Abstract
We have identified a splice-site mutation (IVS6+1G→T) in the RB1 gene, in two unrelated families with incomplete-penetrance retinoblastoma. Analysis of RNA from white blood cells showed that this mutation causes skipping of exon 6. Although this deletion results in a frameshift, most carriers of the mutation did not develop retinoblastoma. Interestingly, the relative abundance of the resultant nonsense messenger RNA varies between members of the same family and is either similar to or considerably lower than the transcript level of the normal allele. Moreover, variation of relative transcript levels is associated with both the sex of the parent that transmitted the mutant allele and phenotypic expression: All eight carriers with similar abundance of nonsense and normal transcript have received the mutant allele from their mother, and only one of them has developed retinoblastoma; by contrast, all eight carriers with reduced abundance of the nonsense transcript have received the mutant allele from their father, and all but two them have retinoblastoma. After treatment with cycloheximide, the relative abundance of transcripts from paternally inherited mutant alleles was partly restored, thus indicating that posttranscriptional mechanisms, rather than transcriptional silencing, are responsible for low levels of mutant messenger RNA. Our data suggest that a specific RB1 mutation can be associated with differential penetrance, on the basis of the sex of the transmitting parent.
Hereditary predisposition to retinoblastoma (MIM 180200) is caused by oncogenic germline mutations in the RB1 gene (Locus Link accession number L11910). Heterozygous carriers can show variable phenotypic expression. This is to be expected, because formation of a tumor focus depends on a chance second mutation, which is a rare event that follows a Poisson distribution (Knudson 1971). Actually, analysis of phenotypic variation within families has shown that the ratio of mutation carriers with bilateral tumors, with unilateral tumors, and without tumors complies with the predictions from such a stochastic model (Lohmann et al. 1994). In most families with retinoblastoma, penetrance is complete, and almost all mutation carriers develop tumors in both eyes. Typically, retinoblastoma predisposition in these families is caused by RB1 mutations that cause premature termination codons in any but the ultimate and penultimate exons of this gene (Lohmann et al. 1996). Analysis of constitutional cells from individuals who are heterozygous for mutations of this kind has shown that transcripts from the mutant allele are considerably less abundant than are those from the normal allele (Dunn et al. 1989). This decreased abundance parallels findings in other genes and is most probably due to nonsense-mediated decay, which is a surveillance mechanism that specifically degrades mutant mRNAs with premature termination codons >50 nt upstream from the ultimate exon-exon junction (Culbertson 1999; Frischmeyer and Dietz 1999; Hentze and Kulozik 1999). Only a few families with retinoblastoma show incomplete penetrance. Most of these families show distinct RB1 mutations that do not result in premature termination codons (Sakai et al. 1991; Onadim et al. 1992; Lohmann et al. 1994; Bremner et al. 1997; Otterson et al. 1997). Here we report an RB1 splice mutation that, although resulting in a nonsense transcript, is associated with incomplete penetrance in two unrelated families with retinoblastoma. Moreover, in both families, this mutation is associated with differential penetrance, on the basis of the sex of the transmitting parent.
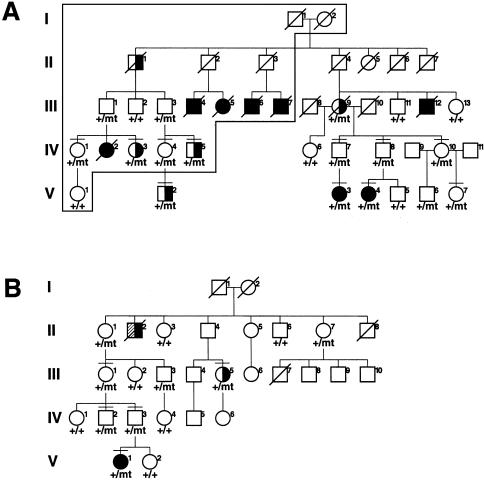
Predictive testing for retinoblastoma predisposition was requested by relatives of a patient with bilateral retinoblastoma (G162 V-4; fig. 1A) whose mutation had been identified in a previous study (Lohmann et al. 1996). Thereby, we learned that retinoblastoma also occurred in a distant branch of her family. Our study was approved by the medical ethics committee of the Universität Essen, and appropriate informed consent was obtained from all human subjects and/or their parents. Molecular analyses including members from this sibship showed the same single-base substitution—namely, g.45867G→T (also known as “IVS6+1G→T”)—with the identical haplotype background, at linked polymorphic loci. Overall, the family comprises 8 and 5 patients with bilateral and unilateral retinoblastoma, respectively; moreover, ⩾12 heterozygous carriers in this family have not developed retinoblastoma. We identified the same mutation with a different haplotype background (at D13S153, RB1.20 [Yandell and Dryja 1989], and D13S1307), in a patient from an unrelated family (M12487 V-1; fig. 1B) who had bilateral retinoblastoma. At the time of diagnosis, she had, in both eyes, more than four tumor foci, a number that is typical for patients from families with complete penetrance. However, mutation testing and pedigree analysis revealed at least eight nonpenetrant carriers in her family.
