Abstract
Children who fail to develop language normally—in the absence of explanatory factors such as neurological disorders, hearing impairment, or lack of adequate opportunity—are clinically described as having specific language impairment (SLI). SLI has a prevalence of ∼7% in children entering school and is associated with later difficulties in learning to read. Research indicates that genetic factors are important in the etiology of SLI. Studies have consistently demonstrated that SLI aggregates in families. Increased monozygotic versus dizygotic twin concordance rates indicate that heredity, not just shared environment, is the cause of the familial clustering. We have collected five pedigrees of Celtic ancestry that segregate SLI, and we have conducted genomewide categorical linkage analysis, using model-based LOD score techniques. Analysis was conducted under both dominant and recessive models by use of three phenotypic classifications: clinical diagnosis, language impairment (spoken language quotient <85) and reading discrepancy (nonverbal IQ minus non-word reading >15). Chromosome 13 yielded a maximum multipoint LOD score of 3.92 under the recessive reading discrepancy model. Simulation to correct for multiple models and multiple phenotypes indicated that the genomewide empirical P value is < .01. As an alternative measure, we also computed the posterior probability of linkage (PPL), obtaining a PPL of 53% in the same region. One other genomic region yielded suggestive results on chromosome 2 (multipoint LOD score 2.86, genomic P value <.06 under the recessive language impairment model). Our findings underscore the utility of traditional LOD-score–based methods in finding genes for complex diseases, specifically, SLI.
Introduction
Specific language impairment (SLI) is clinically defined as failure to develop language normally, given adequate environment for learning language and the absence of hearing deficits, mental retardation, oral motor/structural abnormalities, and neurological or psychiatric impairments affecting language acquisition. This disorder affects ∼7% of children entering school (Tomblin et al. 1997), and, although some children will successfully learn to compensate as adults, many do not (Bishop and Adams 1990; Stothard et al. 1998). Individuals with SLI tend to perform poorly on general assessments of language and reading (Reed 1989; Bishop and Adams 1990; Catts 1993; Snowling et al. 2000, 2001). Research also indicates that many, but not all, have difficulty with higher level phonological processing necessary for the development of both language and reading and also demonstrate concomitant difficulties in processing dynamic (rapidly changing) sensory information within a very brief time range (Tallal and Piercy 1973a, 1973b; Wright et al. 1997, 2000; Witton et al. 1998; Talcott et al. 1999). Many children identified early in life as having SLI will subsequently develop characteristics of dyslexia when entering school (Bishop and Adams 1990; Scarborough 1990; Catts 1993; Stothard et al. 1998; Snowling et al. 2000).
Familial aggregation studies, twin studies, and prospective studies, taken together, suggest that SLI has a genetic component. Several case-control familial aggregation studies of SLI have been reported (Tallal et al. 1989, 2001; Tomblin 1989; van der Lely and Stollwerck 1996; Rice et al. 1998; for review of all familial aggregation studies, see Stromswold 1998). In studies of this type, incidence of SLI will vary depending on the definition of affection status. However, all of the above studies show a significantly increased frequency of impairment in first-degree relatives in families containing a proband (18%–42%) versus control families (3%–26%)
Genetic influences on language delay, a risk factor for SLI, were examined in two-year-old children in a sample of 3,000 twins (Dale et al. 1998). By consideration of both the variance of the control group and the variance of the group with language delay as having separate distributions, genetic factors were found to contribute 73% of the variance for the group with language delay, compared with 25% when all individuals in the sample were considered together. This indicates that individuals with language delay may have some unique genetic component that influences language acquisition, as compared with the general population.
Twin studies using the categorical diagnosis of SLI demonstrate near 100% concordance for MZ twins and ∼50%–70% concordance for DZ twins (Bishop et al. 1995; Tomblin and Buckwalter 1998), indicating that SLI as defined by categorical affection status does have a genetic component. Another twin study systematically explored pre- and perinatal hazards in children with SLI (Bishop 1997). This report examined medical records for relationships with birth weight, Apgar scores, and other obstetrical factors. Although no decisive evidence was found for an association between any of these factors and SLI, suggestive associations of SLI with toxemia of pregnancy, and hypertension were reported. Although it appears that these types of environmental factors are not essential in the development of SLI, this does not exclude the importance of other interactions with the environment over the course of development.
Prospective studies that compare infants who have a positive family history of reading and language problems with infants who have a negative family history may help in the identification of very early stages of the abnormal phenotype. One example of this kind of work used auditory temporal processing measures in infants (Benasich and Tallal 1996). Temporal processing, the ability to discriminate rapid and successive frequency changes in brief intervals, correlates with later language outcomes in infants who have a family history of language problems (Benasich and Spitz 1998). These studies indicate that even before expressive language has developed into easily recognizable words, children at genetic risk for difficulties in learning language perform differently, as a group, from control children on sensory-processing measures that may subsequently be important for phonological development.
Recently, a genome scan for SLI-susceptibility loci performed by use of quantitative-trait analyses based on a sib-pair design was completed by use of a combined clinical/epidemiological sample (SLI Consortium 2002). The authors found genomewide suggestive evidence for loci on 16q and 19q before correction for multiple phenotypes and tests. The locus on chromosome 16 was identified using a children’s test of phonological memory (Gathercole et al. 1994), whereas the locus on 19q was identified using an expressive language score. Although SLI and dyslexia have been postulated to be genetically related, this study did not find any evidence for linkage in regions previously implicated in dyslexia on chromosomes 2, 6, 15, or 18 (Cardon et al. 1994; Grigorenko et al. 1997; Fisher et al. 1999, 2002; Gayán et al. 1999). Furthermore, there was no evidence for linkage to 7q near FOXP2, a gene that is implicated in a severe speech impairment (Lai and Fisher et al. 2001) and is located in a region that may also include a major locus for autism (International Molecular Genetic Study of Autism Consortium 1998; Collaborative Linkage Study of Autism (CLSA) 2001a [originally published in 1999]).
The present study reports the results of a genome scan for SLI-susceptibility loci, using an extended family design. Three different phenotypic classifications were tested for linkage by use of traditional LOD-score–based methods and extensions to this basic approach. Although complex diseases, such as SLI, do not segregate in an apparent Mendelian framework, parametric analysis using a single-locus model has been shown to be an effective method for detection of linkage to oligogenic disorders (for a review of the relevant literature, see, e.g., Vieland 1998; Hodge 2001).
Subjects and Methods
Families and Phenotype Assignment
The sample consisted of branches of five Canadian families of Celtic ancestry that were originally identified during a linkage study of schizophrenia (Brzustowicz et al. 2000) and were noted to have a history of language or reading impairments. A total of 73 subjects were phenotyped with language/reading measures, and these plus 13 additional subjects (86 total) had DNA available. The largest family (n=34 phenotypes and DNA) was not directly part of the schizophrenia study, because they are related to a branch of a family segregating schizophrenia only by a marriage, which should preclude any subject for this study from sharing a schizophrenia locus by descent. A speech/language pathologist screened families, by telephone interview, for a history of language impairment segregating in the family.
Families with a strong history of language impairment were scheduled for assessment. All subjects received a comprehensive battery of tests administered by an experienced tester in their own homes. Assessment tools included the following:
-
1.
The age-appropriate version of the Test of Language Development, which is a comprehensive test of language functioning that addresses specific subtypes of language processes, including comprehension, expression, syntax, grammar, and phonology (either the Test of Adolescent Language [TOAL:2; see Hammill et al. 1987], the Test of Language Development–Primary, 2nd edition [TOLDP:2] [see Newcomer and Hammill 1988], or the Test of Language Development–Intermediate, 2nd edition [TOLDI:2] [see Hammill and Newcomer 1988]).
-
2.
Performance portions of the age-appropriate Wechsler Intelligence Test: either the Wechsler Intelligence Scale for Children (WISC) (Wechsler 1974), the Wechlser Intelligence Scale for Adults (WAIS) (Wechsler 1981), the Wechsler Preschool and Primary Scale of Intelligence (WIPPSI) (Wechsler 1989), or the Wechsler Abbreviated Scale for Intelligence (WASI) (Wechsler 1999).
-
3.
Self report or parental report questionnaire to assess history of hearing difficulties.
-
4.
Word Indentification (single word reading) and Word Attack (single non-word reading) subtests from the Woodcock Reading Mastery Test (Woodcock 1987).
-
5.
The age-appropriate version of the Token Test, which measures a subject’s ability to perform increasingly complex directions (DiSimoni 1978; for the modified version for adults, see Tomblin et al. 1992; for the adult version standardized to the children’s scale, see Tallal et al. 2001).
-
6.
Test of diadochokinesis from the Oral Speech Mechanism Screening Examination (St. Louis and Ruscello 1987), to assess oral structure and motor function.
Subjects were classified as an SLI proband if they met the following inclusionary/exclusionary criteria:
-
1.
Spoken Language Quotient Standard Score (SLQ) ⩽85 on the age-appropriate version of the Test of Language Development.
-
2.
Performance Intelligence Quotient (PIQ) ⩾80 on the age-appropriate version of the Wechsler Intelligence Test, as well as PIQ ⩾ SLQ.
-
3.
Hearing within normal limits (no history of recurrent ear infection or abnormal hearing screen) as assessed by self-report or parental report questionaire.
-
4.
No motor impairments or oral structural deviations affecting speech or non-speech movement of the articulators.
-
5.
No comorbid diagnosis of autism, schizophrenia, psychoses, or neurological disorders.
After all family members who agreed to participate were tested, families were included in the study if at least two members met the criteria for an SLI proband. All subjects were enrolled and tested after giving informed consent that conformed to the guidelines for treatment of human subjects approved by Rutgers University.
Three diagnostic classifications of impairment were employed. The classifications were not mutually exclusive; an individual subject could meet the criteria for more than one of the classifications that follow (see table 1 for the extent of overlap). A subject was classified as language impaired if his or her SLQ was ⩽85. A subject was classified as reading impaired if his or her single nonword reading score (word attack) was 1 SD below their performance IQ (reading discrepancy score). Finally, a subject was classified as clinically impaired if one or more of the following three criteria were met:
-
1.
The subject was language impaired, defined by SLQ ⩽85, or the subject was reading impaired, defined by word identification and/or word attack ⩽85.
-
2.
The subject’s overall SLQ was >85, but the subject scored 1 SD below the mean (⩽7) on three individual subtests of TOLD or scored ⩽85 on the Token Test. This criterion is designed to identify adults who have compensated for their deficit but still show residual language difficulty.
-
3.
The subject had a history of language difficulty defined by at least 2 years of speech/language therapy and/or reading intervention with the label of “dyslexic.”
Table 1.
Overlap Between Phenotypic Classifications
| Phenotypea | n |
| LI only | 0 |
| RI only | 4 |
| CI only | 11 |
| CI+LI | 18 |
| CI+RI | 6 |
| LI+RI+CI | 7 |
LI = language impairment, RI = reading impairment, and CI = clinical impairment. Note that, by definition, individuals classified as LI will also be CI, but the opposite is not necessarily true. The CI-only group represents individuals who were identified by low subtest scores or self-reported history (as outlined in the “Subjects and Methods” section).
All individuals with schizophrenia or schizophrenia spectrum disorders (n=7) were coded as having an unknown phenotype. It was not necessary to exclude any subject from analysis because of mental retardation, abnormal hearing, or oral motor or structural defects.
