Abstract
Bardet-Biedl syndrome (BBS) is an uncommon multisystemic disorder characterized primarily by retinal dystrophy, obesity, polydactyly, and renal dysfunction. BBS has been modeled historically as an autosomal recessive trait, under which premise six independent BBS loci (BBS1–BBS6) have been mapped in the human genome. However, extended mutational analyses of BBS2 and BBS6, the first two BBS genes cloned, suggest that BBS exhibits a more complex pattern of inheritance, in which three mutations at two loci simultaneously are necessary and sufficient in some families to manifest the phenotype. We evaluated the spectrum of mutations in the recently identified BBS4 gene with a combination of haplotype analysis and mutation screening on a multiethnic cohort of 177 families. Consistent with predictions from previous genetic analyses, our data suggest that mutations in BBS4 contribute to BBS in <3% of affected families. Furthermore, integrated mutational data from all three currently cloned BBS genes raise the possibility that BBS4 may participate in triallelic inheritance with BBS2 and BBS1, but not the other known loci. Establishment of the loci pairing in triallelism is likely to be important for the elucidation of the functional relationships among the different BBS proteins.
Introduction
Genetic disorders have historically been divided into Mendelian and complex traits, the former involving mutational events at one locus (or, in the case of contiguous-gene syndromes, genomic rearrangements over a defined region of the genome) and the latter requiring mutations at more than one gene, coupled with environmental and/or stochastic factors, to manifest the phenotype. Recent studies have suggested that some disorders bridge these two categories to establish a continuum of mutational mechanisms, whereby the combinatorial effect of mutations at distinct loci is necessary and sufficient for pathogenicity.
Bardet-Biedl syndrome (BBS [MIM 209900]) appears to be an important example of this model of phenotype transmission, since it can be inherited both as an autosomal recessive trait and as an oligogenic trait. BBS presents as a complex phenotype, with substantial inter- and intrafamilial variability, characterized by the clinical features of retinal dystrophy, obesity, polydactyly, hypogonadism, and renal dysplasia. Less frequent features include short stature, relative or absolute macrocephaly, diabetes mellitus, cardiomyopathy or congenital cardiac malformations, reproductive-tract anomalies, developmental retardation, and speech and behavioral anomalies (Green et al. 1989; Beales et al. 1999; Katsanis et al. 2001a). The segregation of BBS in families and population isolates initially led to the hypothesis that the disorder is inherited as a genetically heterogeneous autosomal recessive trait. On the basis of this model, six BBS loci have been identified: BBS1, on 11q13 (Leppert et al. 1994); BBS2, on 16q21 (Kwitek-Black et al. 1993); BBS3, on 3p12 (Sheffield et al. 1994); BBS4, on 15q22.2-q23 (Carmi et al. 1995); BBS5, on 2q31 (Young et al. 1999); and BBS6, on 20p12 (Katsanis et al. 2000). We have also reported evidence for at least one more locus in the human genome (Beales et al. 2001). However, mutational analysis of the first two cloned BBS genes—BBS2, which encodes a protein of unknown function (Nishimura et al. 2001), and BBS6, which encodes a putative chaperonin (Katsanis et al. 2000; Slavotinek et al. 2000)—suggested that, in some families, the disorder may be inherited in a complex fashion, since the phenotype requires that two “recessive” mutations at one locus be paired with a third mutation at another locus, a phenomenon termed “triallelic inheritance” (Katsanis et al. 2001a, 2001b).
Since the first demonstration of this complex mode of inheritance, a third BBS locus, BBS4, has been identified with homology to the O-linked acetylglucosamine transferase (OGT) family of genes (Mykytyn et al. 2001). Here, we have evaluated, irrespective of locus assignment on the basis of either haplotype or other mutation data for each family, a cohort of 177 families with BBS, to identify mutations in BBS4. As predicted from our earlier genetic analyses, BBS4 contributes the least to BBS, although its prevalence is substantially higher among Arab families than among white families from Europe and North America. We also suggest that BBS4 may exhibit complex inheritance in which both recessive- and dominant-susceptibility mutations at BBS4 are paired with mutations at BBS2, BBS1, or another, as-yet-unidentified locus in the human genome, to cause the disease phenotype.
