Abstract
There is general agreement that the Native American founder populations migrated from Asia into America through Beringia sometime during the Pleistocene, but the hypotheses concerning the ages and the number of these migrations and the size of the ancestral populations are surrounded by controversy. DNA sequence variations of several regions of the genome of Native Americans, especially in the mitochondrial DNA (mtDNA) control region, have been studied as a tool to help answer these questions. However, the small number of nucleotides studied and the nonclocklike rate of mtDNA control-region evolution impose several limitations to these results. Here we provide the sequence analysis of a continuous region of 8.8 kb of the mtDNA outside the D-loop for 40 individuals, 30 of whom are Native Americans whose mtDNA belongs to the four founder haplogroups. Haplogroups A, B, and C form monophyletic clades, but the five haplogroup D sequences have unstable positions and usually do not group together. The high degree of similarity in the nucleotide diversity and time of differentiation (i.e., ∼21,000 years before present) of these four haplogroups support a common origin for these sequences and suggest that the populations who harbor them may also have a common history. Additional evidence supports the idea that this age of differentiation coincides with the process of colonization of the New World and supports the hypothesis of a single and early entry of the ancestral Asian population into the Americas.
There is general agreement that the ancestral populations of Native Americans migrated from Asia into America through Beringia sometime during the Pleistocene (Cavalli-Sforza et al. 1994). Bitter controversies surround the hypotheses as to the ages and the number of these migrations and the size of the ancestral populations. DNA sequence variations of several regions of the genome of Native Americans, especially in the mtDNA control region, have been studied as a tool to help answer these questions. The mtDNA of most Native Americans belongs to four main lineages (A, B, C, and D) (Schurr et al. 1990; Horai et al. 1993; Torroni and Wallace 1995) that show close similarity with modern mtDNA from Asians (Bailliet et al. 1994; Brown et al. 1998), in addition to minor contributions from other lineages (Merriwether et al. 1996; Derenko et al. 2000). Various studies have estimated the time of entry of the ancestral populations into the Americas by means of haplogroup-diversity values, on the basis of both RFLP data (Schurr et al. 1990; Torroni et al. 1992, 1993) and control-region sequence variation (Horai et al. 1993; Forster et al. 1996), and have proposed either a single (Merriwether et al. 1995; Forster et al. 1996; Bonatto and Salzano 1997a, 1997b; Stone and Stoneking 1998) or more than one wave of migration (Torroni et al. 1992, 1994; Horai et al. 1993). Global analyses of the sequence variation of the mtDNA control region in >700 Native Americans and more restricted RFLP studies have revealed surprisingly similar results for all four haplogroups. The average times of diversification estimated on the basis of the hypervariable segment I (HVS-I) values were ∼42,000 and ∼29,000 years before present (BP), depending on whether a slower or a faster nucleotide substitution rate, respectively, was used (Bonatto and Salzano 1997b). Joint calculations for HVS-I+HVS-II did not change the values substantially: ∼43,000 and ∼33,000 years BP, for the slower and faster substitution rates, respectively. These findings support the conclusion of a single early entry of ancestral populations into America. Nevertheless, some RFLP data suggest a late entry for haplogroup B (Torroni et al. 1993).
However, these studies based on the diversity of mtDNA at the control region are complicated by the difficulty in attaining good estimates for the mutation rate and by its extreme variation between sites. In contrast, Ingman et al. (2000) have demonstrated that the molecular-clock hypothesis applies to the segment of ∼15,450 nucleotides (nt) outside the control region in human mtDNA and have estimated a reliable mutation rate by use of the human-chimpanzee divergence time, allowing the estimation of the age of the most recent common ancestor for all human mtDNA and the age for the exodus from Africa.