Genotyping
All family members who were willing to submit DNA samples (n=86) were genotyped. DNA was extracted from peripheral blood samples by the GenePure system (Gentra Systems). Buccal-swab DNA was extracted by use of cell lysis buffer and incubation as described by Laird et al. (1991), followed by NH4OAc precipitation and suspension in tris ethylenediaminetetraacetic acid (TE). Genotyping was conducted in our laboratory and the laboratories of the Center for Inherited Disease Research (CIDR) at Johns Hopkins University in Baltimore. Initial genotyping of 381 markers from the Weber Screening Set, version 6.0, spanning the genome at an average spacing of 9 cM and average heterozygosity of 0.76 was conducted by CIDR by use of automated fluorescent microsatellite analysis (see the CIDR Web site for further details) on 69 subjects. Follow-up genotyping was performed in our laboratory with these and 17 additional subjects, as described elsewhere (Brzustowicz et al. 1997). Two additional markers on chromosome 13 (D13S1317 and D13S1306), one marker on chromosome 2 (D2S352), and one marker on chromosome 17 (D17S809), were also genotyped. PCR primers were ordered from Research Genetics as part of the Human Map Pairs set or were redesigned from the Genome Database locus sequence with the assistance of the Primer 3 program.
Statistical Analysis
Parametric analysis was performed with FASTLINK version 4.1P programs (Cottingham et al. 1993; Schäffer et al. 1994) and the LINKAGE version 5.2 programs (Lathrop and Lalouel 1984; Lathrop et al. 1984). The language impairment, reading impairment, and clinical impairment phenotypes were each analyzed under both a dominant and a recessive model of inheritance, for a total of six analyses. For the dominant models, penetrance for individuals with one or two copies of the susceptibility allele was set to 0.5. For the recessive models, penetrance for individuals with two copies of the susceptibility allele was set to 0.8, and penetrance for individuals with one copy of the susceptibility allele was set to 0.01. For both dominant and recessive models, the penetrance of individuals with no susceptibility alleles was set to 0.001. The disease-allele frequency was set to 0.08 for the dominant model and 0.3 for the recessive model. These models correspond to an ∼7% rate of affection in the population, which is one estimate of the population prevalence of SLI (Tomblin et al. 1997). Although the parameters in our genetic models are almost certainly not correct, it has been demonstrated that use of arbitrary penetrance values with both dominant and recessive modes of inheritance provides a sufficiently powerful test for linkage in complex diseases (Greenberg et al. 1998; Abreu et al. 1999). Two-point linkage analysis was performed by use of the MLINK program, multipoint linkage analysis by use of the LINKMAP program, and heterogeneity testing by use of the HOMOG program. Marker allele frequencies were estimated by use of all available unrelated individuals. Recombination fractions (θ) between markers were taken from the Marshfield map supplied with the screening set. For additional markers, values of θ were also taken from the Marshfield map as D13S1317–.04–D13S800–.04–D13S1306; D2S405–.028–D2S352–.037–D2S1788; D17S2180–.064–D17S809–.057–D17S1290. Haplotypes were generated by a Markov chain–Monte Carlo approach using simulated annealing algorithms implemented in SimWalk2 version 2.82 (Sobel and Lange 1996). Files were analyzed several times by use of slightly different parameters and random number seeds to ensure convergence on a stable solution.
Simulations are useful to determine the proper significance of linkage results either when a sample is unique or when multiple correlated tests have been performed, as in this study. Empirical P values for the complete data set were obtained by simulation of 1,500 sets of 400 markers (representing a genome scan) not linked to a susceptibility gene generated by the SIMULATE program (Terwilliger and Ott 1994). Simulated markers had four alleles of equal frequency, for a heterozygosity of 0.75. Markers were analyzed by the program MSIM and were evaluated for heterogeneity by the program ElodHET. These programs were modified to accommodate analysis of six genetic models and report the maximum homogeneity LOD score and the maximum heterogeneity LOD score across all six models per simulated genome scan. The best homogeneity and heterogeneity LOD scores over each simulated genome scan were extracted and compiled into a single distribution. LOD scores from the real analysis were compared to this distribution from the simulated data sets, to see how often a given result would be expected by chance from an unlinked data set. This is reported as the empirical P value. Pedigree structures, phenotypic classifications, and genetic models were the same as those used in the actual analysis, but marker information was generated without regard to affection status.
To establish the statistical cutoff for follow-up analysis on the initial family set with additional pedigree members, 1,000 replicates were simulated under the assumption of no linkage for the initial sample and were analyzed under all six models, as described above. A LOD score of 1.74 was expected to occur by chance approximately once in every two genome scans, and this score was used as the cutoff for follow-up genotyping with the additional DNA samples.
We have also calculated the posterior probability of linkage (PPL), using the general form of Vieland (1998), which employs two-point LOD scores in lieu of constituent likelihoods (see also Wang et al. 1999, 2000; Vieland et al. 2001). The PPL differs from the LOD score, first, because it directly measures the probability that the genetic distance between the marker and a putative disease gene is <50 cM; and second, because it explicitly incorporates the prior distribution of θ, including the small prior probability of linkage between a trait gene and a random marker. Here we have used a prior probability of linkage of 2% (Elston 1975; Morton 1998) and have modeled the prior density of θ, given linkage, in terms of the random distance of a trait gene to its closest marker on a fixed marker map (Vieland et al. 2001).
We have also implemented a new feature in computing PPLs for the SLI data: rather than fixing the trait parameters at arbitrary values, we have included them as nuisance parameters in the model by assigning them independent uniform prior distributions and then integrating them out, to obtain a marginal posterior density in θ alone (see Appendix B of Vieland et al. 2001). The posterior marginal density of θ was approximated via direct numerical evaluation, by discretizing each parameter, computing two-point LOD scores at each possible combination of parameter values, and then averaging the resultant set of LOD scores (likelihoods) for each value of θ (M. W. Logue, unpublished data). The three penetrances (for the AA, Aa, and aa genotypes) were independently varied from 0 to 1, in increments of 0.10 (but the degenerate case of all penetrances being equal was skipped, and 0.999 was substituted for 1); θ was varied from 0 to .5 in increments of 0.01; and the admixture parameter (α) was varied from 0 to 1, in increments of 0.05. The grid for the disease-gene frequency was 0.001, 0.01, 0.1, 0.3, 0.5, and 0.8. The PPL was computed from the posterior marginal density of θ integrating over θ<.5 by numerical approximation, as described above.
Results
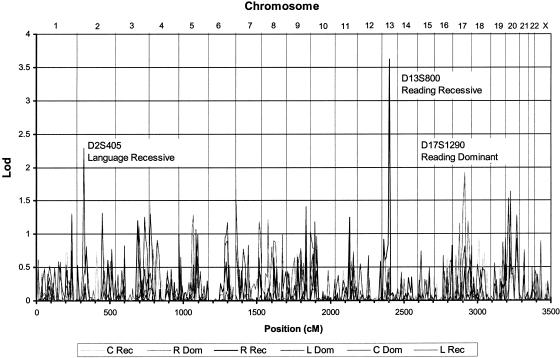
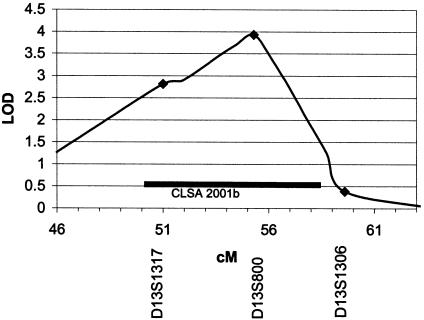
Two-point and three-point analysis was conducted on the initial subject set with follow-up genotyping on markers that produced multipoint heterogeneity LOD (HLOD) scores greater than 1.74. Figure 1 summarizes the two point results for all screening set markers and includes results of follow-up genotyping (see full table of LOD scores in online-only supplementonline-only supplement). After follow-up genotyping, three areas gave two point results over 1.74 (D13S800, 3.62, recessive reading discrepancy; D2S405, 2.28, recessive language impairment; D17S1290, 1.92, dominant reading discrepancy). Two additional markers flanking D13S800 (∼4 cM on each side) also gave positive two-point LOD scores under the recessive reading model (D13S1317, 2.99; D13S1306, 1.00). Four-point analysis done by use of D13S800 and these flanking markers produced a maximum LOD score of 3.92 at 0.9 cM telomeric to D13S800 (fig. 2). Haplotypes generated by SimWalk2 version 2.82 indicated recombination events occur in affected individuals between D13S788 and D13S1317, as well as between D13S800 and D13S1306. Because of the relatively large spacing between the adjacent markers used for this study, the placement of these flanking recombination events cannot be defined with much precision. On the basis of the Marshfield Comprehensive Human Genetic Map, the flanking recombination events are separated by a genetic distance of 4–14 cM. According to the December 2001 assembly of the Human Genome Project Working Draft, this corresponds to physical distance of 6.8–25.9 Mb.
Figure 1.
Maximum two-point heterogeneity LOD scores for all six models, summarized over the entire genome. The three highest peaks are labeled by marker and model. A list of two-point results for all markers and models is located in the online-only supplementonline-only supplement. C = clinical diagnosis; R = reading discrepant; L = language impaired; “Rec” and “Dom” are recessive and dominant modes of inheritance, respectively.
Figure 2.
Four-point analysis of the recessive reading model using markers D13S1317, D13S800, and D13S1306. This graph shows the overlap between our observed linkage and the CLSA (2001b; originally published as Bradford et al. 2001). The horizontal bar indicates the Zmax-1 interval from the CLSA data set.
Since our sample size is relatively small but does have complex pedigree structure and since our phenotypes are moderately correlated, the true false-positive rate may differ from that suggested by traditional guidelines. For a full assessment of the significance of our finding, 1,500 simulated genome scans of unlinked markers were tested under all six models, to determine the empirical significance level. A score ⩾3.92 occurred <1% of the time under homogeneity and heterogeneity analysis, indicating that our genomewide empirical significance level is P<.01.
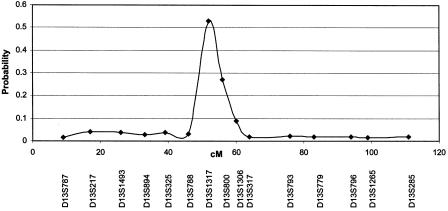
Figure 3 summarizes the PPL results over the length of chromosome 13 for linkage with reading discrepancy. D13S1317 gave a PPL of 0.53%, D13S800 gave a PPL of 27%, and D13S1306 gave a PPL of 9%. Markers that appeared to be unlinked on the basis of LOD scores failed to produce a PPL >0.045.
Figure 3.
PPL across the length of chromosome 13 when the reading discrepancy phenotype is used. Note the height of the linkage peak relative to the background level of linkage across the chromosome.
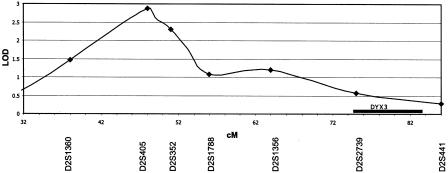
The region with the next highest LOD score was in 2p22 at marker D2S405 (θ=0) with a maximum two-point LOD score of 2.28 under the recessive language model. Markers near D2S405 produced positive LOD scores (1.57 at D2S352; 1.31 at D21788). Four-point analysis of the region that used these three markers gave a maximum LOD score of 2.79 at 0.05 cM distal to D2S405. Overlapping three-point analysis of this region is shown in figure 4. A LOD score of 2.79 corresponds to an empirical P value <.06 in our sample, as determined by simulation. PPL analysis of the markers was only very slightly higher than the prior probability of linkage (for D2S405, PPL = 0.058; for D2S352, PPL = 0.037; for D2S1788, PPL = 0.024). The other region exceeding our screening criteria had a peak near D17S1290 (θ=.01), with a maximum two-point LOD score of 1.92 under the dominant reading model after follow-up genotyping (PPL = 0.036 for reading impairment). Four-point analysis of the region including the two flanking markers (D17S2180 and D17S809), performed at ∼11 cM resolution, gave a maximum LOD score of 2.19, corresponding to an empirical P value of .20.
Figure 4.
Overlapping three-point analysis with the recessive language model of a selected region of chromosome 2. For the purposes of this graph, LOD scores were plotted by use of successive analyses anchored by each marker. The potential localization of DYX3 is shown by the black horizontal bar (Fagerheim et al. 1999).