Patients and Methods
Patients
One hundred seventy-seven families with BBS, of various ethnic origins, were screened for mutations in BBS4. The diagnosis of BBS was based on the established criterion that four of six cardinal features must be present (Beales et al. 1999). In several cases, the diagnosis was ascertained by local physicians and was verified through extensive examination of medical records, by one or more of the coauthors (R.A.L., H.K., and/or P.L.B.). Blood was obtained with consent, in accordance with protocols approved by the appropriate human-subjects ethics committees at each institution, and DNA was extracted by a salting-out process (Puregene and Gentra Systems).
Mutational and Genetic Analysis
The genomic structure of BBS4 was discerned by alignment of the BBS4 cDNA (GenBank accession number XM_027370) to genome assembly scaffold NT_010298 with the nucleotide-nucleotide BLAST (blastn) algorithm and the GCG software package, as described elsewhere (Katsanis et al. 1997, 2000). Primers were designed for the amplification of each of the 17 BBS4 exons (for primers, see the Lupski Lab Web site), and direct sequencing of BBS4 was performed as described elsewhere (Katsanis et al. 2000). For the genetic analyses, fluorescent microsatellite STRPs were genotyped for each family member, and haplotypes were constructed across each BBS locus, as described elsewhere (Beales et al. 1999; Katsanis et al. 1999). Microsatellite sequences were obtained from either the Genome Database or the Whitehead Institute Center for Genome Research.
Identification of BBS4 Homologs
To identify the mouse ortholog of BBS4, we used the human BBS4 peptide sequence to screen the mouse subdivision of the Expressed Sequence Tags Database (dbEST). The sequence of positive cDNAs were downloaded and were assembled into a contig, as described elsewhere (Katsanis and Fisher 1998). Clones were obtained from Research Genetics and were sequenced by dye-terminator chemistry (Applied Biosystems). Gaps in the sequence were closed by a BLAST search of the mouse-genome-sequence database. Validation of the contiguity of the mouse message was achieved by cDNA amplification and the sequencing of PCR products from a mouse-cDNA panel (Clontech).
We identified the Drosophila and thermobacterial homologs of BBS4 by searching Swissprot, Flybase, and the Celera Genomics databases with the human peptide sequence. Sequences were aligned with the Bestfit, Gap, and Pileup programs from the GCG, version 9.0, sequence-analysis computer package (University of Wisconsin).
Results
Mutation Analysis of BBS4
Studies based on haplotype-inferred locus assignment (HILA) have predicted that BBS4 is a minor contributor to BBS, since only 1%–2% of families showed chromosomal inheritance across the BBS4 region consistent with two-allele segregation and linkage to that locus alone (Beales et al. 2001). The recent cloning of BBS4 has enabled us to evaluate this prediction and to investigate the mutational spectrum of this gene. We sequenced, irrespective of HILA, all exons and splice junctions of BBS4, in 177 independent probands of various ethnic origins. We detected numerous alterations, which we evaluated by investigating whether (a) the nucleotide change either substituted a nonconservative amino acid, interrupted translation, or predicted altered mRNA processing; (b) the alteration was not found in a minimum of 192 ethnically matched control chromosomes and 96 random control chromosomes; and (c) the candidate mutation segregated with the disease within the family. The latter criterion was restricted by the possibility that BBS mutations do not necessarily follow a strict Mendelian pattern of inheritance, since it is possible to inherit two BBS mutations at a single locus but fail to manifest a phenotype (Katsanis et al. 2001a). Nevertheless, it seems unlikely that both a conventional recessive model and a triallelic model of disease inheritance would manifest in the same family—that is, a situation in which two mutations at a single locus would be sufficient to cause disease in some but not all affecteds within a single family. Therefore, we reasoned that the same disease-associated mutations, irrespective of the number of BBS loci required for disease manifestation, must be inherited by all affected individuals of any given sibship, and, thus, we restricted segregation analyses only to affected subjects with BBS.