Figure 1.
Pedigrees and segregation of IVS6+1G→T. Blackened symbols denote patients with bilateral retinoblastoma; half-blackened symbols denote patients with unilateral retinoblastoma. Plus signs (+) indicate presence of IVS6+1G; “mt” indicates presence of IVS6+1T; horizontal bar indicates individuals investigated by RNA analysis. A, Family G162. The frame encloses the branch of this family that has been described elsewhere (Czeizel and Gardonyi 1974). B, Family M12487. Individual II-2 died of retinoblastoma and may have had bilateral tumors (half-crosshatched, half-blackened symbol).
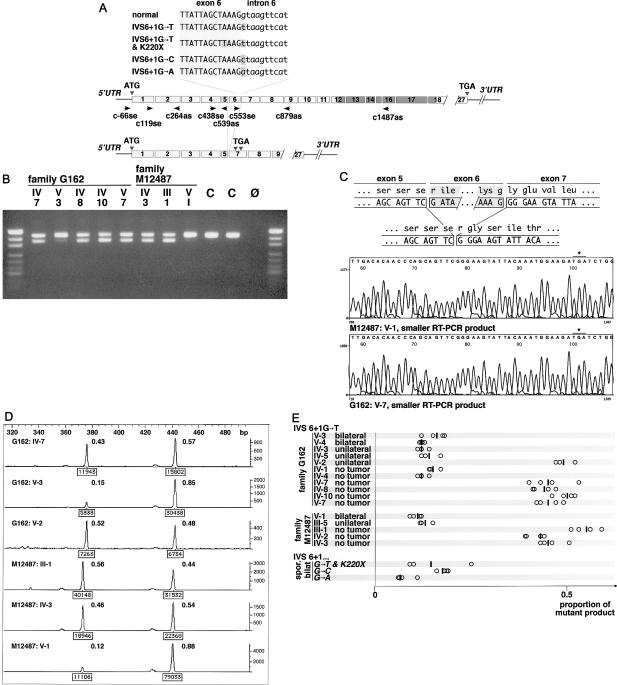
The base substitution that both families share alters position 1 of a GT-AG intron. The most likely effect of this mutation is the skipping of exon 6, which comprises 68 bp, and this mutation results in a frameshift and premature termination codons (the first is expected to be at codon 188 in exon 7). Similar RB1 mutations are associated with complete penetrance and bilateral retinoblastoma. Actually, we know of five unrelated patients with different base substitutions (IVS6+1G→C and IVS6+1G→A; fig. 2A) at the same nucleotide, and all of those patients have sporadic bilateral retinoblastoma (Lohmann et al. 1996 and unpublished data). In light of this, it is astounding that most carriers of the IVS6+1G→T allele developed no tumor. To account for this, we surmised that this mutation might not result in a frameshift and premature termination, possibly because of frame-restoring alternative splicing. To test this, we extracted total RNA from peripheral-blood leukocytes from mutation carriers with (G162 V-3 and M12487 V-1) and without (G162 IV-7, G162 IV-8, G162 IV-10, M12487 III-1, and M12487 IV-3) tumors, as well as from normal controls. We performed RT-PCR with primers c438se and c1487as followed by PCR with primers c438se and c879as (fig. 2A). Agarose gel electrophoresis of PCR products from carriers with retinoblastoma showed a DNA band of the expected (i.e., normal) length and, with considerably less intensity, a smaller product (fig. 2B). In mutation carriers without tumors, the normal and smaller bands had equal intensity. Normal controls showed the normal-length band only. To determine the sequence of these products, we performed RT-PCR, with primers c438se and c1487as, on RNA from mutation carriers with and without tumors (M12487 V-1 and G162 V-7, respectively) and one normal control. To improve separation, we digested the products with EcoRI prior to electrophoresis. DNA was recovered from agarose slices containing individual bands, was amplified by PCR with primers c438se and c879as, and was sequenced. All normal-length products showed a normal sequence; all smaller products from mutation carriers showed the same sequence but without the 68 bp corresponding to exon 6 (fig. 2C). The first in-frame stop codon occurs 23 bp downstream from the exons 5–7 junction. No frame-restoring alteration upstream from the skipped exon was identified by sequencing of RT-PCR products obtained, with primers c−66se and c879as, from RNA from the healthy carrier (M12487 IV-3). To further investigate the differences in the proportion of the mutant transcript between mutation carriers with and without tumors, we obtained leukocyte RNA from additional carriers of the IVS6+1G→T mutation and from patients with similar mutations (i.e., IVS6+1G→C, IVS6+1G→A, and a complex substitution comprising IVS6+1G→T and K220X; fig. 2A). We performed RT-PCR with primers c438se and c879as including a fluorescent label (i.e., FAM) and analyzed the products on a 310 Genetic Analyzer by use of Genescan and Genotyper software (Applied Biosystems) (fig. 2D). The relative abundance of the mutant mRNA