In the present study, we analyzed an 8,829-nt segment of the mitochondrial genome of 40 individuals, most of them Native Americans. Since the analysis of our data together with those from Ingman et al. (2000) for ∼57% of the mtDNA molecule demonstrated that the results are similar to those obtained by sequencing the whole molecule, we restricted our analysis to a segment of 8.8 kb of mtDNA. The segment sequenced extends from nt 7148 to nt 15976 in the reference sequence J01415 (GenBank). The primers used for PCR amplification have been described elsewhere (Reider et al. 1998). Sequencing was performed directly on the PCR products through use of BigDye (Applied Biosystems) chemistry on a ABI377 automatic sequencer. We sequenced both the forward and the reverse strands. Sequence analysis was performed using Sequencing Analysis 3.3 (Applied Biosystems). The sequence validation and the assembly of the region of 8,829 bp were performed using Polyphred software (Nickerson et al. 1997). A 352-nt segment of the HVS-I of the D-loop (nt 16027–16376) was also sequenced by use of primers described elsewhere (Horai et al. 1993). The following sites were analyzed, by PCR amplification and restriction-enzyme digestion, for the 30 Native Americans: presence of HaeIII at nt 663, loss of HincII at nt 13259, loss of AluI at nt 5176, and the 9-bp deletion between COII and tRNALys.
A total of 30 Native Americans of different linguistic stock were selected, to include a well-balanced representation of the four haplogroups based on the HVS-I region and RFLP: Yanomama, Arara, Waiampi, Tyrio, Poturujara, Katuena, Kayapo, and Guarani (all from Brazil); and five Quechua from Peru. In addition, the following non–Native Americans were also included: four African Brazilians, three Brazilians of Japanese origin (Asian Brazilians), and three white Brazilians.
Molecular statistical parameters were estimated using DNAsp3.53 (Rozas and Rozas 1999). For all analyses we used a joint data set that included sequences from the 40 individuals described in the present study (GenBank accession numbers AF465941–AF465980) together with the 53 previously published sequences (Ingman et al. 2000), plus the Anderson reference sequence (Anderson et al. 1981). This 8.8-kb segment is ∼58% more variable than the other half of the molecule and ∼20% more variable than the whole non-control region of the mitochondrial genome. Comparisons between our data and those of Ingman et al. (2000) indicate that the use of this segment instead of the whole 15.4-kb mitochondrial genome is sufficient to give reliable estimates. When only the 8.8-kb region is used, both in Ingman’s data set alone and in our joint data set, the main statistical parameters, such as sequence diversity for Africans being about twice that of non-Africans, are reproduced here (table 1). The mean nucleotide distance (gamma corrected) between the chimpanzee and humans in our data set was estimated at 0.24 substitutions per site. If we assume 5 million years BP as the divergence time between humans and chimpanzees, the mutation rate in this 8.8-kb region is 2.4×10-8 substitutions per site per year, and the time estimate for the exodus from Africa based on this rate is 49,000 years BP, a value that agrees closely with that estimated by Ingman et al. (2000) (52,000 years BP).
Table 1.
