RNA never demonstrated proper splicing of intron 8.
Figure 2.
RT-PCR showing both splicing error due to intronic deletion and correction of defect by restoration of intron size. A, RT-PCR of lymphoblast RNA from proband FCP-102, his mother, and normal control. Primers are within exons 5 and 11 (between the white arrows in fig. 1A). The normal product is expected to be 791 bp. The aberrant larger product is 857 bp. B, RT-PCR of transcripts from NIH3T3 cells transfected with expression constructs containing exons 5–10 of RECQL4 from wild type, FCP-102, or wild type replaced with 11 unrelated base pairs at the deletion site. Primers are within exons 7 and 9 (between the black arrows in fig. 1A). Predicted sizes are 240 bp, for correct splicing of intron 8; 306 bp, for retention of intron 8 containing the deletion; and 317 bp, for the retention of wild-type intron 8. The highest band in each lane represents retention of introns 7 and 8 (389 bp for FCP-102 and 400 bp for wild type and the replaced construct).
To test if intronic size constraint could be responsible for this missplicing, we subcloned exons 5–10, from wild-type and FCP-102 genomic DNA, into a mammalian expression vector (pCDNA3.1/V5-His-TOPO; Invitrogen). Murine NIH3T3 cells were transiently transfected with the constructs. RNA was isolated from cytoplasmic lysates, and DNAse I was treated to minimize contamination by unprocessed transcripts and transfected DNA. RT-PCR using primers specific for the human gene revealed bands representing the proper splicing of intron 8 in RNA from the construct with the wild-type sequence and representing aberrant splicing of intron 8 in RNA from the FCP-102 construct, recapitulating the pattern seen above for the endogenous genes (fig. 2B). We then used site-directed mutagenesis to restore a normal-size intron, by replacing the 11-bp deletion (GCTGGGCTGAG) with an unrelated sequence (TAGATATCATA). The replaced sequence does not contain the consensus for either a branch-point site or an intronic splicing enhancer (Li and Pritchard 2000; McCarthy and Phillips 1998). Transfection and RT-PCR demonstrated restoration of splicing across intron 8 in RNA derived from the replaced 11-bp construct (fig. 2B). Correction of the missplicing defect suggests that intronic size constraint is a mutational mechanism leading to the RTS phenotype in this patient. The ratio of RT-PCR products as shown in figure 2B was observed to remain in the linear range, in 18–30 cycles of PCR (data not shown). The relative amount of correctly spliced product is less in the replaced construct than in the wild type. This may be due to loss of the ISE sequence found in the deleted sequence. However, the fact that correct splicing of intron 8 occurs in the replaced construct when length is restored without recreating an ISE argues that size constraint contributes to the missplicing defect in this patient.
The higher-molecular-weight bands seen in RT-PCR from all three transfectants are misspliced products (that retain introns), as was seen in analysis of endogenous RNA described above. Missplicing of a proportion of wild-type templates presumably occurs because the small introns in RECQL4 are inefficiently spliced.
Our results illustrate that deletions in small introns can be deleterious, because of a constraint on intron size. This has been previously suggested in one case of adult polycystic kidney disease (Peral et al. 1995). These results contrast with a recently reported large intronic deletion in WNKI, which results in increased expression of the protein in pseudohyperaldosteronism type II (Wilson et al. 2001). However, in this case, the resulting intron remains several hundred base pairs in length. Intronic deletions in small introns are likely to be important for other disease genes as well. Analysis of the April 2001 version of the Human Genome Assembly (Lander et al. 2001; also see the UCSC Human Genome Project Working Draft Web site) reveals that 15% of unique mRNAs have at least one intron that is <100 bp. Of these genes with small introns, 48% are located in finished sequence, demonstrating that this is not an artifact of the genomic draft assembly. Thus, full mutational analysis should include assay for deletions in small introns, which, otherwise, may be missed in an exon-by-exon approach that surveys only intron/exon boundaries.
Acknowledgments
We thank the family members reported herein for their willing participation in our research protocol, and we thank Sue Berget and Andrew McCullough for their advice and many helpful discussions. L.L.W. was supported by an American Society of Clinical Oncology Young Investigator Award and by the Children’s Cancer Research Fund.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RTS [MIM #268400] and RECQL4 [MIM #603780])
- UCSC Human Genome Project Working Draft, http://genome.ucsc.edu
References
- Berget SM (1995) Exon recognition in vertebrate splicing. J Biol Chem 270:2411–2414 [DOI] [PubMed] [Google Scholar]
- Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A (1999a) Rothmund-Thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics 61:268–276 [DOI] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y (1999b) Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet 22:82–84 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Li M, Pritchard H (2000) Characterization of the effects of mutations in the putative branchpoint sequence of intron 4 on the splicing within the human lecithin:cholesterol acyltransferase gene. J Biol Chem 275:18079–18084 [DOI] [PubMed] [Google Scholar]
- Lindor NM, Furuichi Y, Kitao S, Shimamoto A, Arndt C, Jalal S (2000) Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am J Med Genet 90:223–228 [DOI] [PubMed] [Google Scholar]
- McCarthy EMS, Phillips JA (1998) Characterization of an intron splice enhancer that regulates alternative splicing of human GH pre-mRNA. Hum Mol Genet 7:1491–1496 [DOI] [PubMed] [Google Scholar]
- Peral B, Gamble V, San Millan JL, Strong C, Sloane-Stanley J, Moreno F, Harris PC (1995) Splicing mutations of the polycystic kidney disease 1 (PKD1) gene induced by intronic deletion. Hum Mol Genet 4:569–574 [DOI] [PubMed] [Google Scholar]
- van Santen VL, Spritz RA (1985) mRNA precursor splicing in vivo: sequence requirements determined by deletion analysis of an intervening sequence. Proc Natl Acad Sci USA 82:2885–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennos EM, Collins M, James WD (1992) Rothmund-Thomson syndrome: review of the world literature. J Am Acad Dermatol 27:750–762 [DOI] [PubMed] [Google Scholar]
- Wang LL, Levy ML, Lewis RA, Chintagumpala MM, Lev D, Rogers M, Plon SE (2001) Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Med Genet 102:11–17 [DOI] [PubMed] [Google Scholar]
- Wang LL, Levy ML, Lewis RA, Gannavarapu A, Stockton D, Lev D, Cunniff CM, Plon SE (2000) Evidence for genetic heterogeneity in Rothmund-Thomson syndrome. Am J Hum Genet 67 Suppl 2:376 [Google Scholar]
- Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, et al (2001) Human hypertension caused by mutations in WNK kinases. Science 293:1107–1112 [DOI] [PubMed] [Google Scholar]