showed a bimodal distribution (fig. 2E): In 8 of 16 carriers of the IVS6+1G→T mutation and all patients with similar mutations, the relative proportion of mutant products was small (mean peak integral of mutant products ± SD was 13%±4% of grand total); in the other 8 carriers, mutant and normal products had about the same intensity (mean peak integral of mutant products ± SD was 47%±5% of grand total). Intriguingly, all eight carriers with similar abundance of nonsense and normal transcript received the mutant allele from their mother, whereas all eight carriers with reduced relative abundance of the nonsense transcript received the mutant allele from their father.
Figure 2.
Results of RNA analysis. A, Genomic organization of RB1. Boxes indicate exons; arrowheads indicate location and orientation of primers that were used for RT-PCR and sequencing. B, Agarose gel electrophoresis of products obtained by RT-PCR with primers c438se and c1487as followed by PCR with primers c438se and c879as. “C” denotes normal control; “Ø” denotes a negative control. The DNA standard used was pUC19/MspI. C, Result of sequence analysis of the smaller products from M12487 V-1 and G162 V-7. The first premature termination codon is created 23 bp downstream from the exons 5–7 junction and is indicated by an asterisk (*). D, Results of quantitative analysis of fluorescent RT-PCR products obtained with primers c438se and c879as. Boxed numeric values below peaks are the peak integrals as determined by the Genotyper software; the ratio of individual peak integrals to the grand total is shown to the right of each peak. E, Graph with summary of results of fluorescent RT-PCR. Each circle indicates the ratio of the mutant peak integral to the grand total and represents the result of a single RT-PCR; vertical bars indicate the arithmetic means.
Reduced abundance of nonsense transcripts is not an unusual finding in heterozygous carriers of mutant alleles. In many genes, this phenomenon was found to be caused by nonsense-mediated decay, which is a posttranscriptional surveillance mechanism (Culbertson 1999; Frischmeyer and Dietz 1999; Hentze and Kulozik 1999). To examine if posttranscriptional mechanisms account for the decreased abundance of the mutant mRNA, we obtained lymphoblastoid cell lines with paternally derived mutant alleles (from G162 V-3 and M12487 V-1) and a cell line with a maternally derived mutant allele (from M12487 IV-3) and incubated them, prior to RNA extraction, with cycloheximide, which is an inhibitor of nonsense-mediated decay (Carter et al. 1995). In parallel experiments, cell lines were harvested after incubation at concentrations of 100 μg/ml for 6 h and 8 h, at 500 μg/ml for 6 h, and without exposure to cycloheximide. We performed fluorescent RT-PCR with primers c438se and c879as and found that, compared to untreated cell lines, the relative abundance of mutant mRNA was increased in all cell lines that were exposed to cycloheximide. (For G162 V-3, abundance in untreated cell lines was 13%, and abundance ± SD in treated cell lines was 45%±1.5%; for M12487 V-1, abundance in untreated cell lines was 7.4%, and abundance ± SD in treated cell lines was 27%±2.5%; for M12487 IV-3, abundance in untreated cell lines was 28%, and abundance ± SD in treated cell lines was 76%±1% [all abundances are given as % grand total].) This suggests that the markedly low relative abundance of nonsense mRNA that is transcribed from paternally inherited alleles is—at least in part—due to posttranscriptional mechanisms. After cycloheximide treatment, the relative level of nonsense mRNA from maternally inherited mutant alleles was also increased, thus indicating that these transcripts are also subject to RNA surveillance.