Comparison of Statistical Parameters for an 8.8-kb Segment of the mtDNA Recalculated for Ingman’s Original Data and in the Joint Data Set Formed by Their and Our Data
| Data Set | No. ofSequences | No. ofSegregatingSites | Mean PairwiseSequenceDifference | GeneticDiversity(π) ×10−3 |
| Ingman et al. (2000): | ||||
| All humans | 53 | 342 | 29.7 | 3.4 |
| Non-Africans | 32 | 183 | 19.1 | 2.2 |
| Africans | 21 | 183 | 36.6 | 4.2 |
| Ingman et al. (2000) plus the present study: | ||||
| All humans | 93 | 395 | 24.5 | 2.8 |
| Non-Africans | 67 | 239 | 16.8 | 1.9 |
| Africans | 26 | 194 | 34.5 | 3.9 |
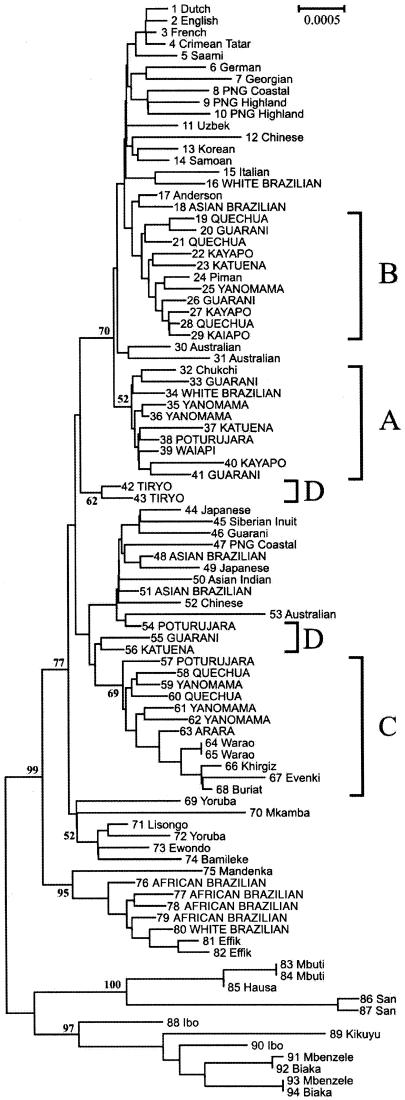
Trees were constructed using several methods, including the neighbor-joining (NJ) method (MEGA software) (Kumar et al. 2001), maximum parsimony (MEGA and PAUP) (Swofford 1998; Kumar et al. 2001), and maximum likelihood (Tree-Puzzle) (Strimmer and von Haeseler 1996), all resulting in essentially identical trees, as well as the median-joining network approach (Bandelt et al. 1995). The phylogenetic tree based on all 94 sequences (fig. 1) shows the same pattern as in the study by Ingman et al. (2000): three main, well-supported exclusively African basal groups; one larger group, including two non-African clades; and a small African clade. Of the 30 Native Americans we studied, 25 (80%) support the same A, B, C haplogroup structure given by analyses of the control region and by the RFLP diagnostic positions (Torroni et al. 1993). Although these clusters did not give high bootstrap values, the confidence values increased consistently when we made a tree with sequence data that included the HVS-I regions, and reached 98%, 71%, and 99%, respectively, when we used the interior branch test (MEGA Software) (results not shown). Haplogroup A is the most homogeneous and best characterized. In addition to the Native Americans, one Siberian Chukchi and one white Brazilian also clustered in haplogroup A. As was found in the studies of the control region, most Chukchi mtDNAs belong to haplogroup A (Bonatto and Salzano 1997a), whereas the white Brazilian is probably of a mixed Native American origin. Three central Asians (Khirgiz, Evenki, and Buriat) grouped in haplogroup C together with the Native Americans. Finally, the five Native Americans who belong to haplogroup D, on the basis of HVS-I sequences and RFLP markers, did not group together in the NJ tree in figure 2: three grouped with other Asian sequences, but two Tyrio were placed in an isolated position. However, in other analyses, such as in the maximum-parsimony tree and in some resolution of the median-joining network (not shown), four or even all five sequences may group together, indicating that additional data are needed to settle the question of whether haplogroup D is monophyletic. One Guarani from the Ingman et al. (2000) study could not be assigned to any known Native American haplogroup, even when we used the D-loop sequence, and probably is an individual of mixed ethnic origin or belonging to a minor haplogroup. The white Brazilian who clustered with the Africans is probably of mixed African origin. Haplogroup B consists only of Native Americans.
Figure 1.
NJ tree based on the 8.8-kb mtDNA segment from 93 humans plus the sequence inferred by Anderson et al. (1981) through use of the proportional distance and rooted with the chimpanzee sequence. Bootstrap percent values (1,000 replicates) are shown on some nodes, and haplogroups A–D are indicated on the right. The population origin of the individuals are given in the twigs, and the 40 new sequences from the present study are capitalized. PNG = Papua New Guinea.
Figure 2.