Alterations in five families fulfilled our three minimal criteria for pathogenicity. We identified two BBS4 mutations in two families (KK021 and PB043), and we detected only a single mutant allele by sequencing in three other families (AR153, AR708, and AR717) (table 1). Because a deletion of exons 2–4 has also been associated with the disease (Mykytyn et al. 2001), we also performed long-range PCR in all 177 patients, to amplify a 7-kb genomic fragment between introns 1 and 4. To investigate the possibility that there are other small deletions at this locus, we also analyzed 12 SNPs across the BBS4 genomic region (table 2). Neither approach suggested the presence of deletions. In all patients, we either amplified only the correct-sized amplicon or inferred, on the basis of heterozygosity at polymorphic positions, that there were two alleles (data not shown).
Table 1.
Summary of BBS4 Mutations
| Family | Mutation 1 | Mutation 2 | Mutation 3 |
| KK021 | IVS3-2A→G | IVS3-2A→G | |
| PB043 | A364E (BBS4) | A364E (BBS4) | T558I (BBS2)a |
| AR153 | L327P | BBS1? | BBS1? |
| AR708 | N165H | Unknown | Unknown |
| AR717 | S457I | Unknown | Unknown |
Although three mutant alleles are listed, the affected individual (PB043-01) in family PB043 carries four potential mutations, since he is homozygous for both A364E and T558I.
Table 2.
Summary of Other Variations
| Exon | Position | Variation | Comment |
| 1 | −17 | C/A | |
| 1 | 42 | G/A | A allele present in 1 patient and in 0/84 control subjects |
| 3 | −6 | A/G | G allele present in 1 patient and in 0/81 control subjects |
| 4 | 91 | G/A | |
| 5 | 8 | A/C | C allele present in 1 patient and in 0/93 control subjects |
| 5 | 28 | insA | |
| 6 | 17 | C/T | |
| 13a | c.1061 | C/T | |
| 20 | C/T | ||
| 15 | 18 | C/T | C allele present in 1 patient and in 0/93 control subjects |
| 16 | −45 | C/T | |
| 16a | c.1561 | G/C | C allele present in 1 patient and in 0/87 control subjects |
SNPs, annotated by the position in the cDNA sequence, where the first base of the start methionine is base 1.
Recessive Inheritance of Mutations in BBS4
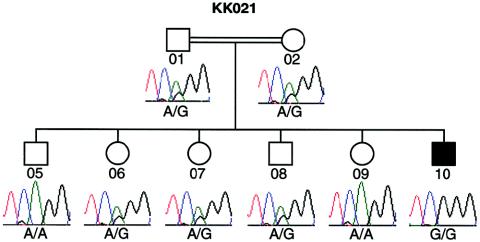
Affected individual KK021-10, from consanguineous Saudi Arabian family KK021, is homozygous for an A→G transition at the acceptor splice site of exon 4 (IVS3-2A→G) (fig. 1 and table 1). This alteration is likely to be a pathogenic mutation, since it was not found in 108 ethnically matched control chromosomes and is predicted to result in a null allele due to missplicing and subsequent nonsense-mediated RNA decay (Losson and Lacroute 1979). Direct sequencing of the DNA from all available members of this family revealed that individual KK021-10 inherited one mutant allele from each parent. Consistent with a recessive model of disease inheritance, we found that all other available unaffected family members either were heterozygous or had the wild-type allele at this site (fig. 1). These data concur with our genetic prediction that BBS in family KK021 is due to a homozygous BBS4 mutation, since this family both exhibited identity-by-descent (IBD) across the BBS4 region and was excluded from all other known BBS loci by haplotype analysis.
Figure 1.
Recessive inheritance of IVS3-2A→G, an exon 4 splice-acceptor mutation, in Saudi Arabian family KK021.