We observed that relative transcript abundance and parental origin are also associated with penetrance: Only one of the eight carriers with similar abundance of nonsense and normal transcript developed retinoblastoma, whereas, among the eight carriers with reduced abundance, all but two are affected. The link between parental origin and penetrance is still valid if family members from whom RNA was not obtained are included. By pedigree analysis, the parental origin of the mutation can be resolved for 27 family members (14 patients with retinoblastoma and 13 unaffected mutation carriers). In 13 of 14 patients with retinoblastoma but in only 4 of 13 unaffected carriers, the mutant allele is of paternal origin. This is a very unlikely (P<.002, by Fisher's exact-probability test) observation, if the null hypothesis of no differential penetrance is assumed. Statistical significance (P<.00002, by Fisher's exact-probability test) is even higher when evaluating the diseased-eye ratio, which is a parameter that takes into account both penetrance and expressivity (Lohmann et al. 1994): The 17 carriers of paternally inherited mutant alleles have 34 eyes (22 affected and 12 without tumor); among the 10 carriers of maternally inherited mutant alleles, only 1 of 20 eyes is affected.
The association between incomplete penetrance of retinoblastoma and the presence of nonsense transcripts suggests that this mutant mRNA may have a residual function. It has been suggested that translation initiation at internal AUG codons may modulate disease phenotypes (Chang and Gould 1998). Intriguingly, N-terminal truncated retinoblastoma proteins (pRbs)—including p56RB, which may result from translation initiation at codon 379 (located 528 bp downstream from the deletion in the mutant transcript)—may be constitutively active and thus may have strong growth-suppression potential (Hamel et al. 1992; Antelman et al. 1997). To identify N-terminal truncated pRbs that result from translation initiation at internal AUG codons, we analyzed cytoplasmic and nuclear extracts from lymphoblastoid cell lines from mutation carriers with (G162 V-3) and without (G162 IV-7 and G162 IV-10) tumors. Western blot analyses were performed using a mouse monoclonal anti-human pRb antibody, G3-245 (recognizing an epitope located at amino acids 332–344 [exon 10]; PharMingen). These experiments demonstrated the presence of wild type but not of a truncated form of pRb. We used a second mouse monoclonal anti-pRb antibody, IF8 (epitope encoded by exons 21–27; Santa Cruz Biotechnology), but again did not detect any N-terminal truncated pRb (data not shown).
To our knowledge, differential penetrance of retinoblastoma, on the basis of the sex of the transmitting parent, has not previously been reported in offspring. In particular, this inheritance pattern was not observed in published analyses of parent-of-origin effects in retinoblastoma (Munier et al. 1992; Naumova and Sapienza 1994; Seminara and Dryja 1994), thus indicating that it is not apparent in retinoblastoma as a whole (also see the Imprinted Gene Catalogue Web site). Nevertheless, it may be restricted to some rare families. Surveying the literature, we found that an extended family that was part of Macklin's (1960) “Study of Retinoblastoma in Ohio” also shows differential penetrance: All nine patients with retinoblastoma received the mutant allele via the paternal germline (see fig. 4 of Macklin 1960). Referring to this family, Macklin noted that “the degree of penetrance varies widely not only in different families but in different sibships in the same family” (1960, p. 32). In fact, variation between sibships in the same family is to be expected if penetrance varies based on the sex of the transmitting parent.