Discussion
This study has demonstrated significant evidence for linkage between 13q21 and susceptibility to SLI, by use of a reading-based phenotype. Our analysis also suggests two additional loci, on 2p22 and 17q23, that may play a role in the overall phenotype associated with SLI. Although the families in this study were initially ascertained for schizophrenia, all persons with a diagnosis of schizophrenia (present in three of five families), were coded as phenotypically unknown. Furthermore, the largest pedigree in our data set, which contributed ∼50% of the linkage signal on 13q21 and 2p22, was connected to an original family with schizophrenia only by a marriage so the portion of the family in our data set should not segregate a schizophrenia locus by descent. As schizophrenia and SLI are two common disorders, it would be expected to find families that segregate both these disorders independently. Thus it seems most likely that our results relate to SLI and not schizophrenia.
The locus on 13q21 was identified using a reading discrepancy phenotype that might be considered a dyslexia phenotype. To date, no studies have strongly implicated 13q21 or the surrounding region in dyslexia. One possible explanation for this lack of overlap may lie within the ascertainment definitions of SLI and dyslexia. Traditionally, for a subject to be classified with dyslexia or specific reading disability, language (receptive and expressive grammar, syntax, and vocabulary) would have to be within the developmentally normal range. The diagnosis of SLI, however, requires difficulty in the acquisition of language skills that, by necessity, would make learning to read more difficult and thus reduce reading scores. Our reading phenotype in a population selected for SLI is most likely measuring the resultant reading outcome of an underlying language deficit as opposed to a reading deficit in isolation. It is interesting to note that reading discrepancy as a quantitative trait was found to have significant heritability (h2g=.46; SE=.15) in twins selected for reading disability (Pennington et al. 1992). Although the relationship between a similar, categorical reading discrepancy and SLI are still unclear, our data suggest that further work should be done to clarify why such a discrepancy shows utility for finding genes in SLI, whereas language-IQ discrepancy, as demonstrated by Bishop et al. (1995), does not (h2g=0–0.17±0.5–0.91 with four language measures in 90 twin pairs).
The 13q21 region has also been suggestively implicated in autism (MIM 209850) by the CLSA (2001a, 2001b [originally published in 1999, 2001]). When the authors of this study divided their autism sample in subsets, on the basis of language delay in the probands (onset of phrase speech >36 mo), and coded the parents as affected/unaffected according to questionnaire information on the parents’ history of language, reading, and spelling ability, the evidence for linkage was increased. Virtually all of the linkage signal came from the group with language-delayed probands. The maximum multipoint HLOD score was 2.54 at D13S800, the same marker where the peak LOD score occurs in the present study. Figure 2 shows the extent of overlap between our study and the study by the CLSA (2001b, originally published in 2001). Both autism and SLI appear to follow complex patterns of inheritance, so, if these disorders do share a common gene, then it would not be responsible for the entire phenotypic presentation of either disorder. Furthermore, since human disease genes have been documented to have many separate mutations, even implication of the same gene in both disorders does not necessarily imply the same molecular mechanism (Noone and Knowles 2001). A recent report by Kjelgaard and Tager-Flusberg (2001) indicates that a subset of children with autism show language deficits that are very similar to SLI. Since SLI is a common disorder, genes that negatively modulate language may be segregating independently from autism but still having an impact on the phenotypic presentation of that disorder. The use of special populations with circumscribed deficits may prove useful for the elucidation of other complex disorders, through determination of the genetic mechanisms for phenotypic components, such as the language component of autism.
PPL analysis has been used to further characterize our higher LOD scores. The PPL differs from the HLOD in a few notable respects. First, it already has incorporated into it the small prior probability of linkage (which is not used in calculation of the HLOD, although it is often invoked as a consideration in judgment of whether the observed HLOD is large enough to constitute strong evidence). Second, it incorporates a prior probability distribution for θ (again, the HLOD does not; whereas the PPL is integrated over θ, the HLOD is maximized over it). Third, whereas the HLOD has been calculated under the assumption of a particular trait model, the PPL is written as a function of the parameters of a (marginal) single-locus trait model, including admixture, and then these parameters are integrated out of the final statistic (so, again, the PPL is integrated over the trait parameter-space, whereas the HLOD, in this case, is taken at two particular fixed points in that space, one dominant and one recessive). Although all of these features introduce some differences between the statistics, it is this last item that is most likely to explain the fact that the HLODs maximize at a slightly different location than does the PPL. Localization on the basis of PPL would indicate that the disease gene is closer to D13S1317 than to D13S800, in contrast to the multipoint results. Calculation of the PPL is based solely on two-point analysis, whereas multipoint analysis has the ability to recover some of the power lost to marker uninformativeness, which is especially important in small data sets. Since the two localizations differ only by <5 cM, it is reasonable to conclude that the 13q21 region is implicated in the etiology of SLI, but further analysis will be required to refine the location estimate in this sample.
The PPL has an additional advantage over LOD scores in that it can be interpreted directly as a measure of the probability of linkage, given the data. A 53% PPL means that, more likely than not, there is a susceptibility gene at this location, and failure to follow up on this result is likely to miss a valid finding. It is important to note as well that the prior distribution for all nuisance parameters was taken to be uniform, which is likely to be conservative. The uniform distribution for penetrance will weight a model that has a 90% phenocopy rate equally with one that has a very low phenocopy rate. However, a 90% phenocopy rate would not be consistent with the behavioral genetics of SLI, which suggests a strong genetic component and therefore would indicate use of a very small prior distribution for such unlikely models. Yet, definition of the genetically likely parameter space is not a straightforward undertaking, and thus the observed PPL can be considered conservative. Figure 3 graphically demonstrates the signal-to-noise ratio of the PPL across chromosome 13, which indicates both the conservative nature of the statistic and the relative strength of a 53% PPL.
Our findings on 2p22 are plotted in figure 4, with the location of the dyslexia locus, DYX3, indicated (Fagerheim et al. 1999). The maximum LOD score in our sample is ∼40 cM from the location of DYX3. Since 40 cM approaches the approximate limit of detectable linkage, it is very unlikely that our locus represents linkage to DYX3. The PPLs in this region were also low (3.7% and 2.4%) but were higher than the prior probability of linkage. Thus, it would be premature to entirely exclude this locus for SLI. The locus on 2p22 was suggested only with the language impairment model, which is analogous to the 13q21 locus being found only under the reading impairment model. Although the two models do correlate to a modest extent (see table 1), they are not derived from the same phenotypic measures and are likely to have somewhat differential sensitivities for the respective domains of reading and language, which may account for our results. Further, the clinical impairment classification may have been unconservative in that it would increase the phenocopy rate for either language or reading impairments, since it also included self-reported history information, which—although increasing sensitivity to detect compensated adults—may not be as reliable as direct testing (Tallal et al. 2001).
This study has used a population with Celtic ancestry to map genes that either are involved in a limited but distinct disruption of language acquisition (SLI) or represent part of the lower tail of normal variation in language ability. There remains an unresolved controversy in the field of language learning with regard to the exact etiology of SLI (Aram 1991; Johnston 1991; Leonard 1991; Bishop 1994, 2001; Fitch et al. 1997; Dale et al. 1998; Leonard 1998). This debate may be resolvable, within limits, once the gene at 13q21 and other SLI-susceptibility genes are successfully found and characterized.
Lai and Fisher et al. (2001) recently cloned the FOXP2 gene (MIM 606354) at the SPCH1 locus (MIM 602081) responsible for a more severe and complex speech/language abnormality. The presumably causal gene was shown to have a point mutation in one extended pedigree (the KE family) and not in 364 ethnically matched controls. The only other case this group reports resulted from a disruption of FOXP2 due to a de novo translocation event. As a result, the rather unique phenotype of the KE kindred (severe oral motor dyspraxia and, in some cases, very low IQ), which encompassed more than the isolated language deficits defining SLI, does not appear to be part of normal genetic variation in language ability. Conducting linkage studies in samples that show only language and reading deficits may prove to be more useful in further definition of the etiology of SLI and may illuminate molecular mechanisms that are part of normal genetic variation.
The present study differs from the genome scan by the SLI Consortium (2002) in several ways. The SLI Consortium presented suggestive evidence for linkage on 16q and 19q, whereas our study did not. Such differences may reflect the statistical difficulty of the replication of loci or may reflect locus heterogeneity. It is possible that susceptibility alleles within the fairly homogeneous sample of Celtic ancestry that we studied (although it was not a population isolate) do not segregate within the United Kingdom as a whole with great enough frequency for the SLI Consortium study to detect the 13q21 locus, particularly because nuclear families do not provide very much power to detect admixture. Furthermore, the statistical approach used by the SLI Consortium was based solely on quantitative genetic analysis. Although quantitative traits in sib pairs/nuclear families overcomes certain constraints inherent to research of disorders based on quantitative measures and have been proven useful by the SLI Consortium’s suggestion of SLI loci on chromosomes 16 and 19, our study represents another demonstration of the utility of using categorical phenotypes with the traditional LOD-score–based method for implication of loci in complex diseases. It is important to keep in mind that different statistical methods require different assumptions and require different data structures/dependencies. This study indicates that using categorical techniques in extended pedigrees may reveal important genetic factors in disease etiology that could potentially be missed by sib-pair analysis.
Acknowledgments
We would like to thank the participating families, who contributed their time and patience to make this study possible; Linda Hirsch, Teresa Realpe, and Jason Nawyn, for managing the phenotype database in Newark; Jared Hayter, for technical assistance in the laboratory; the experienced testers associated with the laboratory of Paula Tallal; and Dawn Waterworth, for useful comments on earlier revisions. This research was funded by research grant 12-FY98-0008 from the March of Dimes Birth Defects Foundation (support to L.M.B.) and the National Alliance for Autism Research (with research partner The Sidgmore Family Foundation; support to C.W.B. and L.M.B.). P.T., L.M.B., and J.F.F. were supported by National Institute on Deafness and Other Communication Disorders grant RO1 DC01654. V.J.V. is supported by National Institutes of Health grants K02-MH01432 and MH52841. M.W.L. is supported by National Institutes of Health grant 5 T32 MH14620 (support to Dr. Raymond Crowe). A.S.B. is supported by the Medical Council of Canada and The Bill Jefferies Schizophrenia Endowment Fund. Genotyping services were provided by CIDR. CIDR is fully funded through contract N01-HG-65403 from the National Institutes of Health to The Johns Hopkins University.