A homozygous alteration was also detected in family PB043, a consanguineous sibship of Kurdish origin. The only affected individual (PB043-01) inherited one 1091C→A mutation from each parent, which, on conceptual translation, would result in an A364E alteration, a potentially deleterious change in the local polarity of the polypeptide. Consistent with our hypothesis that this alteration is a disease-causing mutation, we did not detect the A364E allele in 384 control chromosomes (including 84 Kurdish chromosomes). Furthermore, segregation of this allele, both by sequencing and by restriction analysis, indicated that only the affected individual (PB043-01) was homozygous at this locus, whereas two unaffected sibs who were tested were carriers. These observations are consistent with a traditional model of autosomal recessive transmission and are concordant with previous genetic data that had excluded family PB043 from all other BBS loci by haplotype analysis.
In North American families AR153, AR708, and AR717, we were able to identify a single alteration in BBS4 (table 1). Without functional data on the BBS4 protein, the pathogenicity of these alterations is unclear. However, each mutation is predicted to result in a nonconservative amino acid substitution that would change either the local charge (N168H in AR708), the hydrophobicity (S457I in AR717), or the secondary structure (L327P in AR153). Also, each mutation occurred in both affected family members and did not occur in 384 ethnically matched control chromosomes and 96 random control chromosomes.
Evolutionary Conservation of BBS4 Mutation Sites
To evaluate further the pathogenic potential of each missense alteration in BBS4, we ascertained the evolutionary tolerance for variation at each amino acid position. Using the BBS4 amino acid sequence, we performed a BLAST search of all subdivisions of both dbEST and the Drosophila genome database. We identified multiple bovine, mouse, and Drosophila ESTs, which we assembled into discrete contigs and compared to the human genomic sequence. Conceptual translation of each contig and alignment to the human BBS4 amino acid sequence suggested that we had identified the complete coding sequence of the mouse (89.9% identity and 92.1% similarity) ortholog of BBS4 and partial sequences of Drosophila (36.7% identity and 49.1% similarity) and bovine (92.4% identity and 93.5% similarity) orthologs.
We compared all BBS4 peptide sequences to each other and to the amino acid sequence of a more distantly related Methanothermobacter thermautotrophicus OGT (GenBank accession number NC_000916; 35.7% similarity to the human BBS4 peptide sequence). Evaluation of the four mutated residues revealed substantial interspecies conservation (fig. 2): the asparagine residue at position 165 of the human sequence was conserved in both mammalian species, as well as M. thermautotrophicus; the leucine residue at position 327 and the alanine residue at position 364 were conserved in mammals and insects; and the serine residue at position 457 could be evaluated only in mouse, which also contains a serine at this position.
Figure 2.
Evolutionary conservation of residues involved in complex inheritance of BBS. A, Alignment of BBS4 amino acid sequences of human (Hsa), mouse (Mmu), bovine (Bta), and Drosophila (Dme). Mutant alleles, indicated by asterisks (*), are named above the sequence. Because of a substantial sequence divergence, the amino acid sequence of an M. thermautotrophicus (Mth) OGT is shown only at the residues mutated in BBS. The predicted tetratricopeptide domain is boxed. B, Alignment, at residues 547–566, between human BBS2 peptide sequence and mouse, rat (Rno), and zebrafish (Dre) BBS2 peptide sequence, to illustrate the evolutionary conservation of the threonine residue at position 558. Note that T558I has previously been reported as T560I (Katsanis et al. 2001a).
Does BBS4 Also Exhibit Complex Inheritance?
