Abstract
In the present study, we report a kindred with hearing loss, congenital heart defects, and posterior embryotoxon, segregating as autosomal dominant traits. Six of seven available affected patients manifested mild-to-severe combined hearing loss, predominantly affecting middle frequencies. Two patients were diagnosed with vestibular pathology. All patients had congenital heart defects, including tetralogy of Fallot, ventricular septal defect, or isolated peripheral pulmonic stenosis. No individual in this family met diagnostic criteria for any previously described clinical syndrome. A candidate-gene approach was undertaken and culminated in the identification of a novel Jagged 1 (JAG1) missense mutation (C234Y) in the first cysteine of the first epidermal-growth-factor–like repeat domain of the protein. JAG1 is a cell-surface ligand in the Notch signaling pathway. Mutations in JAG1 have been identified in patients with Alagille syndrome. Our findings revealed a unique phenotype with highly penetrant deafness, posterior embryotoxon, and congenital heart defects but with variable expressivity in a large kindred, which demonstrates that mutation in JAG1 can cause hearing loss.
Congenital heart disease is the most common birth defect, occurring in ∼1% of live births. Although multifactorial inheritance has been postulated for the majority of cases, single-gene transmission is demonstrated by the observation of mutations in NKX2.5 or Jagged 1 (JAG1 [MIM 601920]; GenBank accession number XM_056118), within families segregating congenital heart defects (Schott et al. 1998; Eldadah et al. 2001). JAG1 encodes a highly conserved ligand within the Notch family, which consists of components of an intercellular signaling pathway shown to be crucial for cell-fate decisions in organisms spanning the phylogenetic spectrum (Lindsell et al. 1996; Lissemore and Starmer 1999). JAG1 mRNA is abundantly expressed during development, and targeted disruption of Jag1 in mice results in vascular defects and embryonic lethality (Loomes et al. 1999; Xue et al. 1999). The JAG1 protein has several domains that show high interspecies conservation, including an N-terminal “DSL” motif found in the Delta, Serrate, and Lag-2 ligands in the Notch family; 16 tandemly repeated epidermal-growth-factor (EGF)–like domains; and a transmembrane region (Lindsell et al. 1995; Oda et al. 1997). Among the 16 EGF-like domains, the first and second ones have been shown to be important for the formation of a high-affinity complex with Notch (Shimizu et al. 1999). Mutations in JAG1 have been identified in ∼70% of patients with Alagille syndrome (AGS [MIM 118450]), a rare autosomal dominant condition (Krantz et al. 1998; Yuan et al. 1998; Crosnier et al. 1999, 2001; Onouchi et al. 1999; Pilia et al. 1999; Heritage et al. 2000; Colliton et al. 2001; Giannakudis et al. 2001; Spinner et al. 2001) that comprises three of these five main features: paucity of intrahepatic bile ducts, congenital heart defects (predominantly peripheral pulmonic stenosis), skeletal defects, ocular anomalies, and characteristic facies (Alagille et al. 1975; Krantz et al. 1997). The absence of phenotypic differences between total gene deletions and protein-truncating mutations suggests that haploinsufficiency is a pathogenic mechanism causing AGS (Krantz et al. 1998; Crosnier et al. 1999). To date, 26 unique missense mutations have been identified in JAG1, most of them leading to AGS. A missense mutation (G274D) in JAG1 has also been identified, in a kindred with right-heart obstructive disease and characteristic facies, that is distinct from that seen in AGS (Eldadah et al. 2001). In addition, two mouse mutants, Slalom and Headturner, showed that missense mutations (P269S and G289D, respectively) in the second EGF-like domain of the Jag1 gene lead to disturbances of the patterning of the organ of Corti in the inner ear (Kiernan et al. 2001; Tsai et al. 2001).