Supplemental Online-Only Table.
|
Clinical Impairment |
Reading Impairment |
Language Impairment |
|||||||||||
| Dominant |
Recessive |
Dominant |
Recessive |
Dominant |
Recessive |
||||||||
| Chromosomeand Marker | cM | LOD | θ | LOD | θ | LOD | θ | LOD | θ | LOD | θ | LOD | θ |
| Chromosome 1: | |||||||||||||
| D1S468 | 4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S2660 | 11 | .616 | .05 | 0 | .5 | .569 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S1612 | 16 | 0 | .5 | 0 | .5 | .287 | 0 | .0242 | 0 | 0 | .5 | .009 | .4 |
| D1S1597 | 30 | 0 | .5 | 0 | .5 | 0 | .5 | .0272 | 0 | 0 | .5 | .022 | .4 |
| D1S3669 | 37 | .0725 | 0 | 0 | .5 | .413 | 0 | 0 | .5 | .225 | 0 | 0 | .5 |
| D1S552 | 45 | .308 | .1 | 0 | .5 | .08 | .2 | .002 | .4 | 0 | .5 | 0 | .5 |
| D1S1622 | 57 | .478 | .05 | .0023 | .1 | .113 | .3 | .051 | .3 | 0 | .5 | 0 | .5 |
| D1S255 | 65 | .003 | .4 | 0 | .5 | 0 | .5 | .001 | .4 | 0 | .5 | 0 | .5 |
| D1S3721 | 73 | 0 | .5 | 0 | .5 | .239 | .1 | .002 | .3 | .111 | .1 | .028 | .3 |
| D1S2134 | 76 | .398 | .1 | .107 | .2 | 0 | .5 | .029 | .2 | .1512 | 0 | .298 | .1 |
| D1S3728 | 89 | 0 | .5 | 0 | .5 | .052 | 0 | .213 | .1 | .352 | 0 | .516 | .05 |
| D1S1665 | 102 | .093 | .3 | .048 | .3 | 0 | .5 | 0 | .5 | .123 | .1 | .326 | .2 |
| D1S1728 | 109 | .0313 | .05 | 0 | .5 | 0 | .5 | .013 | .3 | .0091 | 0 | .028 | .3 |
| D1S551 | 114 | .021 | .3 | 0 | .4 | 0 | .5 | .093 | .2 | .268 | 0 | .051 | .3 |
| D1S1588 | 126 | .5 | 0 | .0775 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S1631 | 137 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .005 | .4 | .012 | .4 |
| D1S3723 | 141 | .0163 | 0 | 0 | .5 | .176 | .1 | 0 | .5 | .0103 | 0 | 0 | .5 |
| D1S534 | 152 | .019 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .583 | .1 | .03 | .4 |
| D1S1653 | 164 | .5764 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | .333 | .2 | .005 | .4 |
| D1S1679 | 171 | .1687 | .05 | .005 | .4 | 0 | .5 | 0 | .5 | .254 | .2 | 0 | .5 |
| D1S1677 | 176 | 0 | .5 | .003 | .4 | 0 | .5 | 0 | .5 | .158 | .3 | .05 | .4 |
| D1S1619 | 189 | .0024 | 0 | .01 | .4 | .0001 | 0 | 0 | .5 | .069 | .2 | .011 | .4 |
| D1S1589 | 193 | .022 | .3 | .0293 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S518 | 203 | .538 | 0 | .717 | .1 | 0 | .5 | 0 | .5 | .374 | .2 | .228 | .3 |
| D1S1660 | 213 | .2475 | .1 | .718 | .2 | .139 | .1 | 0 | .5 | .45 | .2 | .181 | .3 |
| D1S1647 | 217 | 0 | .5 | 0 | .5 | .306 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| GATA124F08 | 227 | .303 | 0 | .1982 | .1 | .227 | 0 | 0 | .5 | .146 | .2 | .038 | .3 |
| D1S2141 | 234 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .231 | .2 | .192 | .3 |
| D1S549 | 240 | .01 | .3 | .153 | .3 | .129 | .05 | .746 | 0 | .435 | .2 | 1.3046 | 0 |
| D1S3462 | 248 | 0 | .5 | 0 | .5 | .262 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S235 | 255 | 0 | .5 | 0 | .5 | .0133 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S547 | 268 | .002 | .4 | .17 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D1S1609 | 275 | 0 | .5 | .159 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .035 | .4 |
| Chromosome 2: | |||||||||||||
| D2S2976 | 4 | 0 | .5 | 0 | .5 | 0 | .5 | .5264 | 0 | 0 | .5 | 0 | .5 |
| D2S2952 | 18 | .0492 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S1400 | 28 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S1360 | 38 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .088 | .3 | .417 | .2 |
| D2S405 | 48 | .015 | .3 | .0878 | .3 | .2172 | .5 | .061 | .3 | .38 | .1 | 2.285 | 0 |
| D2S1788 | 56 | .1206 | .1 | .076 | .3 | 0 | .5 | .2575 | .05 | .5 | .2 | .391 | .05 |
| D2S1356 | 64 | 0 | .5 | .0021 | .4 | 0 | .5 | 0 | .5 | .814 | .1 | .775 | .1 |
| D2S2739 | 74 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .209 | .2 | .452 | .1 |
| D2S441 | 87 | 0 | .5 | .003 | .4 | .042 | .4 | 0 | .5 | 0 | .5 | .051 | .3 |
| D2S1394 | 91 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .0472 | .05 |
| D2S1777 | 99 | 0 | .5 | .058 | .4 | .016 | .4 | 0 | .5 | 0 | .5 | .044 | .3 |
| D2S1790 | 103 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S2972 | 114 | 0 | .5 | .038 | 0 | .156 | 0 | .022 | .4 | 0 | .5 | 0 | .5 |
| D2S410 | 125 | 0 | .5 | .0231 | 0 | .006 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S1328 | 133 | .0052 | 0 | .719 | .1 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S1334 | 145 | .011 | .4 | .014 | .4 | 0 | .5 | .026 | .3 | 0 | .5 | 0 | .5 |
| D2S442 | 147 | .004 | .4 | .367 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .03 | .4 |
| D2S1399 | 152 | 0 | .5 | .0228 | 0 | .005 | .4 | .009 | .4 | 0 | .5 | 0 | .5 |
| D2S1353 | 165 | 0 | .5 | .045 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S1776 | 173 | .132 | .2 | .202 | 0 | 0 | .5 | 0 | .5 | 1.312 | 0 | 0 | .5 |
| D2S1391 | 186 | .441 | .1 | .684 | .05 | 0 | .5 | 0 | .5 | .0583 | 0 | .161 | 0 |
| D2S1384 | 200 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D2S2944 | 210 | 0 | .5 | .076 | .2 | .4643 | 0 | .606 | 0 | 0 | .5 | .096 | .3 |
| D2S434 | 216 | 0 | .5 | .325 | .1 | .5442 | 0 | .4098 | 0 | 0 | .5 | 0 | .5 |
| D2S1363 | 227 | 0 | .5 | .004 | .4 | .002 | .4 | .083 | .2 | 0 | .5 | .5838 | 0 |
| D2S427 | 237 | .778 | 0 | .667 | .05 | .0003 | 0 | 0 | .5 | .443 | .01 | .591 | 0 |
| D2S2968 | 252 | .028 | .3 | .013 | .4 | 0 | .5 | 0 | .5 | .014 | .4 | .012 | .4 |
| D2S125 | 261 | .003 | .4 | .013 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | .304 | .2 |
| Chromosome 3: | |||||||||||||
| D3S2387 | 6 | 0 | .5 | 0 | .5 | 0 | .5 | .008 | .4 | 0 | .5 | 0 | .5 |
| D3S1560 | 19 | .386 | 0 | .195 | .2 | .053 | 0 | .085 | 0 | .002 | .4 | 0 | .5 |
| D3S1304 | 22 | .064 | .1 | 0 | .5 | 0 | .5 | .111 | 0 | .0067 | 0 | .08 | 0 |
| D3S4545 | 26 | .094 | .2 | .0077 | .3 | 0 | .5 | 0 | .5 | .076 | .3 | 0 | .5 |
| D3S1259 | 37 | 0 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | .052 | .3 | .007 | .4 |
| D3S3038 | 45 | 0 | .5 | .168 | .2 | 0 | .5 | 0 | .5 | .456 | .1 | .128 | .3 |
| D3S2432 | 58 | .328 | .1 | .0125 | .1 | 0 | .5 | 0 | .4 | .0391 | .1 | .0991 | .05 |
| D3S1768 | 62 | 0 | .5 | .019 | .4 | .205 | .2 | .0721 | .2 | 0 | 0 | .826 | .05 |
| D3S2409 | 71 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S1766 | 79 | .009 | .4 | .006 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S4542 | 90 | .005 | .4 | .032 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S2406 | 103 | 0 | .5 | .0097 | .2 | .091 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S4529 | 112 | 0 | .5 | .005 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S2459 | 119 | 0 | .5 | 0 | .5 | .188 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S3045 | 124 | 0 | .5 | 0 | .5 | .058 | .2 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S2460 | 135 | 0 | .5 | 0 | .5 | .033 | .2 | .001 | .4 | 0 | .5 | 0 | .5 |
| D3S4523 | 138 | .0024 | 0 | .1068 | 0 | .043 | .2 | .131 | .2 | .0141 | .05 | .1003 | .2 |
| D3S1764 | 153 | .105 | .1 | .0141 | 0 | 0 | .5 | 1.201 | 0 | 0 | .5 | .0012 | 0 |
| D3S1744 | 161 | .031 | .3 | .0262 | .05 | .2954 | 0 | .648 | .1 | .024 | .4 | 1.117 | 0 |
| D3S1763 | 177 | .114 | .1 | .264 | .1 | 0 | .5 | .048 | .2 | .235 | 0 | .1802 | 0 |
| D3S3053 | 182 | 0 | .5 | 0 | .5 | .0083 | 0 | .1155 | 0 | .0382 | 0 | .311 | .1 |
| D3S2427 | 188 | .03 | .3 | .032 | .4 | .002 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| D3S1262 | 201 | 1.257 | 0 | .203 | .2 | .093 | .3 | .024 | .3 | .469 | .1 | .152 | .3 |
| D3S2398 | 209 | .911 | 0 | .661 | .01 | .15 | .1 | .087 | .2 | .246 | 0 | .1488 | .1 |
| D3S2418 | 216 | .768 | 0 | .08 | .3 | .115 | .3 | .012 | .3 | .0053 | 0 | .318 | .2 |
| D3S1311 | 225 | .164 | .2 | .13 | .3 | .001 | .4 | .032 | .3 | .306 | .1 | .163 | .3 |
| Chromosome 4: | |||||||||||||
| D4S403 | 26 | .734 | .05 | .508 | .1 | .346 | .1 | 0 | .5 | .127 | .2 | .164 | .2 |
| D4S2639 | 33 | .1849 | .2 | .1679 | 0 | .769 | 0 | .1882 | .1 | .03 | .4 | 0 | .5 |
| D4S2397 | 43 | .009 | .4 | 0 | .5 | .321 | .05 | 0 | .5 | .004 | .4 | 0 | .5 |
| D4S2632 | 51 | .603 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S1627 | 60 | .9102 | 0 | .8404 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S3248 | 73 | .6675 | .01 | .023 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S2367 | 78 | .384 | 0 | .019 | .3 | 0 | .5 | .17 | 0 | 0 | .5 | 0 | .5 |
| D4S3243 | 88 | .0476 | 0 | .3 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .013 | .4 |
| D4S2361 | 93 | .023 | .3 | .2973 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S1647 | 105 | .005 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S2623 | 114 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S2394 | 130 | .15 | .3 | .014 | .4 | .452 | 0 | .0043 | .2 | .38 | .1 | .242 | .2 |
| D4S1644 | 143 | 0 | .5 | .012 | .4 | .008 | .4 | 0 | .5 | .235 | 0 | .03 | .3 |
| D4S1625 | 146 | 0 | .5 | 0 | .5 | .025 | .3 | 0 | .5 | 0 | .5 | .036 | .3 |
| D4S1629 | 158 | .475 | 0 | 0 | .5 | 0 | .5 | .31 | .05 | 0 | .5 | .318 | 0 |
| D4S2368 | 168 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D4S2431 | 176 | .119 | .2 | .023 | .4 | 0 | .5 | .272 | .2 | .038 | .4 | 0 | .5 |
| D4S2417 | 182 | .106 | .1 | 0 | .5 | 0 | .5 | .025 | .2 | .114 | .2 | .023 | .4 |