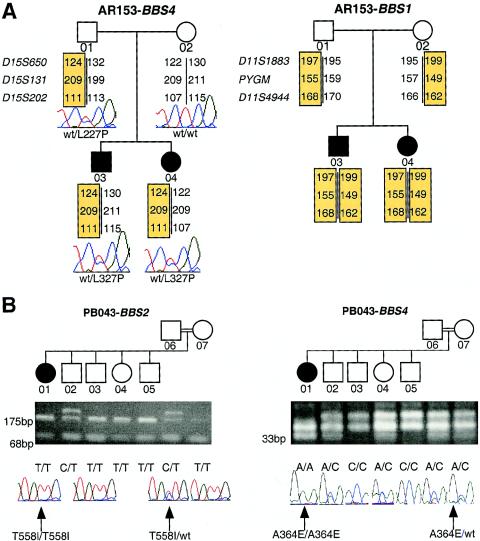
Both the conservation of the mutated amino acids in lower organisms and the presence of these alterations in all affected individuals (but not in 384 control chromosomes) suggest that these changes represent pathogenic alleles, rather than benign polymorphisms. Although we detected no deletions by long-range PCR and SNP analysis, we cannot exclude the possibility that a second mutant BBS4 allele may map to a regulatory region of the genomic locus or may occur with another local rearrangement that is not detectable by these techniques. We therefore examined the haplotypes at all six known BBS loci in all five families in which we found at least one mutant BBS4 allele, including the two sibships in which both alleles were found. The segregation of the extended haplotypes in families KK021 and PB043 is consistent with linkage to BBS4, but AR717 is too small for definitive assignment by HILA. However, we were able to exclude recessive inheritance of BBS4 mutations in families AR153 and AR708. In family AR708, both the affected sib and the unaffected sib inherited the same parental BBS4 chromosomes. In family AR153, the two affected sibs had inherited the same paternal chromosome (carrying the L327P mutation) but a different maternal chromosome (fig. 3A). Therefore, if the mother (AR153-02) was a carrier for a BBS4 mutation that was undetectable by sequencing, then she could not have transmitted the mutant allele to both of her children. Further haplotype analysis indicated that family AR153 could be excluded from all loci except BBS1, raising the possibility that a single L327P mutation in BBS4 may act in conjunction with two BBS1 mutations (fig. 3A).
Figure 3.
Examples of complex inheritance involving BBS4 and other loci. “wt” denotes wild type. A, Potential triallelic inheritance between BBS1 and BBS4, in family AR153. This family is genetically excluded from the inheritance of recessive BBS4 mutations and bears haplotypes consistent with linkage across BBS1, yet both patients inherited a heterozygous L327P BBS4 mutation. B, Potential tetra-allelic inheritance between BBS2 and BBS4, in family PB043. The affected individual (PB043-01) has inherited a homozygous T558I BBS2 mutation and a homozygous A364E BBS4 mutation. Confirmation, by restriction analysis, of mutations in family PB043 is also shown: the T558I allele generates 68-bp and 175-bp SspI fragments, whereas the A364E allele generates a 33-bp MboI fragment.
We have previously reported several instances in which mutations that, at first, appear to segregate in a traditional recessive fashion are determined, when additional genes become available for testing, to be triallelic (Katsanis et al. 2001a). Therefore, we integrated all sequence data from our earlier mutation screenings at BBS2 and BBS6, irrespective of the initial HILA for each of the five families with BBS4 mutations. Although four of the families had the wild-type allele for each of the other BBS genes, the affected individual (PB043-01) in consanguineous family PB043 was homozygous for a T558I mutant BBS2 allele. Segregation of this allele by both sequencing and restriction analysis suggested that three unaffected sibs (PB043-03, PB043-04, and PB043-05) and the mother (PB043-07) were homozygous for this alteration in BBS2. These data suggest either that T558I is a benign, albeit rare, polymorphism, in which case family PB043 would represent an example of classical Mendelian inheritance, or that the disease is inherited in this family in a tetra-allelic fashion (fig. 3B). Both an unaffected sib (PB043-04) and the mother (PB043-07) carry three BBS mutations (a homozygous T558I mutation, in BBS2, and a heterozygous A364E mutation, in BBS4), thereby implying that, if the T558I mutation in BBS2 is pathogenic, then four mutant alleles may be both necessary and sufficient for the manifestation of BBS in this family. Despite both the absence of the T558I alteration from 192 control chromosomes and the evolutionary conservation of the threonine residue at position 558 (fig. 2B), functional assays will be required to clarify the potentially pathogenic role of this alteration and, consequently, the mode of inheritance in this family.
Discussion
We report a mutation screening of BBS4, the third of at least six BBS genes in the human genome, in a large, multiethnic patient cohort. As predicted from earlier genetic linkage/haplotype analyses, BBS4 is a minor contributor to BBS, since only 5/177 (∼2.8%) families harbor BBS4 mutations. The substantial size and diversity of the North American/European cohort that we studied suggest that BBS4 contributes less in these populations than in populations of Middle Eastern origin. Only 3/146 (2.05%) North American/European families carry at least one BBS4 mutation—compared to 2/31 (6.45%) Arab and Kurdish families (although the sample size of the latter is not large enough to draw statistically significant conclusions).