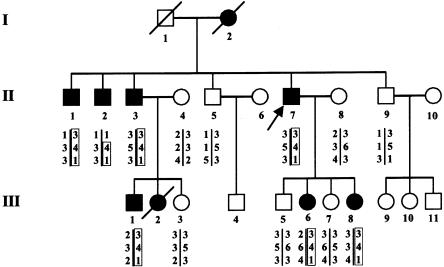
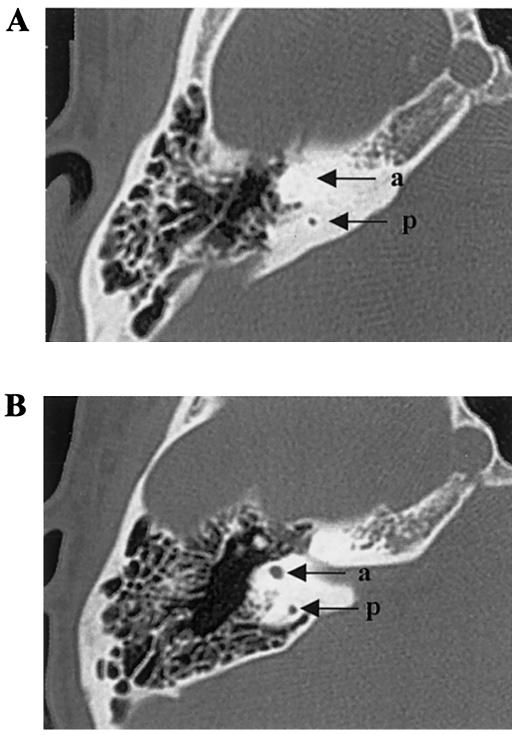
We studied a kindred (eight affected patients and seven unaffected individuals) with mild-to-severe combined deafness and congenital heart defects segregating as autosomal dominant traits (fig. 1). Auditory thresholds were determined by standard pure-tone audiometry with air and bone conductions at 250–8,000 Hz. In addition, previous audiological tests were collected. The phenotype of affected patients in this family was variable (table 1). Six of seven available affected individuals manifested mild-to-severe combined hearing loss, predominantly affecting middle frequencies. Two patients were diagnosed with vestibular pathology consistent with unstable equilibrium most prevalent during walking. To characterize vestibular dysfunction, a computed tomography (CT) scan was performed on patient III-6 and revealed bilateral aplasia of the anterior semicircular canal and hypoplasia of the posterior semicircular canal (fig. 2). All patients, who were evaluated in the Service de Cardiologie at CHU, exhibited congenital heart disease, including tetralogy of Fallot (ToF [MIM 187500]), ventricular septal defect (VSD), and/or peripheral pulmonic stenosis (PPS). Patient II-7 had ToF that did not allow surgical correction, because of severe PPS. He also had severe combined hearing loss that required a hearing aid (fig. 3), as well as vestibular dysfunction. One of his daughters (III-8) had very severe ToF, defined as the presence of pulmonary atresia and ventricular septal defect, which had been successfully surgically corrected in early childhood. Her audiogram was normal. His second daughter (III-6) had a VSD that spontaneously closed, as well as moderate combined hearing loss that required surgical correction and a hearing aid, associated with vestibular dysfunction. Patient II-1 had VSD, PPS, and moderate combined hearing loss. Patients II-2, II-3, and III-1 had isolated PPS and mild-to-moderate combined hearing loss. Patient III-2 had acute lymphoblastic leukemia and died at age 2.5 years; autopsy showed isolated PPS. All individuals had normal growth and cognition. No history of hepatic dysfunction was noted for any of the patients.
Figure 1.
Pedigree of the family with deafness and congenital heart defects, showing three-locus genotypes and inferred haplotypes. Unblackened symbols denote unaffected individuals; blackened symbols denote affected individuals. Genotypes are shown in the following order, from top to bottom: D20S115, D20S186, and D20S112. The arrow indicates the proband.
Table 1.
Comparison of the Clinical Features of Affected Individuals in the Family, with the Frequencies of Clinical Criteria in Patients with AGS
|
Clinical Featurea |
||||||||||
| Patientor Sample | CardiacDisease | Deafness | VestibularPathology | ChronicCholestasis | OcularAbnormalities | CaracteristicFacies | VertebralAnomalies | GrowthRetardation | RenalDysfunction | MentalRetardation |
| II-7 | ToF | Severe | + | − | + | − | − | − | − | − |
| III-6 | VSD | Moderate | + | − | + | − | − | − | − | − |
| III-8 | ToF | − | − | − | + | − | − | − | − | − |
| II-1 | VSD, PPS | Moderate | − | − | + | − | − | − | − | − |
| II-2 | PPS | Mild | − | − | + | − | − | − | − | − |
| II-3 | PPS | Mild | − | − | + | − | − | − | − | − |
| III-1 | PPS | Mild | − | − | + | − | − | − | − | − |
| III-2 | PPS | NA | NA | NA | NA | − | NA | − | − | NA |
| Patients with AGSb | 92% | Rare | Rare | 95% | 78% | 91% | 70% | 50%–90% | 23%–74% | 0%–16% |
+ = Present; − = absent; NA = not available.