The inheritance pattern in both the family described by Macklin (1960) and the families described herein is similar to that in families with hereditary glomus tumors and suggests the effect of genomic imprinting (van der Mey et al. 1989). However, the SDHD gene, which is mutated in patients with hereditary glomus tumors, did not show parent-of-origin–specific expression in any of the normal tissues that were investigated (Baysal et al. 2000). To date, there is no model that clearly explains the inheritance pattern for hereditary glomus tumors (Taschner et al. 2001). Likewise, RB1 is not known to be imprinted (Morison et al. 2001). The families with the IVS6+1G→T mutation are no exception, because the effect of cycloheximide treatment suggests that posttranscriptional mechanisms, rather than transcriptional silencing, accounts for the low relative abundance of transcripts from paternally inherited mutant alleles.
It has been reported that loci in the immediate vicinity of RB1 show parent-of-origin effects (Kato et al. 1996; Bhattacharyya et al. 2000). It is tempting to speculate that the unusual inheritance pattern is not a consequence of the IVS6+1G→T mutation but is due to allelic association with a mutation at an imprinted locus in cis. The example of the callipyge phenotype in sheep shows that a mutation can modify the expression of genes in cis without altering their imprinting status (Charlier et al. 2001). However, since the IVS6+1G→T in the two families are in phase with distinct alleles at linked polymorphic loci, they are not identical by descent, and it is unlikely that they share a mutation in cis, unless this is a relatively frequent variant.
It is intriguing that we observed the unusual inheritance pattern only in association with the IVS6+1G→T mutation, whereas several carriers with other substitutions at the same nucleotide show sporadic bilateral retinoblastoma. It must be noted, however, that most new germline mutations in RB1 arise on the paternally derived chromosome (Dryja et al. 1997 and literature cited therein); consequently, there is a bias against the observation of new mutations on maternally derived chromosomes. Therefore, a parent-of-origin effect that is associated with other mutations in RB1 may have gone undetected.
Acknowledgments
We thank the families, for their cooperation; Dr. N. Bornfeld and Dr. A. Schüler, for referral of patients; U. Röhrig and S. Plambeck, for establishing and maintaining lymphoblastoid cell lines; H. Hirche and J. Hüsing, for discussion of statistical issues; and Dr. B. Horsthemke, for discussion and continuous support. We are also indebted to Dr. L. Timár, from the Genetic Counseling Clinic at the National Center of Public Health, Budapest, Hungary. He has helped us to verify phenotypic information and to obtain informed consent and samples from the Hungarian branch of family G162. Parts of this work were funded by a grant of the Deutsche Forschungsgemeinschaft (KFO 109/1-1).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Imprinted Gene Catalogue, http://cancer.otago.ac.nz/IGC/Web/home.html (for parent-of-origin effects associated with retinoblastoma)
- Locus Link, http://www.ncbi.nlm.nih.gov/LocusLink/ (for RB1 [accession number L11910])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for retinoblastoma [MIM 180200])
References
- Antelman D, Perry S, Hollingsworth R, Gregory RJ, Driscoll B, Fung YK, Bookstein R (1997) Engineered mutants of pRB with improved growth suppression potential. Oncogene 15:2855–2866 [DOI] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW III, Cornelisse CJ, Devilee P, Devlin B (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287:848–851 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Leaves NI, Wiltshire S, Cox R, Cookson WO (2000) A high-density genetic map of the chromosome 13q14 atopy locus. Genomics 70:286–291 [DOI] [PubMed] [Google Scholar]
- Bremner R, Du DC, Connolly-Wilson MJ, Bridge P, Ahmad KF, Mostachfi H, Rushlow D, Dunn JM, Gallie BL (1997) Deletion of RB exons 24 and 25 causes low-penetrance retinoblastoma. Am J Hum Genet 61:556–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270:28995–29003 [DOI] [PubMed] [Google Scholar]