| D4S408 | 195 | .18 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .0288 | 0 | 0 | .5 |
| D4S1652 | 208 | 0 | .5 | .0164 | 0 | .0004 | 0 | .117 | 0 | .0476 | 0 | .0511 | 0 |
| Chromosome 5: | |||||||||||||
| D5S2488 | 0 | 0 | .5 | 0 | .5 | .059 | 0 | 1 | 0 | 0 | .5 | .334 | .2 |
| D5S2849 | 8 | 0 | .5 | 0 | .5 | .046 | .2 | .025 | .3 | 0 | .5 | 0 | .5 |
| D5S2505 | 14 | .593 | .05 | .395 | .2 | 0 | .5 | .021 | .3 | .017 | .4 | .656 | .1 |
| D5S807 | 19 | .0346 | 0 | .3502 | 0 | 0 | .5 | .12 | .2 | .0555 | 0 | .4249 | 0 |
| D5S817 | 23 | 0 | .5 | .1 | .2 | 0 | .5 | .223 | .1 | 0 | .5 | 0 | .5 |
| D5S2845 | 36 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .012 | .4 | 0 | .5 |
| D5S2848 | 40 | 0 | .5 | .011 | .4 | .0011 | 0 | .114 | .2 | .022 | .4 | .006 | .4 |
| D5S1470 | 45 | 0 | .5 | .025 | .4 | 0 | .5 | .121 | .2 | 0 | .5 | .026 | .4 |
| D5S1457 | 59 | .0429 | 0 | .006 | .4 | 0 | .5 | 0 | .5 | .015 | .4 | .1052 | 0 |
| D5S2500 | 69 | 0 | .5 | .05 | .3 | .026 | .3 | 0 | .5 | 0 | .5 | .276 | .1 |
| D5S424 | 82 | .078 | .01 | .038 | .1 | .235 | 0 | .272 | 0 | .086 | .01 | 0 | .5 |
| D5S641 | 92 | .125 | .1 | .0429 | 0 | 1.083 | 0 | .085 | 0 | 0 | .5 | .0658 | 0 |
| D5S1725 | 98 | .178 | .1 | 1.016 | .05 | 1.284 | 0 | .3219 | 0 | .0498 | 0 | .5417 | 0 |
| D5S1503 | 108 | 0 | .5 | 0 | .5 | .001 | .4 | 0 | .5 | .09 | 0 | 0 | .5 |
| D5S1453 | 115 | .397 | .1 | .993 | 0 | .682 | 0 | 0 | .5 | .1406 | 0 | .6107 | 0 |
| D5S2501 | 117 | .3336 | .05 | .4989 | 0 | 0 | .5 | .002 | .3 | .3264 | 0 | 1.066 | 0 |
| D5S1505 | 130 | .664 | 0 | .2807 | 0 | .604 | 0 | 0 | .5 | 1.004 | 0 | .195 | .2 |
| D5S816 | 139 | .103 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .181 | .2 |
| D5S1480 | 147 | 0 | .5 | .002 | .4 | .05 | .3 | .039 | .3 | 0 | .5 | .434 | .2 |
| D5S820 | 160 | .0929 | 0 | .1802 | .05 | 0 | .5 | .004 | .4 | .0018 | 0 | 0 | .5 |
| D5S1471 | 172 | .0297 | .2 | .237 | .1 | 0 | .5 | 0 | .5 | .213 | 0 | .0253 | .05 |
| D5S1456 | 175 | 0 | .5 | .0016 | .3 | .002 | .4 | 0 | .5 | .0508 | 0 | 0 | .3 |
| D5S211 | 183 | .016 | .4 | .0857 | 0 | .116 | .1 | .015 | .2 | 0 | .5 | .0013 | 0 |
| D5S408 | 195 | 0 | .5 | 0 | .5 | .2647 | 0 | .0296 | .05 | 0 | .5 | 0 | .5 |
| Chromosome 6: | |||||||||||||
| F13A1 | 9 | .0062 | .01 | 0 | .5 | .0019 | .2 | .0137 | .2 | 0 | .5 | 0 | .5 |
| D6S2434 | 25 | .0011 | .3 | .0073 | 0 | 0 | .5 | 0 | .5 | .012 | .4 | 0 | .5 |
| D6S1959 | 34 | .004 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | .004 | .4 | 0 | .5 |
| D6S2439 | 42 | 0 | .5 | 0 | .5 | .004 | .3 | .003 | .3 | 0 | .5 | 0 | .5 |
| D6S2427 | 54 | .002 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | .002 | .4 | .031 | .4 |
| D6S1017 | 63 | 0 | .5 | 0 | .5 | 0 | .5 | .008 | .3 | 0 | .5 | 0 | .5 |
| D6S2410 | 73 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D6S1053 | 80 | 0 | .5 | 0 | .5 | 0 | .5 | .022 | .3 | 0 | .5 | 0 | .5 |
| D6S1031 | 89 | 0 | .5 | 0 | .5 | .099 | .2 | .2547 | 0 | 0 | .5 | .003 | .4 |
| D6S1056 | 103 | 0 | .5 | 0 | .5 | .088 | .3 | .128 | .2 | .009 | .4 | .02 | .4 |
| D6S1021 | 112 | 0 | .5 | 0 | .5 | .088 | .1 | .191 | .2 | 0 | .5 | 0 | .5 |
| D6S474 | 119 | 0 | .5 | .0881 | 0 | .003 | .4 | .406 | .05 | .941 | 0 | .819 | .1 |
| D6S1040 | 129 | .1336 | 0 | .4865 | 0 | 0 | .5 | .577 | .1 | .99 | 0 | .882 | .1 |
| D6S1009 | 138 | .075 | .2 | .147 | .3 | 0 | .5 | .155 | .2 | 1.175 | 0 | .903 | .1 |
| GATA184A08 | 146 | 0 | .5 | 0 | .5 | .1312 | 0 | .533 | .05 | .234 | .01 | .0108 | .3 |
| D6S2436 | 155 | .221 | .05 | .482 | .1 | .0123 | 0 | 0 | .5 | 0 | .5 | .058 | .3 |
| D6S1277 | 173 | .012 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D6S1027 | 187 | .088 | .3 | .024 | .4 | 0 | .5 | 0 | .5 | .167 | .2 | .197 | .3 |
| Chromosome 7: | |||||||||||||
| D7S3056 | 7 | .826 | .05 | 1.671 | 0 | 0 | .5 | .076 | .2 | .764 | .1 | 1.525 | 0 |
| D7S513 | 17 | .4321 | 0 | .7441 | 0 | 0 | .5 | 0 | .5 | .504 | .1 | .7377 | 0 |
| D7S3051 | 29 | .005 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | .454 | 0 | .1495 | 0 |
| D7S1802 | 33 | 0 | .5 | 0 | .5 | .146 | 0 | 0 | .5 | .007 | .4 | 0 | .5 |
| D7S1808 | 42 | .1398 | 0 | .0462 | .2 | 0 | .5 | 0 | .5 | .454 | .2 | .3048 | 0 |
| D7S817 | 50 | .346 | .1 | .174 | .2 | .012 | .4 | .013 | .4 | .356 | .2 | .419 | .05 |
| D7S2846 | 58 | .0096 | 0 | .071 | .2 | 0 | .5 | 0 | .5 | .771 | 0 | 0 | .5 |
| D7S1818 | 70 | .098 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .529 | .05 | .333 | .1 |
| D7S3046 | 79 | .0039 | 0 | .004 | .4 | 0 | .5 | 0 | .5 | .099 | .3 | .041 | .4 |
| D7S2204 | 91 | .829 | 0 | .096 | 0 | .0004 | 0 | .003 | .4 | .093 | .2 | 0 | .5 |
| D7S2212 | 95 | .0275 | 0 | .524 | 0 | 0 | .5 | 0 | .5 | .033 | .2 | .303 | 0 |
| D7S821 | 109 | 0 | .5 | .02 | .3 | 0 | .5 | 0 | .5 | .021 | .4 | .021 | .4 |
| D7S1799 | 114 | 0 | .5 | .003 | .4 | .183 | .2 | .002 | .4 | .033 | .4 | .024 | .4 |
| D7S3061 | 128 | 0 | .5 | 0 | .5 | .26 | .1 | 0 | .5 | .009 | .4 | 0 | .5 |
| D7S1804 | 137 | 0 | .5 | 0 | .5 | .174 | .2 | 0 | .5 | 0 | .5 | 0 | .5 |
| D7S1824 | 150 | 0 | .5 | 0 | .5 | .321 | 0 | .005 | .4 | 0 | .5 | 0 | .5 |
| D7S2195 | 155 | 0 | .5 | 0 | .5 | .175 | .1 | 0 | .5 | .004 | .4 | 0 | .5 |
| D7S3070 | 163 | 0 | .5 | 0 | .5 | 1.184 | 0 | .448 | 0 | .64 | .05 | 0 | .5 |
| D7S3058 | 174 | .078 | .2 | 0 | .5 | 1.012 | 0 | .375 | 0 | .221 | .2 | 0 | .5 |
| D7S559 | 182 | .004 | .4 | 0 | .5 | .431 | .05 | .0021 | .3 | .126 | .2 | 0 | .5 |
| Chromosome 8: | |||||||||||||
| D8S264 | 1 | .012 | .4 | 0 | .5 | .202 | .2 | .022 | .3 | 0 | .5 | 0 | .5 |
| D8S1469 | 16 | .007 | .3 | .0001 | .4 | .1 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1130 | 22 | .1274 | 0 | 0 | .5 | .1117 | 0 | .005 | .4 | .089 | .05 | 0 | .5 |
| D8S1106 | 26 | 0 | .5 | .014 | .4 | .4089 | 0 | 0 | .5 | 0 | .5 | .029 | .4 |
| D8S1145 | 37 | 0 | .5 | 0 | .5 | .348 | .05 | .115 | .2 | 0 | .5 | 0 | .5 |
| D8S136 | 44 | .193 | .2 | .0732 | 0 | 1.209 | 0 | .006 | .3 | 0 | .5 | 0 | .5 |
| D8S1771 | 50 | .106 | 0 | 0 | .5 | .061 | .05 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1477 | 60 | 0 | .5 | 0 | .5 | .372 | 0 | .1145 | 0 | 0 | .5 | 0 | .5 |
| D8S1110 | 67 | .024 | .3 | 0 | .5 | .806 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1113 | 78 | .018 | .4 | 0 | .5 | .473 | .05 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1136 | 82 | .3946 | 0 | 0 | .5 | .901 | 0 | .647 | 0 | .084 | .3 | 0 | .5 |
| D8S2324 | 94 | .027 | .3 | 0 | .5 | .873 | 0 | .161 | .1 | .195 | .2 | 0 | .5 |
| D8S1119 | 101 | .364 | .1 | .0004 | .4 | .418 | .05 | 0 | .5 | .5153 | 0 | 0 | .5 |
| GAAT1A4 | 110 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1132 | 119 | 0 | .5 | 0 | .5 | .2665 | 0 | 0 | .5 | .026 | .3 | 0 | .5 |
| D8S592 | 125 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1179 | 135 | 0 | .5 | 0 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D8S1128 | 140 | .023 | .2 | 0 | .5 | 1 | 0 | .131 | 0 | .456 | .1 | 0 | .5 |
| D8S256 | 148 | .0347 | .2 | .038 | .3 | 0 | .5 | 0 | .5 | .184 | .05 | 0 | .5 |
| D8S373 | 164 | .027 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .431 | 0 | 0 | .5 |
| Chromosome 9: | |||||||||||||
| D9S2169 | 14 | 0 | .2 | .459 | .01 | .133 | .1 | .007 | .4 | .136 | .2 | .434 | 0 |
| D9S921 | 22 | .002 | .5 | .016 | .5 | .42 | .5 | .185 | .5 | 0 | .5 | .297 | .5 |
| D9S925 | 32 | 0 | .5 | 0 | 0 | 0 | .05 | 0 | .2 | 0 | .5 | 0 | .5 |
| D9S1121 | 44 | 0 | .1 | .1518 | .2 | .383 | .01 | .07 | .5 | 0 | .2 | 0 | .5 |
| D9S1118 | 58 | .227 | .2 | .201 | .3 | .644 | .5 | 0 | .5 | .097 | 0 | 0 | .5 |
| D9S301 | 66 | .036 | .05 | .003 | .05 | 0 | 0 | 0 | .2 | .145 | .5 | 0 | .5 |
| D9S1122 | 76 | .184 | .2 | .704 | .4 | .604 | 0 | .138 | 0 | 0 | .1 | 0 | .01 |
| D9S922 | 80 | .425 | 0 | .47 | .05 | .508 | 0 | .657 | 0 | .502 | 0 | .585 | .01 |
| D9S283 | 95 | 0 | .5 | 0 | .5 | .673 | 0 | .017 | .4 | .143 | .1 | 0 | .5 |
| D9S910 | 104 | .534 | 0 | .442 | .1 | .784 | 0 | .01 | .3 | .732 | 0 | 0 | .5 |
| D9S938 | 111 | .285 | 0 | 0 | .5 | .394 | 0 | .073 | .3 | 0 | .5 | 0 | .5 |
| D9S930 | 120 | .462 | 0 | .3326 | 0 | .557 | .05 | .036 | .3 | .138 | .2 | .0595 | 0 |
| D9S934 | 128 | .047 | .2 | .0542 | .2 | 0 | .5 | 0 | .5 | .031 | .2 | 0 | .5 |
| D9S1825 | 136 | 1.412 | 0 | .3428 | .05 | .457 | .01 | .99 | 0 | .2383 | 0 | .751 | .1 |
| D9S2157 | 147 | .0481 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | .0091 | .2 | 0 | .5 |
| D9S1826 | 160 | 0 | .5 | 0 | .5 | 0 | .5 | .006 | .4 | 0 | .5 | 0 | .5 |
| D9S1838 | 164 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| Chromosome 10: | |||||||||||||
| D10S1435 | 4 | .407 | 0 | .533 | .2 | .01 | .4 | 0 | .5 | .394 | .1 | 1.029 | .1 |
| D10S189 | 19 | .157 | .2 | .695 | 0 | .607 | 0 | .2287 | 0 | .702 | 0 | .789 | .1 |
| D10S1412 | 28 | .167 | .2 | .277 | .2 | .599 | .1 | .777 | 0 | .183 | .1 | .787 | .05 |
| D10S2325 | 33 | 1.186 | 0 | .344 | .2 | .016 | .3 | .137 | .2 | .0433 | 0 | .303 | .1 |
| D10S1423 | 46 | 0 | .5 | 0 | .5 | .966 | 0 | .758 | 0 | .448 | 0 | .0443 | 0 |
| D10S1426 | 59 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S1208 | 63 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .04 | .3 |
| D10S1221 | 76 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .0012 | 0 | 0 | .5 |