Data are from Krantz et al. (1997) and are expressed as percentages of patients showing each clinical feature.
Figure 2.
A, Axial CT scan of patient III-6, showing aplasia of the anterior semicircular canal (a) and hypoplasia of the posterior semicircular canal (p). B, Comparison with an unaffected individual.
Figure 3.
Representative audiogram for the family (individual II-7)
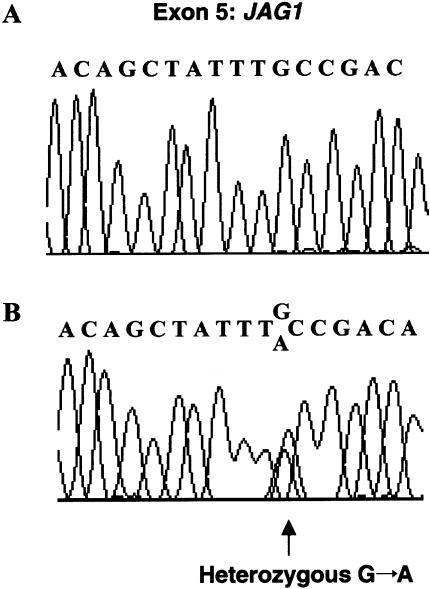
Our protocol was approved by the appropriate institutional review boards. After informed consent was obtained, blood samples were collected from 14 family members. FISH analysis excluded microdeletion at 22q11, a common cause of syndromic and isolated conotruncal heart defects. Genomic DNA was prepared from peripheral lymphocytes. Linkage analysis was performed by standard methods, through use of fluorescence-labeled polymorphic STRs (ABI Prism Linkage Mapping Set version 2). Genotypes were analyzed with an ABI 377 automated sequencer. Alleles were assigned using the Genotyper version 2.0 program. Two-point linkage analysis was calculated using the MLINK program version 5.1, assuming a disease penetrance of 0.95. Because congenital heart defects are linked to mutations in JAG1 or NKX2.5 in humans and in TBX1 or NTF3 in mice, we considered these genes as suitable candidates (Donovan et al. 1996; Schott et al. 1998; Eldadah et al. 2001; Jerome and Papaioannou 2001; Lindsay et al. 2001; Merscher et al. 2001). Linkage analysis excluded the NKX2.5, TBX1, and NTF3 loci but was conclusive for marker D20S186, in the vicinity of the JAG1 gene (maximum LOD score [Zmax] 3.01 at recombination fraction 0.00) (Lathrop and Lalouel 1984). Mutation analysis was conducted by direct sequencing of the JAG1 gene through use of an ABI 377 automated sequencer. All exons were amplified using previously described primers (Krantz et al. 1998). Direct sequencing of JAG1 was performed on an affected family member (II-7). An amplicon spanning JAG1 exon 5 revealed heterozygosity for a G→A transition at nucleotide 701 of the coding sequence, predicting substitution of cysteine (C) for tyrosine (Y) at position 234 (C234Y) of the JAG1 protein (fig. 4). The mutation occurred within the first of the 16 tandemly repeated EGF-like domains of JAG1. This sequence change was confirmed in all affected individuals with available genomic DNA (n=7) in this family but was absent in unaffected family members. No DNA was available for the deceased sibling with PPS (III-2). C234Y was not found in 120 chromosomes from unrelated and unaffected individuals. Since the JAG1 gene has been shown to cause AGS, a clinical geneticist (C.L.C.) reexamined all affected patients for subclinical manifestations of this syndrome. In all gene carriers, no characteristic facies or vertebral defects were identified, but all had asymptomatic posterior embryotoxon. Blood examination showed normal electrolytes, calcium, urea nitrogen, creatinine, total and direct bilirubin, transaminases, gamma-glutamyl transferase, and alkaline phosphatase consistent with normal renal and hepatic functions.
Figure 4.
Sequence chromatograms showing the mutation found in the kindred. A, Sequencing data from individual II-8, representing the normal JAG1 allele. B, Sequencing data from an affected patient (II-7), representing the normal and the mutant JAG1 alleles. A G→A transition changes the codon for cysteine (TGC) in position 234 of the JAG1 protein to the codon for tyrosine (TAC).