Data matrix showing the informative nucleotide positions for the 8.8-kb mtDNA segment. The trees on the left are cladograms with the same topology and numbering of individuals as the tree in figure 1. Gray blocks denote groups of nucleotide changes that are identical in several sequences of the four major Native American haplogroups and the two mutations common to many Asians.
Figure 2 presents the mutations within this 8.8-kb region that are characteristic for each haplogroup. The two mutations at 14783T→C and at 15543G→A define a cluster that contains only Asians and one Australian, and it subdivides into two groups: (a) Native American haplogroup C, which contains the two additional mutations 9545A→G and 13263A→G (the latter mutation causes the HincII site loss that identifies Native American group C by RFLP); and (b) other Asians, which show no additional consistent change. Haplogroup A is characterized by the 8794C→T transition, and haplogroup B is characterized by the 13590G→A transition in lineages that lack the 12705C→T change.
Nucleotide distances and diversity values were estimated using the Kimura two-parameter distance (Kimura 1980), with gamma correction and SEs estimated using 1,000 bootstraps (MEGA). A gamma parameter α=0.14, describing site-to-site rate heterogeneity, was estimated using a maximum-likelihood approach (Tree-Puzzle). Haplogroup diversification times and 95% CIs were calculated as described by Bonatto and Salzano (1997b), taking into account both mutation rate and nucleotide-diversity SEs. The nucleotide diversity in these three Native American haplogroup clades and in the five sequences from haplogroup D are very similar (table 2). If we include the three central Asians in haplogroup C, the diversity values increase a little but still are not significantly different from the values obtained for the other three haplogroups. Using the substitution rate estimated for this region, we calculated very similar values for the time of diversification of the four haplogroups, with a weighted mean of 21,000 years BP and a 95% CI of 18,600–23,400 years BP. If we exclude the nonmonophyletic haplogroup D, we find a weighted mean of 20,000 (95% CI 17,800–22,400) years BP. These values are slightly lower than some previous estimates, made by us and others on the basis of the diversity of the control region and RFLP, in the range of 26,000–42,000 years BP, and they agree closely with the estimate of 20,000–25,000 years BP proposed by Forster et al. (1996). Since our tests demonstrate that the estimates based on this 8.8-kb segment do not differ significantly from those obtained by the analysis of the whole mitochondrial genome, and since we have studied Native Americans from a wide range of geographic and linguistic origins in South America, it is not likely that this conclusion can be substantially changed by the analysis of either a larger sample or a longer DNA segment.
Table 2.
π and Age Estimates for mtDNA Belonging to the Four Founder Haplogroups of Native Americans
| Haplogroup | No. ofSequences | π × 10−3[SE] | Mean Age [95% CI](years) |
| A | 10 | .97 [.18] | 20,500 [16,400–24,600] |
| B | 11 | .86 [.16] | 18,100 [14,600–21,700] |
| C | 9 | 1.02 [.19] | 21,600 [17,300–25,900] |
| D | 5 | 1.12 [.20] | 23,800 [19,300–28,300] |
| Weighted mean | 35 | .99 [.09] | 21,000 [18,600–23,400] |
This high similarity in the nucleotide diversity within all four haplogroups, despite the small sample size and the nonmonophyletic haplogroup D, suggests that they have the same age of diversification and share a common history. These results are similar to those found previously, through use of the mtDNA control region (Forster et al. 1996; Bonatto and Salzano 1997b; Lorenz and Smith 1997). Even though it cannot be directly determined where the diversification of the four Native American haplogroups began, we can use these and other similar findings (Merriwether et al. 1995; Kolman et al. 1996)—such as the presence of the four Native American haplogroups throughout the Americas but a restricted distribution in Asia, as well as their pattern of population expansion—to suggest that the differentiation is correlated with the colonization process. We argue that, together with Y-chromosome evidence (Santos et al. 1999), our mtDNA data support a single and early wave of migration for the peopling of the Americas.