- Chang CC, Gould SJ (1998) Phenotype-genotype relationships in complementation group 3 of the peroxisome-biogenesis disorders. Am J Hum Genet 63:1294–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Segers K, Karim L, Shay T, Gyapay G, Cockett N, Georges M (2001) The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet 27:367–369 [DOI] [PubMed] [Google Scholar]
- Culbertson MR (1999) RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet 15:74–80 [DOI] [PubMed] [Google Scholar]
- Czeizel A, Gardonyi J (1974) Retinoblastoma in Hungary, 1960–1968. Humangenetik 22:153–158 [PubMed] [Google Scholar]
- Dryja TP, Morrow JF, Rapaport JM (1997) Quantification of the paternal allele bias for new germline mutations in the retinoblastoma gene. Hum Genet 100:446–449 [DOI] [PubMed] [Google Scholar]
- Dunn JM, Phillips RA, Zhu X, Becker A, Gallie BL (1989) Mutations in the RB1 gene and their effects on transcription. Mol Cell Biol 9:4596–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8:1893–1900 [DOI] [PubMed] [Google Scholar]
- Hamel PA, Gallie BL, Phillips RA (1992) The retinoblastoma protein and cell cycle regulation. Trends Genet 8:180–185 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310 [DOI] [PubMed] [Google Scholar]
- Kato MV, Shimizu T, Nagayoshi M, Kaneko A, Sasaki MS, Ikawa Y (1996) Genomic imprinting of the human serotonin-receptor (HTR2) gene involved in development of retinoblastoma. Am J Hum Genet 59:1084–1090 [PMC free article] [PubMed] [Google Scholar]
- Knudson AG (1971) Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 68:820–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann DR, Brandt B, Höpping W, Passarge E, Horsthemke B (1994) Distinct RB1 gene mutations with low penetrance in hereditary retinoblastoma. Hum Genet 94:491–496 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Spectrum of RB1 germ-line mutations in hereditary retinoblastoma. Am J Hum Genet 58:940–949 [PMC free article] [PubMed] [Google Scholar]
- Macklin MT (1960) A study of retinoblastoma in Ohio. Am J Hum Genet 12:1–43 [PMC free article] [PubMed] [Google Scholar]
- Morison IM, Paton CJ, Cleverley SD (2001) The imprinted gene and parent-of-origin effect database. Nucleic Acids Res 29:275–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munier F, Spence MA, Pescia G, Balmer A, Gailloud C, Thonney F, van Melle G, Rutz HP (1992) Paternal selection favoring mutant alleles of the retinoblastoma susceptibility gene. Hum Genet 89:508–512 [DOI] [PubMed] [Google Scholar]
- Naumova A, Sapienza C (1994) The genetics of retinoblastoma, revisited. Am J Hum Genet 54:264–273 [PMC free article] [PubMed] [Google Scholar]
- Onadim Z, Hogg A, Baird PN, Cowell JK (1992) Oncogenic point mutations in exon 20 of the RB1 gene in families showing incomplete penetrance and mild expression of the retinoblastoma phenotype. Proc Natl Acad Sci USA 89:6177–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterson GA, Chen WD, Coxon AB, Khleif SN, Kaye FJ (1997) Incomplete penetrance of familial retinoblastoma linked to germ-line mutations that result in partial loss of RB function. Proc Natl Acad Sci USA 94:12036–12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Ohtani N, McGee TL, Robbins PD, Dryja TP (1991) Oncogenic germ-line mutations in Sp1 and ATF sites in the human retinoblastoma gene. Nature 353:83–86 [DOI] [PubMed] [Google Scholar]
- Seminara SB, Dryja TP (1994) Unbiased transmission of mutant alleles at the human retinoblastoma locus. Hum Genet 93:629–634 [DOI] [PubMed] [Google Scholar]
- Taschner PE, Jansen JC, Baysal BE, Bosch A, Rosenberg EH, Brocker-Vriends AH, van der Mey AG, van Ommen GJ, Cornelisse CJ, Devilee P (2001) Nearly all hereditary paragangliomas in the Netherlands are caused by two founder mutations in the SDHD gene. Genes Chromosomes Cancer 31:274–281 [DOI] [PubMed] [Google Scholar]
- van der Mey AG, Maaswinkel-Mooy PD, Cornelisse CJ, Schmidt PH, van de Kamp JJ (1989) Genomic imprinting in hereditary glomus tumours: evidence for new genetic theory. Lancet 2:1291–1294 [DOI] [PubMed] [Google Scholar]
- Yandell DW, Dryja TP (1989) Detection of DNA sequence polymorphisms by enzymatic amplification and direct genomic sequencing. Am J Hum Genet 45:547–555 [PMC free article] [PubMed] [Google Scholar]