Deafness has been described in a few patients with AGS and cytogenetically visible deletions on 20p (Anad et al. 1990) but in none of the studies of large samples of patients with AGS. Therefore, hearing loss has been reported in patients with AGS who have chromosomal abnormalities on 20p (Krantz et al. 1997), suggesting a contiguous gene syndrome. First, however, the JAG1 gene is expressed in the sensory regions of the ear (Crosnier et al. 2000). Second, the mouse mutant Headturner showed dominant head-shaking behavior indicative of a vestibular defect. It also demonstrates disturbances of the patterning of the organ of Corti in the inner ear, characterized by reduced numbers of outer hair cells, as well as loss of sensory structures in the vestibular system, including missing posterior and sometimes anterior ampulae and a truncation of the respective semicircular canals (Kiernan et al. 2001). Despite these patterning anomalies in the organ of Corti, Headturner mutants were not deaf but had slightly raised thresholds for compound action potentials, which is an indication of cochlear nerve responses, although these differences were not significant. A similar phenotype has also been seen in the mouse mutant Slalom, which also carries a mutation in Jag1 (Tsai et al. 2001). Third, severe anomalies have been observed in temporal bones of four individuals with AGS. These patients exhibited partially or totally absent posterior and anterior semicircular canals. The cochlea was observed to be shortened in one case. An audiogram of one patient showed a bilateral moderate combined hearing loss (Okuno et al. 1990). Finally, hearing loss was described in a large kindred with classical AGS (LaBrecque et al. 1982). Patients were found to have normal karyotypes. The father and two children demonstrated a mild conductive hearing loss. At least two other family members were known to have hearing difficulties. The present study describes six of seven C234Y carriers with mild-to-severe combined hearing loss. JAG1 C234Y is the first mutation described in a large kindred with hearing loss.
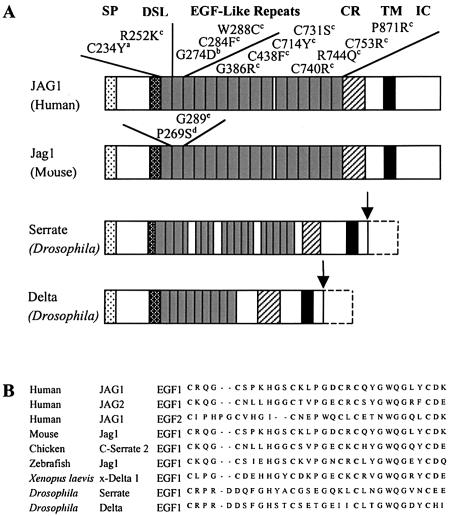
The vast majority of JAG1 mutations that cause AGS create premature termination codons, which predict an unstable message or the expression of truncated polypeptides that lack essential domains (Krantz et al. 1998; Crosnier et al. 1999). AGS is also caused by hemizygosity for JAG1, establishing haploinsufficiency as the relevant pathogenetic mechanism. However, the mechanism by which missense mutations in AGS lead to disease is unknown, and a dominant negative mechanism cannot be ruled out. In Drosophila, mutant Delta and Serrate (Notch ligands that are highly homologous to JAG1) act in a dominant negative fashion (fig. 5A) (Rebay et al. 1991; Sun and Artavanis-Tsakonas 1996). Among the 26 unique missense mutations identified in JAG1, 12 occurred in the EGF repeats (fig. 5A). Eight result in loss or gain of a conserved cysteine residue that forms intramolecular disulfide linkages, an event known to perturb folding and to target proteolytic degradation in other proteins (Schrijver et al. 1999). Each EGF-like domain contains six cysteine residues that form stable disulfide bridges creating stable three-dimensional conformations. Cysteine substitutions disrupt one of three disulfide bridges that covalently connect three pairs of cysteine residues that are highly conserved in EGF-like domains (Aoyama et al. 1993; Downing et al. 1996). This has a predictable detrimental effect on the domain itself, as shown in structural studies of similar domains in other proteins, but it also interferes with calcium binding even when the required consensus sequence is intact (Reinhardt et al. 1997). The novel missense mutation we describe implicates the first cysteine in the first EGF-like domain that disrupts the C1-C3 disulfide bridge and probably affects calcium binding. The mutation alters a residue that is conserved through evolution within this and the other EGF-like domains of JAG1, as well as other proteins, including Jagged 2, Delta, and Serrate (fig. 5B ).
Figure 5.