We also report that BBS4 mutations may be inherited in a complex, as well as autosomal recessive, pattern. Two families that harbor a single heterozygous BBS4 mutation could be excluded genetically from the inheritance of recessive BBS4 mutations, whereas the affected individual in a third family had inherited four mutations (a homozygous T558I mutation, in BBS2, and a homozygous A364E mutation, in BBS4). Unlike the BBS2 and BBS6 mutational pairs, in which at least one family segregated two null alleles at one locus and a third null allele at the second locus (Katsanis et al. 2001a), unequivocal establishment of triallelism for BBS4 is confounded by the fact that all non-Mendelian candidate susceptibility mutations detected in our screenings are missense. Other than the similarities between OGTs and BBS4 alterations and the predicted presence of a tetratricopeptide motif toward the amino-terminus of the BBS4 protein (Mykytyn et al. 2001), no functional data assist the evaluation of the effect of these BBS4 alterations. Both the conservation across phyla (including the evolutionarily remote insect and thermobacterial species) of the mutated residues and the absence of these alterations from control chromosomes support the hypothesis that these amino acid substitutions are pathogenic mutations. The probability that we have detected BBS4 carriers by chance alone is negligible. Given BBS's frequency of 1:125,000–1:150,000 and the ∼3% contribution of BBS4, the detection of three heterozygous carriers in 177 families represents a statistically significant deviation from the expected carrier frequency of 1:5900–1:6400 for BBS4 (χ2=299.1; P<10-4). These data therefore suggest that BBS4 may participate in complex inheritance with other BBS loci, such as BBS1 (in family AR153), BBS2 (in family PB043), and another, unmapped BBS locus (in family AR708). Nevertheless, this genetic model requires independent substantiation by the establishment of the biochemical and/or cellular effect(s) of these alterations.
Our mutation analyses of BBS4 also indicate that, in some instances, more than three mutant alleles may be required for the manifestation of disease. In family PB043, the affected individual (PB043-01) carries two homozygous mutations and is IBD across both the BBS2 and the BBS4 genomic regions. An unaffected sib (PB043-04) and the mother (PB043-07) are carriers for three mutations (two BBS2 mutations and one BBS4 mutation) and are clinically normal, thereby suggesting that a fourth mutation, in this instance, may be necessary and sufficient to cause the phenotype. If the BBS2 T558I allele is a benign polymorphism, then this family segregates the A364E allele in a Mendelian recessive fashion. Alternatively, however, perhaps the T558I mutation is mild, but its effect is unmasked and/or exacerbated by the presence of additional mutations. Such examples are likely to be rare, because any offspring has only a 1/16 chance to inherit all four mutant chromosomes; however, consanguinity would escalate that likelihood.
Assessment of complex inheritance in BBS is essential both to understand the molecular pathogenesis of this syndrome and to construct models for disorders that bridge Mendelian and complex traits. Our observation of potential multiallelism between BBS4 and two other known BBS loci, BBS1 and BBS2, may have mechanistic importance, although these pairings occur between BBS4 and the two most common loci, thereby suggesting that these associations may have been detected by chance alone. The two most plausible molecular explanations for the genetic phenomenon of triallelism are (1) that the different BBS proteins interact directly with each other and mutations in more than one member of a multisubunit complex are required for disease or (2) that the different BBS proteins operate in distinct but complementary pathways. Dysfunction of one pathway may be rescued by an alternative route, which, in turn, would be compromised by the occurrence of additional mutations. Regardless of which model may be shown to be supported by further experimentation, description of exclusive mutation pairings may assist in the elucidation of important functional aspects of the different BBS proteins.