| D10S1225 | 81 | .002 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| GATA121A08 | 88 | .062 | .3 | .0051 | .3 | 0 | .5 | .02 | .2 | .143 | .2 | 0 | .5 |
| D10S1432 | 94 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .028 | .4 | 0 | .5 |
| D10S2327 | 101 | .081 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | .059 | .4 | .041 | .4 |
| D10S2470 | 113 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S677 | 117 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S1239 | 125 | .033 | .3 | 0 | .5 | .45 | 0 | .07 | 0 | .005 | .4 | 0 | .5 |
| D10S1237 | 135 | 0 | .5 | .0021 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S1230 | 143 | 0 | .5 | 0 | .5 | .027 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S1213 | 148 | 0 | .5 | 0 | .5 | .017 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S217 | 158 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D10S212 | 171 | .319 | 0 | .1348 | 0 | .067 | .05 | .068 | .1 | 0 | .5 | 0 | .5 |
| Chromosome 11: | |||||||||||||
| D11S1984 | 2 | .014 | .4 | 0 | .5 | 0 | .5 | .122 | .2 | 0 | .5 | 0 | .5 |
| D11S2362 | 9 | .0003 | 0 | .028 | .4 | 0 | .5 | 0 | .5 | .213 | .2 | .064 | .3 |
| D11S1999 | 17 | 0 | .5 | .031 | .4 | 0 | .5 | .088 | .2 | .3 | .2 | .113 | .3 |
| D11S1981 | 21 | 0 | .5 | .023 | .4 | .0178 | 0 | .006 | .3 | .378 | .1 | .042 | .4 |
| ATA34E08 | 33 | .0555 | 0 | 0 | .5 | 0 | .5 | .007 | .4 | 0 | .5 | 0 | .5 |
| D11S1392 | 43 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D11S1344 | 58 | .196 | .2 | .452 | .4 | 0 | .5 | 0 | .5 | .305 | .1 | 0 | .5 |
| D11S2371 | 76 | 0 | .5 | 0 | .5 | 0 | .5 | .2 | .1 | .096 | .3 | 0 | .5 |
| D11S2002 | 85 | 0 | .5 | 0 | .5 | .021 | .4 | .02 | .4 | 0 | .5 | 0 | .5 |
| D11S2000 | 101 | .547 | .01 | .3651 | 0 | .739 | 0 | 1.246 | 0 | .033 | .3 | .574 | .1 |
| D11S1391 | 105 | 0 | .5 | 0 | .5 | .058 | .1 | .064 | .2 | .062 | .2 | .139 | .3 |
| D11S1998 | 113 | .1686 | 0 | 0 | .5 | .591 | 0 | .291 | .1 | .0601 | 0 | 0 | .5 |
| D11S4464 | 123 | .1517 | .1 | .209 | .2 | 0 | .5 | .0658 | 0 | .494 | 0 | .729 | 0 |
| D11S912 | 131 | 0 | .5 | 0 | .5 | .519 | .05 | 0 | .5 | .167 | .2 | .286 | .2 |
| D11S968 | 148 | .193 | 0 | .2124 | 0 | .04 | .3 | 0 | .5 | 0 | .5 | 0 | .5 |
| Chromosome 12: | |||||||||||||
| D12S372 | 6 | 0 | .5 | 0 | .5 | .012 | .4 | .02 | .4 | 0 | .5 | .0022 | .4 |
| GATA49D12 | 18 | 0 | .5 | .049 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | .452 | .2 |
| D12S391 | 26 | 0 | .5 | .151 | .3 | .15 | .05 | 0 | .5 | .05 | .3 | .551 | .2 |
| D12S373 | 36 | .0896 | 0 | .1548 | .1 | 0 | .5 | 0 | .5 | .0529 | 0 | .1474 | 0 |
| D12S1042 | 49 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .0018 | 0 | 0 | .5 |
| GATA91H06 | 56 | .009 | .4 | 0 | .5 | .0316 | .2 | 0 | .5 | 0 | .5 | 0 | .5 |
| D12S398 | 68 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .0607 | .2 |
| D12S1294 | 78 | .0988 | .2 | .2458 | 0 | 0 | .5 | 0 | .5 | .049 | .3 | .6717 | 0 |
| D12S375 | 81 | .002 | .4 | 0 | .5 | .015 | .3 | 0 | .5 | .066 | .3 | 0 | .5 |
| D12S1052 | 83 | .0766 | 0 | 0 | .5 | .339 | 0 | .0093 | 0 | .078 | .1 | 0 | .5 |
| D12S1064 | 95 | 0 | .5 | .036 | .3 | 0 | .5 | .016 | .4 | 0 | .5 | 0 | .5 |
| D12S1300 | 104 | 0 | .5 | 0 | .5 | .005 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| PAH | 109 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D12S2070 | 125 | 0 | .5 | 0 | .5 | .038 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| D12S395 | 137 | 0 | .5 | .0484 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D12S2078 | 150 | 0 | .5 | 0 | .5 | 0 | .5 | .007 | .4 | 0 | .5 | 0 | .5 |
| D12S1045 | 161 | 0 | .5 | 0 | .5 | 0 | .5 | .5869 | 0 | .048 | .4 | 0 | .5 |
| D12S392 | 166 | 0 | .5 | 0 | .5 | 0 | .5 | .0173 | 0 | .041 | .4 | 0 | .5 |
| Chromosome 13: | |||||||||||||
| D13S787 | 9 | 0 | .5 | 0 | .5 | .017 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| D13S217 | 17 | .005 | .3 | .193 | .01 | .744 | 0 | .915 | 0 | .018 | .2 | .0001 | .4 |
| D13S1493 | 26 | .147 | .05 | .0264 | .1 | .645 | 0 | .6034 | 0 | 0 | .5 | 0 | .5 |
| D13S894 | 33 | .01 | .4 | 0 | .5 | .203 | .2 | .647 | 0 | 0 | .5 | 0 | .5 |
| D13S325 | 39 | .1728 | .1 | .1936 | 0 | .284 | .2 | .725 | .05 | 0 | .5 | .0124 | 0 |
| D13S788 | 46 | .472 | .1 | 0 | .5 | .083 | .2 | .844 | 0 | .052 | .3 | .265 | .2 |
| D13S800 | 56 | .144 | .1 | 0 | .5 | .4999 | 0 | 3.624 | 0 | 0 | .5 | 0 | .3 |
| D13S317 | 64 | 0 | .5 | 0 | .5 | 0 | .5 | .077 | .2 | 0 | .5 | 0 | .5 |
| D13S793 | 76 | .005 | .4 | .013 | .4 | 0 | .5 | .112 | .2 | 0 | .5 | 0 | .5 |
| D13S779 | 83 | .1 | .2 | .089 | .3 | .01 | .3 | .011 | .4 | 0 | .5 | .0001 | .4 |
| D13S796 | 94 | 0 | .5 | 0 | .5 | .017 | .2 | 0 | .5 | 0 | .5 | 0 | .5 |
| D13S1265 | 99 | 0 | .5 | .0697 | .2 | 0 | .5 | 0 | .5 | .084 | .3 | .078 | .1 |
| D13S285 | 111 | 0 | .5 | .168 | .2 | .1838 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| Chromosome 14: | |||||||||||||
| D14S742 | 12 | 0 | .5 | 0 | .5 | 0 | .5 | .0161 | .3 | 0 | .5 | 0 | .5 |
| D14S1280 | 26 | .402 | .05 | .2977 | .01 | .43 | .05 | .153 | .2 | .0089 | 0 | 0 | .5 |
| D14S608 | 28 | 0 | .5 | .046 | .2 | .37 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D14S599 | 41 | .025 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | .017 | .4 | 0 | .5 |
| D14S306 | 44 | .067 | .3 | .006 | .4 | .323 | 0 | .023 | 0 | 0 | .5 | 0 | .5 |
| D14S587 | 56 | 0 | .5 | 0 | .5 | 0 | .5 | .101 | .2 | 0 | .5 | 0 | .5 |
| D14S592 | 67 | .0107 | .01 | 0 | .5 | .0255 | 0 | .005 | .4 | 0 | .5 | 0 | .5 |
| D14S588 | 76 | 0 | .5 | 0 | .5 | 0 | .5 | .3441 | 0 | 0 | .5 | .1154 | .2 |
| D14S53 | 86 | 0 | .5 | 0 | .5 | 0 | .5 | .157 | 0 | 0 | .5 | .004 | .4 |
| D14S606 | 92 | .011 | .3 | .039 | .3 | 0 | .5 | .1168 | .2 | 0 | .5 | .065 | .3 |
| GATA193A07 | 96 | 0 | .5 | 0 | .5 | .005 | .4 | .3533 | 0 | .003 | .4 | .019 | .4 |
| D14S617 | 106 | 0 | .5 | 0 | .5 | .005 | .4 | .035 | .4 | 0 | .5 | 0 | .5 |
| D14S1434 | 113 | 0 | .5 | .017 | .4 | 0 | .5 | .0024 | 0 | 0 | .5 | 0 | .5 |
| D14S1426 | 126 | .0057 | .3 | .0749 | 0 | .0478 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| Chromosome 15: | |||||||||||||
| D15S822 | 12 | .018 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D15S165 | 20 | .303 | 0 | 0 | .5 | .106 | 0 | .091 | .05 | 0 | .5 | 0 | .5 |
| D15S1012 | 36 | .017 | .3 | .023 | .3 | 0 | .5 | .02 | .1 | .005 | .4 | .735 | 0 |
| D15S659 | 43 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .116 | .2 | 0 | .5 |
| D15S643 | 52 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .016 | .4 | 0 | .5 |
| D15S1507 | 60 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .3091 | 0 | 0 | .5 |
| D15S980 | 72 | .144 | .2 | 0 | .5 | .306 | .05 | 0 | .5 | .17 | .2 | 0 | .5 |
| D15S655 | 83 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .043 | .4 | 0 | .5 |
| D15S652 | 90 | .357 | .1 | .298 | .2 | 0 | .5 | 0 | .5 | .495 | .05 | .031 | .4 |
| D15S816 | 101 | .0003 | .2 | .12 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D15S657 | 105 | 0 | .5 | .037 | .3 | 0 | .5 | 0 | .5 | .076 | .3 | 0 | .5 |
| D15S966 | 112 | 0 | .5 | .016 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | .051 | .2 |
| D15S642 | 121 | .022 | .3 | .353 | .2 | 0 | .5 | 0 | .5 | .001 | .4 | 0 | .5 |
| Chromosome 16: | |||||||||||||
| D16S2616 | 11 | 0 | .5 | 0 | .5 | .086 | 0 | .0033 | 0 | .0882 | 0 | .193 | 0 |
| D16S748 | 23 | 0 | .5 | .2414 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D16S764 | 30 | 0 | .5 | .0001 | .2 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D16S403 | 44 | 0 | .5 | 0 | .5 | 0 | .5 | .005 | .4 | 0 | .5 | 0 | .5 |
| D16S769 | 51 | 0 | .5 | 0 | .5 | .046 | .3 | 0 | .5 | 0 | .5 | 0 | .5 |
| D16S540 | 58 | 0 | .5 | .013 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D16S3396 | 64 | .021 | .4 | .081 | .3 | 0 | .5 | 0 | .5 | .092 | .2 | .673 | .1 |
| D16S3253 | 72 | 0 | .5 | 0 | .5 | .431 | .01 | .05 | .3 | 0 | .5 | 0 | .5 |
| D16S2620 | 81 | 0 | .5 | 0 | .5 | .099 | .1 | .039 | .2 | 0 | .5 | 0 | .5 |
| D16S2624 | 88 | 0 | .5 | .63 | 0 | .515 | 0 | .198 | 0 | 0 | .5 | .351 | 0 |
| D16S516 | 100 | .097 | .2 | .358 | .2 | .201 | 0 | 0 | .5 | 0 | .5 | .004 | .4 |
| D16S3091 | 111 | .592 | 0 | .33 | .1 | .023 | .4 | .035 | .3 | .0539 | .1 | .08 | .3 |
| D16S539 | 125 | .556 | 0 | .015 | .3 | .581 | 0 | .824 | 0 | 0 | .5 | 0 | .5 |
| D16S2621 | 130 | .0029 | 0 | .292 | .1 | .1019 | 0 | .3937 | 0 | .075 | .2 | .083 | .3 |
| Chromosome 17: | |||||||||||||
| D17S1308 | 1 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D17S1298 | 11 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D17S974 | 22 | .152 | .2 | 0 | .5 | .685 | 0 | .054 | .2 | .051 | .3 | 0 | .5 |
| D17S1303 | 24 | .054 | .3 | 0 | .5 | .1337 | 0 | 0 | .5 | 0 | .5 | 0 | .5 |
| D17S969 | 28 | .366 | 0 | .371 | 0 | 0 | .5 | 0 | .5 | .187 | 0 | .176 | .05 |
| D17S2196 | 45 | .019 | .4 | 0 | .5 | 1.152 | 0 | .039 | .3 | .004 | .4 | 0 | .5 |
| D17S975 | 51 | 0 | .5 | 0 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .4 |
| D17S1293 | 56 | 0 | .5 | 0 | .5 | .604 | 0 | .263 | 0 | 0 | .5 | 0 | .5 |
| D17S1299 | 62 | 0 | .5 | .194 | .2 | .81 | 0 | .002 | .4 | .39 | .05 | .284 | .2 |