A, Schematic diagram of human JAG1, mouse Jag1, and Drosophila Delta and Serrate protein structures, and the locations of missense mutations identified in EGF-like domains of human JAG1 and in the second EGF-like domain of mouse Jag1. The arrows indicate C-terminal deletions of Delta and Serrate. Superscripts associated with mutations are as follows: a = missense mutation described in the present study; b = mutation described in a kindred segregating ToF and characteristic facies; c = mutations described in patients with AGS; d = mutation described in mouse Slalom mutant; e = mutation described in mouse Headturner mutant. SP = signal peptide; DSL = conserved domain shared by Delta, Serrate, and Lag2; CR = cysteine-rich region; TM = transmembrane domain; IC = intracellular domain. B, Alignment of the amino acid sequence of the first EGF-like domain of human JAG1 with the corresponding sequences of the first EGF-like domain of human JAG2, the second EGF-like domain of human JAG1, and those from other species. Note the first cysteine is conserved in all of the wild-type sequences.
In the present study, we describe a kindred segregating a unique phenotype with highly penetrant deafness, posterior embryotoxon, and congenital heart defects. We have identified a novel missense mutation (C234Y) in the extracellular domain of JAG1 in the first of the 16 EGF-like domains, which demonstrates that mutations in JAG1 can cause hearing loss. In the presence of a congenital heart defect suggestive of AGS-associated cardiopathies, a audiological test in infancy should be proposed. Further studies of the missense mutations in the EGF-like repeats of JAG1 may provide new insights in JAG1-related diseases.
Acknowledgments
The authors would especially like to thank the family members, for participating in this study, Dr. C. Albouy, for ophthalmological examination, and Dr. A. David and Dr. J. M. Rival, for valuable suggestions and discussions. This work was supported by the Direction de la Recherche Clinique du Centre Hospitalo-Universitaire de Nantes (protocol number BRD 01/12-A).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the JAG1 gene [accession number XM_056118])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AGS [MIM 118450], JAG1 [MIM 601920], and ToF [MIM 187500])
References
- Alagille D, Odievre M, Gautier M, Dommergues JP (1975) Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. J Pediatr 86:63–71 [DOI] [PubMed] [Google Scholar]
- Anad F, Burn J, Matthews D, Cross I, Davison BC, Mueller R, Sands M, Lillington DM, Eastham E (1990) Alagille syndrome and deletion of 20p. J Med Genet 27:729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Tynan K, Dietz HC, Francke U, Furthmayr H (1993) Missense mutations impair intracellular processing of fibrillin and microfibril assembly in Marfan syndrome. Hum Mol Genet 2:2135–2140 [DOI] [PubMed] [Google Scholar]
- Colliton RP, Bason L, Lu FM, Piccoli DA, Krantz ID, Spinner NB (2001) Mutation analysis of Jagged1 (JAG1) in Alagille syndrome patients. Hum Mutat 17:151–152 [DOI] [PubMed] [Google Scholar]
- Crosnier C, Attie-Bitach T, Encha-Razavi F, Audollent S, Soudy F, Hadchouel M, Meunier-Rotival M, Vekemans M (2000) JAGGED1 gene expression during human embryogenesis elucidates the wide phenotypic spectrum of Alagille syndrome. Hepatology 32:574–581 [DOI] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, Bernard O, Hadchouel M, Meunier-Rotival M (1999) Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology 116:1141–1148 [DOI] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, Hadchouel M, Meunier-Rotival M (2001) Fifteen novel mutations in the JAGGED1 gene of patients with Alagille syndrome. Hum Mutat 17:72–73 [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Hahn R, Tessarollo L, Hempstead BL (1996) Identification of an essential nonneuronal function of neurotrophin 3 in mammalian cardiac development. Nat Genet 14:210–213 [DOI] [PubMed] [Google Scholar]
- Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, Handford PA (1996) Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85:597–605 [DOI] [PubMed] [Google Scholar]
- Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, Elkins R, Dietz HC (2001) Familial tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet 10:163–169 [DOI] [PubMed] [Google Scholar]
- Giannakudis J, Ropke A, Kujat A, Krajewska-Walasek M, Hughes H, Fryns JP, Bankier A, Amor D, Schlicker M, Hansmann I (2001) Parental mosaicism of JAG1 mutations in families with Alagille syndrome. Eur J Hum Genet 9:209–216 [DOI] [PubMed] [Google Scholar]