Acknowledgments
We sincerely thank the families reported here for their willing and continued cooperation in these investigations. This study was supported in part by National Eye Institute, National Institutes of Health, grant EY12666 (to N.K.) and grants from the March of Dimes (to N.K. and J.R.L.), the Foundation Fighting Blindness (to R.A.L. and J.R.L.), the National Kidney Research Fund (to B.E.H.), the Research to Prevent Blindness (to R.A.L.), the Wellcome Trust (to P.L.B.), and the Birth Defects Foundation (to P.L.B.). R.A.L. is a Research to Prevent Blindness Senior Scientific Investigator; P.L.B. is an Advanced Wellcome Trust Fellow.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/ (for blastn)
- Expressed Sequence Tags Database, http://www.ncbi.nlm.nih.gov/dbEST/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BBS4 cDNA [accession number XM_027370] and M. thermautotrophicus genome [accession number NC_000916])
- Genome Database, The, http://www.gdb.org/
- Lupski Lab, The, http://www.imgen.bcm.tmc.edu/molgen/lupski/index.html (for primers)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BBS [MIM 209900])
- Whitehead Institute Center for Genome Research, http://www-genome.wi.mit.edu/
References
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA (1999) New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 36:437–446 [PMC free article] [PubMed] [Google Scholar]
- Beales PL, Katsanis N, Lewis RA, Ansley SJ, Elcioglu N, Raza J, Woods MO, Green JS, Parfrey PS, Davidson WS, Lupski JR (2001) Genetic and mutational analyses of a large multiethnic Bardet-Biedl cohort reveal a minor involvement of BBS6 and delineate the critical intervals of other loci. Am J Hum Genet 68:606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi R, Rokhlina T, Kwitek-Black AE, Elbedour K, Nishimura D, Stone EM, Sheffield VC (1995) Use of a DNA pooling strategy to identify a human obesity syndrome locus on chromosome 15. Hum Mol Genet 4:9–13 [DOI] [PubMed] [Google Scholar]
- Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O'Leary E, Pryse-Phillips W (1989) The cardinal manifestations of Bardet-Biedl syndrome, a form of Lawrence-Moon-Biedl syndrome. N Engl J Med 321:1002–1009 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR (2001a) Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293:2256–2259 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26:67–70 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Fisher EMC (1998) A novel C-terminal binding protein (CTBP2) is closely related to CTBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics 47:294–299 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Lewis RA, Stockton DW, Mai PM, Baird L, Beales PL, Leppert M, Lupski JR (1999) Delineation of the critical interval of Bardet-Biedl syndrome 1 (BBS1) to a small region of 11q13, through linkage and haplotype analysis of 91 pedigrees. Am J Hum Genet 65:1672–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Lupski JR, Beales PL (2001b) Exploring the molecular basis of Bardet-Biedl syndrome. Hum Mol Genet 10:2293–2299 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Yaspo M-L, Fisher EMC (1997) Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mamm Genome 8:526–529 [DOI] [PubMed] [Google Scholar]
- Kwitek-Black AE, Carmi R, Duyk GM, Buetow KH, Eldebour K, Parvari R, Yandava CN, et al (1993) Linkage of Bardet-Biedl syndrome to chromosome 16q and evidence for non-allelic genetic heterogeneity. Nat Genet 5:392–396 [DOI] [PubMed] [Google Scholar]
- Leppert M, Baird L, Anderson KL, Otterud B, Lupski JR, Lewis RA (1994) Bardet-Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat Genet 7:108–112 [DOI] [PubMed] [Google Scholar]
- Losson R, Lacroute F (1979) Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA 76:5134–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10:865–874 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Carmi R, Kwitek-Black A, Rokhlina T, Nishimura D, Duyk GM, Elbedour K, Sunden SL, Stone EM (1994) Identification of a Bardet-Biedl syndrome locus on chromosome 3 and evaluation of an efficient approach to homozygosity mapping. Hum Mol Genet 3:1331–1335 [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG (2000) Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet 26:15–16 [DOI] [PubMed] [Google Scholar]
- Young TL, Penney L, Woods MO, Parfrey PS, Green JS, Hefferton D, Davidson WS (1999) A fifth locus for Bardet-Biedl syndrome maps to 2q31. Am J Hum Genet 64:900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]