Acknowledgments
The authors thank Adriana A. Marques, Cristiane A. Ferreira, Rafaela M. Maia, Bruno M. Carvalho, and Ana Cecilia F. Santos, for expert technical assistance, and Israel T. Silva and Rodrigo M. Souza, for their contribution to the bioinformatics analysis. This work was supported by Grants of the Research Foundation of the State of São Paulo (FAPESP) and the National Research Council of Brazil (CNPq).
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for the chimpanzee [accession number D38113] and human mtDNA sequences from the study by Ingman et al. [2000] and for the 40 new sequences described in the present study [accession numbers AF465941–AF465980])
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Bailliet G, Rothhammer F, Carnese FR, Bravi CM, Bianchi NO (1994) Founder mitochondrial haplotypes in Amerindian populations. Am J Hum Genet 55:27–33 [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatto SL, Salzano FM (1997a) Diversity and age of the four major mtDNA haplogroups, and their implications for the peopling of the New World. Am J Hum Genet 61:1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1997b) A single and early origin for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc Natl Acad Sci USA 94:1866–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC (1998) mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am J Hum Genet 63:1852–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Piazza A, Menozzi P (1994) History and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Derenko MV, Denisova GA, Malyarchuk BA, Dambueva IK, Dorzhu CM, Stolpovski YM, Lotosh EA, Luzina FA, Zakharov IA (2000) Mitochondrial DNA variability in Turkic-speaking populations of the Altai and Sayan region from South Siberia. Am J Hum Genet Suppl 67:A1161 [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Horai S, Kondo R, Nakagamma-Hattori Y, Hayashi S, Sonoda S, Tajima K (1993) Peopling of the Americas founder by four major lineages of mitochondrial DNA. Mol Biol Evol 10:23–47 [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Kimura MA (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120 [DOI] [PubMed] [Google Scholar]
- Kolman CJ, Sambuughin N, Bermingham E (1996) Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics 142:1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Arizona State University, Tempe [DOI] [PubMed] [Google Scholar]
- Lorenz JG, Smith DG (1997) Distribution of sequence variation in the mtDNA control region of Native North Americans. Hum Biol 69:749–776 [PubMed] [Google Scholar]
- Merriwether DA, Hall WW, Vahlne A, Ferrell RE (1996) mtDNA variation indicates Mongolia may have been the source for the founding population for the New World. Am J Hum Genet 59:204–212 [PMC free article] [PubMed] [Google Scholar]
- Merriwether DA, Rothhammer F, Ferrell RE (1995) Distribution of the four founding lineage haplotypes in Natives Americans suggests a single wave of migration for the New World. Am J Phys Anthropol 98:411–430 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider MJ, Taylor SL, Tobe VO, Nickerson DA (1998) Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res 26:967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 [DOI] [PubMed] [Google Scholar]
- Santos FR, Pandya A, Tyler-Smith C, Pena SD, Schanfield M, Leonard WR, Osipova L, Crawford MH, Mitchell RJ (1999) The central Siberian origin for native American Y chromosomes. Am J Hum Genet 64:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Ballinger SW, Gan Y-Y, Hodge JA, Merriwether DA, Lawrence DN, Knowler WC, Weiss KW, Wallace DC (1990) Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet 46:613–622 [PMC free article] [PubMed] [Google Scholar]
- Stone AC, Stoneking M (1998) mtDNA analysis of a prehistoric Oneota population: Implications for the peopling of the New World. Am J Hum Genet 62:1153–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A (1996) Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol 13:964–969 [Google Scholar]
- Swofford DL (1998) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- Torroni A, Neel JV, Barrantes R, Schurr TG, Wallace DC (1994) Mitochondrial DNA “clock” for the Amerinds and its implications for timing their entry into North America. Proc Natl Acad Sci USA 91:1158–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang C-C, Szathmary EJE, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM, Wallace DC (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Wallace DC (1995) mtDNA haplogroups in Native Americans. Am J Hum Genet 56:1234–1236 [PMC free article] [PubMed] [Google Scholar]