- Heritage ML, MacMillan JC, Colliton RP, Genin A, Spinner NB, Anderson GJ (2000) Jagged1 (JAG1) mutation detection in an Australian Alagille syndrome population. Hum Mutat 16:408–416 [DOI] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE (2001) DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet 27:286–291 [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Ahituv N, Fuchs H, Balling R, Avraham KB, Steel KP, Hrabe de Angelis M (2001) The Notch ligand Jagged1 is required for inner ear sensory development. Proc Natl Acad Sci USA 98:3873–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Colliton RP, Genin A, Rand EB, Li L, Piccoli DA, Spinner NB (1998) Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet 62:1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Piccoli DA, Spinner NB (1997) Alagille syndrome. J Med Genet 34:152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBrecque DR, Mitros FA, Nathan RJ, Romanchuk KG, Judisch GF, El-Khoury GH (1982) Four generations of arteriohepatic dysplasia. Hepatology 2:467–474 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A (2001) Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410:97–101 [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G (1996) Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci 8:14–27 [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G (1995) Jagged: a mammalian ligand that activates Notch1. Cell 80:909–917 [DOI] [PubMed] [Google Scholar]
- Lissemore JL, Starmer WT (1999) Phylogenetic analysis of vertebrate and invertebrate Delta/Serrate/LAG-2 (DSL) proteins. Mol Phylogenet Evol 11:308–319 [DOI] [PubMed] [Google Scholar]
- Loomes KM, Underkoffler LA, Morabito J, Gottlieb S, Piccoli DA, Spinner NB, Baldwin HS, Oakey RJ (1999) The expression of Jagged1 in the developing mammalian heart correlates with cardiovascular disease in Alagille syndrome. Hum Mol Genet 8:2443–2449 [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R (2001) TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104:619–629 [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Meltzer PS, Chandrasekharappa SC (1997) Identification and cloning of the human homolog (JAG1) of the rat Jagged1 gene from the Alagille syndrome critical region at 20p12. Genomics 43:376–379 [DOI] [PubMed] [Google Scholar]
- Okuno T, Takahashi H, Shibahara Y, Hashida Y, Sando I (1990) Temporal bone histopathologic findings in Alagille's syndrome. Arch Otolaryngol Head Neck Surg 116:217–220 [DOI] [PubMed] [Google Scholar]
- Onouchi Y, Kurahashi H, Tajiri H, Ida S, Okada S, Nakamura Y (1999) Genetic alterations in the JAG1 gene in Japanese patients with Alagille syndrome. J Hum Genet 44:235–239 [DOI] [PubMed] [Google Scholar]
- Pilia G, Uda M, Macis D, Frau F, Crisponi L, Balli F, Barbera C, Colombo C, Frediani T, Gatti R, Iorio R, Marazzi MG, Marcellini M, Musumeci S, Nebbia G, Vajro P, Ruffa G, Zancan L, Cao A, DeVirgilis S (1999) Jagged-1 mutation analysis in Italian Alagille syndrome patients. Hum Mutat 14:394–400 [DOI] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S (1991) Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67:687–699 [DOI] [PubMed] [Google Scholar]
- Reinhardt DP, Mechling DE, Boswell BA, Keene DR, Sakai LY, Bachinger HP (1997) Calcium determines the shape of fibrillin. J Biol Chem 272:7368–7373 [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG (1998) Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281:108–111 [DOI] [PubMed] [Google Scholar]
- Schrijver I, Liu W, Brenn T, Furthmayr H, Francke U (1999) Cysteine substitutions in epidermal growth factor-like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am J Hum Genet 65:1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H (1999) Mouse jagged1 physically interacts with notch2 and other notch receptors: assessment by quantitative methods. J Biol Chem 274:32961–32969 [DOI] [PubMed] [Google Scholar]
- Spinner NB, Colliton RP, Crosnier C, Krantz ID, Hadchouel M, Meunier-Rotival M (2001) Jagged1 mutations in Alagille syndrome. Hum Mutat 17:18–33 [DOI] [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S (1996) The intracellular deletions of Delta and Serrate define dominant negative forms of the Drosophila Notch ligands. Development 122:2465–2474 [DOI] [PubMed] [Google Scholar]
- Tsai H, Hardisty RE, Rhodes C, Kiernan AE, Roby P, Tymowska-Lalanne Z, Mburu P, Rastan S, Hunter AJ, Brown SD, Steel KP (2001) The mouse slalom mutant demonstrates a role for Jagged1 in neuroepithelial patterning in the organ of Corti. Hum Mol Genet 10:507–512 [DOI] [PubMed] [Google Scholar]
- Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T (1999) Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet 8:723–730 [DOI] [PubMed] [Google Scholar]
- Yuan ZR, Kohsaka T, Ikegaya T, Suzuki T, Okano S, Abe J, Kobayashi N, Yamada M (1998) Mutational analysis of the Jagged 1 gene in Alagille syndrome families. Hum Mol Genet 7:1363–1369 [DOI] [PubMed] [Google Scholar]