| D17S2180 | 67 | .069 | .2 | 0 | .5 | 1.368 | 0 | .055 | 0 | .602 | .05 | .12 | .1 |
| D17S1290 | 82 | 0 | .5 | 0 | .5 | 1.922 | .01 | .496 | 0 | .816 | 0 | 0 | .5 |
| D17S2193 | 89 | 0 | .5 | 0 | .5 | .181 | .1 | .055 | .4 | 0 | .5 | 0 | .5 |
| D17S1301 | 100 | .137 | .2 | .413 | .1 | 1.178 | 0 | .085 | .1 | .419 | .1 | .079 | .3 |
| D17S784 | 117 | .678 | 0 | .296 | .1 | .6709 | 0 | .007 | .4 | .495 | 0 | .066 | .2 |
| D17S928 | 126 | .237 | .2 | .119 | .2 | .277 | .2 | .201 | .2 | .05 | .3 | .826 | .05 |
| Chromosome 18: | |||||||||||||
| GATA178F11 | 3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D18S976 | 13 | .151 | .2 | .0501 | 0 | .099 | .2 | .25 | .1 | .0572 | 0 | .3514 | 0 |
| D18S843 | 28 | .0042 | 0 | 0 | .5 | 0 | .5 | .0006 | .4 | 0 | .5 | 0 | .5 |
| D18S542 | 41 | .005 | .4 | .595 | .1 | .002 | .4 | .103 | 0 | .1585 | 0 | .536 | .1 |
| D18S877 | 54 | .023 | .3 | .92 | .1 | 0 | .5 | 0 | .5 | .052 | 0 | .307 | .2 |
| D18S535 | 64 | .0281 | 0 | .152 | .2 | 0 | .5 | 0 | .5 | .451 | 0 | .1025 | 0 |
| D18S851 | 75 | .14 | .2 | .641 | .1 | 0 | .5 | 0 | .5 | .0914 | 0 | .4932 | 0 |
| D18S858 | 80 | .126 | .2 | .355 | .2 | .004 | .4 | .0392 | .1 | .0007 | 0 | .452 | .2 |
| D18S862 | 89 | .012 | .3 | .6984 | 0 | .002 | .4 | 0 | .5 | .011 | .3 | .452 | .1 |
| D18S1364 | 99 | 0 | .5 | .0388 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| ATA82B02 | 107 | 0 | .5 | .4442 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | .094 | .3 |
| D18S1371 | 116 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .039 | .3 | 0 | .5 |
| D18S70 | 126 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| Chromosome 19: | |||||||||||||
| D19S591 | 10 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .059 | .4 | .0136 | .2 |
| D19S1034 | 21 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D19S586 | 33 | .556 | 0 | .41 | 0 | 0 | .5 | 0 | .5 | .243 | 0 | .025 | .3 |
| D19S714 | 42 | .016 | 0 | .17 | .3 | 0 | .5 | 0 | .5 | .1933 | 0 | .313 | .2 |
| D19S433 | 52 | 0 | .5 | 0 | .5 | 0 | .5 | .005 | .4 | 0 | .5 | 0 | .5 |
| D19S245 | 59 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D19S178 | 68 | .324 | .05 | .529 | .1 | 0 | .5 | 0 | .5 | .083 | .2 | .1 | .3 |
| D19S246 | 78 | 0 | .5 | .0177 | .3 | 0 | .5 | .274 | 0 | 0 | .5 | 0 | .5 |
| D19S589 | 88 | .874 | .05 | .588 | .2 | 0 | .5 | .1997 | 0 | .1175 | 0 | .1168 | .05 |
| D19S254 | 101 | .2067 | 0 | .3065 | 0 | .007 | .4 | 0 | .5 | .288 | .1 | .5383 | 0 |
| Chromosome 20: | |||||||||||||
| D20S103 | 2 | .518 | .05 | .16 | .2 | .5608 | 0 | 0 | .5 | .933 | 0 | 0 | .5 |
| D20S482 | 12 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .293 | 0 | 0 | .5 |
| D20S851 | 25 | .1709 | .1 | 0 | .5 | .018 | .4 | 0 | .5 | 1.531 | 0 | .032 | .4 |
| D20S604 | 33 | .852 | 0 | 1.266 | 0 | 0 | .5 | 0 | .5 | .3322 | 0 | .4561 | 0 |
| D20S470 | 39 | .689 | .1 | .51 | .1 | .005 | .4 | .009 | .3 | .539 | .05 | 1.64 | 0 |
| D20S477 | 48 | .388 | .1 | .259 | .2 | .013 | .4 | .0001 | .4 | .4579 | 0 | .4196 | 0 |
| D20S478 | 54 | .384 | .05 | .515 | .2 | 0 | .5 | 0 | .5 | .496 | 0 | .392 | .2 |
| D20S481 | 62 | 0 | .5 | .059 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| D20S480 | 80 | .915 | .05 | 1.228 | .1 | .017 | .4 | 0 | .5 | .893 | 0 | 1.278 | .1 |
| D20S171 | 96 | .1482 | 0 | .2114 | 0 | .004 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| Chromosome 21: | |||||||||||||
| D21S1432 | 3 | .0024 | .3 | .011 | .4 | 0 | .5 | 0 | .5 | .038 | .3 | .127 | .3 |
| D21S1437 | 13 | 0 | .5 | 0 | .5 | 0 | .5 | .0136 | 0 | .052 | .3 | 0 | .5 |
| D21S2052 | 25 | 0 | .5 | 0 | .5 | .003 | .4 | .001 | .4 | 0 | .5 | 0 | .5 |
| D21S1440 | 37 | 0 | .5 | .07 | .2 | 0 | .5 | 0 | .5 | .378 | 0 | .755 | 0 |
| D21S2055 | 40 | 0 | .5 | 0 | .5 | 0 | .5 | .0022 | 0 | 0 | .5 | 0 | .5 |
| D21S1446 | 58 | 0 | .5 | .0695 | .2 | .028 | .2 | .118 | .1 | 0 | .5 | 0 | .5 |
| Chromosome 22: | |||||||||||||
| D22S420 | 4 | .0353 | 0 | .1392 | 0 | 0 | .5 | 0 | .5 | .037 | .4 | .079 | .4 |
| MFD313 | 19 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | .003 | .4 | 0 | .5 |
| D22S689 | 29 | .011 | .4 | .011 | .4 | .016 | .4 | .111 | .2 | .163 | .2 | .055 | .3 |
| D22S685 | 32 | .0571 | 0 | .0193 | .01 | 0 | .5 | 0 | .5 | .496 | 0 | .0454 | .1 |
| D22S683 | 36 | .007 | .4 | 0 | .5 | .023 | .4 | .1768 | 0 | .159 | .3 | 0 | .5 |
| D22S445 | 46 | .0432 | 0 | .0434 | 0 | .0003 | 0 | .101 | .1 | .294 | .1 | .0156 | 0 |
| Chromosome X: | |||||||||||||
| DXS9900 | 4 | .112 | .1 | .49 | .1 | .32 | 0 | .0093 | 0 | 0 | .5 | .4469 | .1 |
| DXS9895 | 16 | 0 | .5 | .2048 | .1 | 0 | .5 | 0 | .5 | .0439 | 0 | .3789 | 0 |
| DXS9902 | 22 | .026 | .4 | .039 | .4 | 0 | .5 | 0 | .5 | .387 | 0 | .049 | .3 |
| DXS9896 | 31 | .0023 | 0 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| DXS1068 | 37 | 0 | .5 | 0 | .5 | .009 | .4 | 0 | .5 | 0 | .5 | 0 | .5 |
| DXS6810 | 43 | .885 | 0 | .0908 | .1 | 0 | .5 | 0 | .5 | .0277 | 0 | .018 | .4 |
| GATA144D04 | 47 | .1472 | 0 | .38 | .2 | 0 | .5 | 0 | .5 | .1825 | .1 | .095 | .3 |
| DXS7132 | 53 | 0 | .5 | .183 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| DXS6800 | 58 | 0 | .5 | .0165 | 0 | 0 | .5 | 0 | .5 | .006 | .4 | 0 | .5 |
| DXS6789 | 63 | .001 | .4 | .166 | .3 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| DXS6797 | 67 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| GATA172D05 | 69 | 0 | .5 | .027 | .4 | 0 | .5 | 0 | .5 | 0 | .5 | 0 | .5 |
| GATA165B12 | 77 | .086 | .3 | .259 | .3 | 0 | .5 | 0 | .5 | .008 | .4 | .025 | .4 |
| DXS1047 | 82 | .013 | .4 | .219 | .3 | .056 | .3 | 0 | .5 | .023 | .4 | .016 | .4 |
| CXS318 | 88 | .1232 | 0 | .0525 | 0 | 0 | .5 | 0 | .5 | .126 | 0 | .0929 | 0 |
| DXS9908 | 93 | 0 | .5 | 0 | .5 | .031 | .2 | 0 | .5 | .189 | .1 | .287 | .01 |
| DXYS154 | 105 | 0 | .5 | 0 | .5 | .005 | .3 | 0 | .5 | 0 | .5 | 0 | .5 |
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- CIDR, http://www.cidr.jhmi.edu/ (for genotyping protocols)
- Genome Database, http://www.gdb.org/ (for initial STS sequences used to redesign primers)
- Human Genome Project Working Draft, http://genome.ucsc.edu
- Marshfield Medical Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for order and genetic distances of microsatellite markers)
- National Center for Biotechnology Information (NCBI) Genetic Analysis Software, ftp://fastlink.nih.gov/pub/fastlink/ (for FASTLINK versions of MLINK and LINKMAP)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autistic disorder [MIM 209850], FOXP2 [MIM 606354], and SPCH1 [MIM 602081])
- Primer 3, http://www-genome.wi.mit.edu/genome_software/other/primer3.html (for designing primers)
- Rockefeller University, ftp://linkage.rockefeller.edu/software/ (for SIMULATE, MSIM, ElodHet, HOMOG, and linkage utility programs)
- University of Pittsburgh, http://watson.hgen.pitt.edu/register/soft_doc.html (for SimWalk2 version 2.82)
References
- Abreu PC, Greenberg DA, Hodge SE (1999) Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet 65:847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aram DM (1991) Comments on specific language impairment as a clinical category. Lang Speech Hear Serv School 22:84–87 [Google Scholar]
- Benasich AA, Spitz RV (1998) Insights from infants: temporal processing abilities and genetics contribute to language development. In: Willems G, Whitmore K (eds) A neurodevelopmental approach to specific learning disorders. MacKeith Press, London, pp 191–210 [Google Scholar]
- Benasich AA, Tallal P (1996) Auditory temporal processing thresholds, habituation, and recognition memory over the first year. Infant Behav Dev 19:339–357 [Google Scholar]
- Bishop DVM (1994) Is specific language impairment a valid diagnostic category? Genetic and psycholinguistic evidence. Philos Trans R Soc Lond B Biol Sci 346:105–111 [DOI] [PubMed] [Google Scholar]
- ——— (1997) Pre- and perinatal hazards and family background in children with specific language impairments: a study of twins. Brain Lang 56:1–26 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Genetic and environmental risks for specific language impairment in children. Philos Trans R Soc Lond B Biol Sci 356:369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DVM, Adams C (1990) A prospective study of the relationship between specific language impairment, phonological disorders and reading retardation. J Child Psychol Psychiatry 31:1027–1050 [DOI] [PubMed] [Google Scholar]
- Bishop DVM, North T, Donlan C (1995) Genetic basis of specific language impairment: evidence from a twin study. Dev Med Child Neurol 37:56–71 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EWC, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-1q22. Science 288:678–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow WC, Hogan J, Hodgkinson K, Bassett AS (1997) Use of a quantitative trait to map a locus associated with severity of positive symptoms in familial schizophrenia to chromosome 6p. Am J Hum Genet 61:1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed] [Google Scholar]
- Catts HW (1993) The relationship between speech-language and reading disabilities. J Speech Hear Res 36:948–958 [DOI] [PubMed] [Google Scholar]
- CLSA (2001a) An autosomal genomic screen for autism. Am J Med Genet 105:805; originally published as Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- CLSA (2001b) Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet 105:805; originally published as Bradford Y, Haines J, Hutcheson H, Gardiner M, Braun T, Sheffield V, Cassavant T, et al (2001) Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet 105:539–547 [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Dale PS, Simonoff E, Bishop DVM, Eley TC, Oliver B, Price TS, Purcell S, Stevenson J, Plomin R (1998) Genetic influence on language delay in two-year-old children. Nat Neurosci 1:324–328 [DOI] [PubMed] [Google Scholar]
- DiSimoni F (1978) The Token Test for children. Teaching Resources, Boston [Google Scholar]
- Elston RC (1975) The prior probability of autosomal linkage. Ann Hum Genet 38:341–350 [DOI] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tønnessen FE, Pedersen M, Tranebjærg L, Lubs HA (1999) A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet 36:664–669 [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Franks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Olson RK, Pennington BF, Smith SD, DeFries JC, Stein JF, Monaco AP (2002) Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet 30:86–91 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lanb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch RH, Miller S, Tallal P (1997) Neurobiology of speech perception. Annu Rev Neurosci 20:331–353 [DOI] [PubMed] [Google Scholar]
- Gathercole SE, Willis CS, Baddeley AD, Emslie H (1994) The Children's test of nonword repetition: a test of phonological working memory. Memory 2:103–127 [DOI] [PubMed] [Google Scholar]
- Gayán J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL (1997) Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet 60:27–39 [PMC free article] [PubMed] [Google Scholar]
- Hammill DD, Brown VL, Larsen SC, Wiederhodl JL (1987) Test of Adolescent Language—2. Pro-Ed, Austin, TX [Google Scholar]
- Hammill DD, Newcomer PL (1988) Test of Language Development—2, Intermediate. Pro-Ed, Austin, TX [Google Scholar]
- Hodge SE (2001) Model-free vs. model-based linkage analysis: a false dichotomy? Am J Med Genet 105:62–64 [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- Johnston JR (1991) The continuing relevance of cause: a reply to Leonard’s “Specific language impairment as a clinical category.” Lang Speech Hear Serv School 22:75–79 [Google Scholar]
- Kjelgaard MM, Tager-Flusberg H (2001) An investigation of language impairment in autism: implications for genetic subgroups. Lang Cognit Process 16:287–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 [DOI] [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A (1991) Simplified mammalian DNA isolation procedure. Nucleic Acids Res 19:4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus analysis in humans. Proc Natl Acad Sci 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard LB (1991) Comments on specific language impairment as a clinical category. Lang Speech Hear Serv School 22:84–87 [Google Scholar]
- ——— (1998) Children with specific language impairment. MIT Press, Cambridge, MA [Google Scholar]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62:690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer PL, Hammill DD (1988) Test of language development–2, Primary. Pro-Ed, Austin, TX [Google Scholar]
- Noone PG, Knowles MR (2001) “CFTR-opathies”: disease phenotypes associated with cystic fibrosis transmembrane regulator gene mutations. Respir Res 2:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Gilger JW, Olson RK, DeFries JC (1992) The external validity of age- versus IQ-discrepancy definitions of reading disability: lessons from a twin study. J Learn Disabil 25:562–573 [DOI] [PubMed] [Google Scholar]
- Reed MA (1989) Speech perception and the discrimination of brief auditory cues in reading disabled children. J Exp Child Psychol 48:270–292 [DOI] [PubMed] [Google Scholar]
- Rice ML, Honey KJ, Wexler K (1998) Family histories of children with SLI who show extended optional infinitives. J Speech Lang Hear Res 41:419–432 [DOI] [PubMed] [Google Scholar]
- Scarborough H, Dobrich W (1990) Development of children with early language delay. J Speech Hear Res 33:70–83 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW Jr (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- SLI Consortium (2002) A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet 70:384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowling M, Adams JW, Bishop DV, Stothard SE (2001) Educational attainments of school leavers with a preschool history of speech-language impairments. Int J Lang Commun Disord 36:173–183 [DOI] [PubMed] [Google Scholar]
- Snowling M, Bishop DVM, Stothard SE (2000) Is preschool language impairment a risk factor for dyslexia in adolescence? J Speech Lang Hear Res 41:587–600 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- St. Louis KO, Ruscello DM (1987) Oral speech mechanism screening examination. Pro-Ed, Austin, TX [Google Scholar]
- Stothard SE, Snowling MJ, Bishop DVM, Chipchase BB, Kaplan CA (1998) Language impaired preschoolers: a follow-up into adolescence. J Speech Lang Hear Res 41:407–418 [DOI] [PubMed] [Google Scholar]
- Stromswold K (1998) Genetics of spoken language disorders. Hum Biol 70:297–324 [PubMed] [Google Scholar]
- Talcott JB, Witton C, McClean M, Hansen PC, Rees A, Green GG, Stein JF (1999) Can sensitivity to auditory frequency modulation predict children's phonological and reading skills? Neuroreport 10:2045–2050 [DOI] [PubMed] [Google Scholar]
- Tallal P, Hirsch LS, Realpe-Bonilla T, Miller S, Brzustowicz LM, Bartlett CW, Flax JF (2001) Familial aggregation in specific language impairment. J Speech Lang Hear Res 44:1172–1182 [DOI] [PubMed] [Google Scholar]
- Tallal P, Piercy M (1973a) Defects of non-verbal auditory perception in children with developmental aphasia. Nature 241:468–469 [DOI] [PubMed] [Google Scholar]
- ——— (1973b) Developmental aphasia: impaired rate of non-verbal processing as a function of sensory modality. Neuropsychologia 11:389–398 [DOI] [PubMed] [Google Scholar]
- Tallal P, Ross R, Curtiss S (1989) Familial aggregation in specific language impairment. J Speech Hear Disord 54:167–173 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. Johns Hopkins University Press, Baltimore [Google Scholar]
- Tomblin JB (1989) Familial concentration of developmental language impairment. J Speech Hear Disord 54:287–295 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Buckwalter PR (1998) Heritability of poor language achievement among twins. J Speech Lang Hear Res 41:188–199 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Freese PR, Records NL (1992) Diagnosing specific language impairment in adults for the purpose of pedigree analysis. J Speech Hear Res 35:832–844 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O’Brien M (1997) Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res 40:1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lely H, Stollwerck K (1996) A grammatical specific language impairment in children: an autosomal dominant inheritance? Brain Lang 52:484–504 [DOI] [PubMed] [Google Scholar]
- Vieland VJ (1998) Bayesian linkage analysis, or: how I learned to stop worrying and love the posterior probability of linkage. Am J Hum Genet 63:947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieland VJ, Wang K, Huang J (2001) Power to detect linkage based on multiple sets of data in the presence of locus heterogeneity: comparative evaluation of model-based linkage methods for ASP data. Hum Hered 51:199–208 [DOI] [PubMed] [Google Scholar]
- Wang K, Huang J, Vieland VJ (1999) A Bayesian approach to replication of linkage studies. Genet Epidemiol Suppl 17:S749–S754 [DOI] [PubMed] [Google Scholar]
- ——— (2000) The consistency of the posterior probability of linkage. Ann Hum Genet 64:533–553 [DOI] [PubMed] [Google Scholar]
- Wechsler D (1974) Wechsler intelligence scale for children—revised. Psychological Corporation, San Antonio [Google Scholar]
- ——— (1981) Wechsler adult intelligence scale—revised. Psychological Corporation, San Antonio [Google Scholar]
- ——— (1989) Wechsler pre-school and primary scale of intelligence—revised. Psychological Corporation, San Antonio [Google Scholar]
- ——— (1999) Wechsler abbreviated scale of intelligence. Psychological Corporation, San Antonio [Google Scholar]
- Witton C, Talcott JB, Hansen PC, Richardson AJ, Griffiths TD, Rees A, Stein JF, Green GG (1998) Sensitivity to dynamic auditory and visual stimuli predicts nonword reading ability in both dyslexic and normal readers. Curr Biol 8:791–797 [DOI] [PubMed] [Google Scholar]
- Woodcock RW (1987) Woodcock reading mastery tests—revised. American Guidance Service, Circle Pines, MN [Google Scholar]
- Wright BA, Bowen RW, Zecker SG (2000) Nonlinguistic perceptual deficits associated with reading and language disorders. Curr Opin Neurobiol 10:482–486 [DOI] [PubMed] [Google Scholar]
- Wright BA, Lombardino LJ, King WM, Puranik CS, Leonard LM, Merzenich MM (1997) Deficits in auditory temporal and spectral resolution in language-impaired children. Nature 387:176–178 [DOI] [PubMed] [Google Scholar]