Abstract
Variants of different Class I alcohol dehydrogenase (ADH) genes have been shown to be associated with an effect that is protective against alcoholism. Previous work from our laboratory has shown that the two sites showing the association are in linkage disequilibrium and has identified the ADH1B Arg47His site as causative, with the ADH1C Ile349Val site showing association only because of the disequilibrium. Here, we describe an initial study of the nature of linkage disequilibrium and genetic variation, in population samples from different regions of the world, in a larger segment of the ADH cluster (including the three Class I ADH genes and ADH7). Linkage disequilibrium across ∼40 kb of the Class I ADH cluster is moderate to strong in all population samples that we studied. We observed nominally significant pairwise linkage disequilibrium, in some populations, between the ADH7 site and some Class I ADH sites, at moderate values and at a molecular distance as great as 100 kb. Our data indicate (1) that most ADH-alcoholism association studies have failed to consider many sites in the ADH cluster that may harbor etiologically significant alleles and (2) that the relevance of the various ADH sites will be population dependent. Some individual sites in the Class I ADH cluster show Fst values that are among the highest seen among several dozen unlinked sites that were studied in the same subset of populations. The high Fst values can be attributed to the discrepant frequencies of specific alleles in eastern Asia relative to those in other regions of the world. These alleles are part of a single haplotype that exists at high (>65%) frequency only in the eastern-Asian samples. It seems unlikely that this haplotype, which is rare or unobserved in other populations, reached such high frequency because of random genetic drift alone.
Introduction
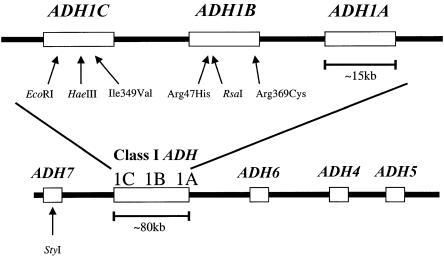
The seven known alcohol dehydrogenase (ADH) genes encode enzymes that catalyze the conversion of alcohols to aldehydes. All seven genes exist in a cluster extending ∼380 kb on the long arm of chromosome 4 (i.e., 4q21-23 [GenBank accession numbers AP002026, AP002027, AP002028, and AC097530]). The Class I ADH genes (ADH1A [MIM 103700], ADH1B [MIM 103720], and ADH1C [MIM 103730]) exist in a tighter cluster of ∼77 kb, flanked upstream by ADH7 (MIM 600086) and downstream by ADH6 (MIM 103735), ADH4 (MIM 103740), and ADH5 (MIM 103710), in that order (fig. 1). Although the greatest similarity seen is among the Class I genes, all seven ADH enzymes are very similar in amino acid sequence and structure but differ in preferred substrates (Edenberg 2000). Two of the three Class I genes are known to have alleles that produce enzymes that catalyze the oxidation of ethanol at different rates (Edenberg and Bosron 1997).
Figure 1.
Relative map of SNP sites studied. A detailed map of the Class I cluster is expanded above the map of the gene cluster as a whole. In the map of the whole gene cluster, the names of the Class I ADH genes are abbreviated to “1A,” “1B,” and “1C,” for “ADH1A,” “ADH1B,” and “ADH1C,” respectively.
A note on nomenclature is warranted. Allelic nomenclature based on the protein differences is not adequate for analyses of individual polymorphic sites in the genomic sequence of the gene cluster. For example, at the protein level, the allelic series for ADH1B (previously called “ADH2”) is generated by variation at two different sites at the genomic level: the ADH1B*1 allele is composed of 47Arg and 369Arg, the ADH1B*2 allele is composed of 47His and 369Arg, and the ADH1B*3 allele is composed of 47Arg and 369Cys. We have not seen the “double variant” (composed of 47His and 369Cys), but we assume that it could exist. For our present purposes, we consider the sites separately.
The functional variants in the corresponding metabolic enzymes make the Class I ADH genes obvious candidates for risk of developing alcoholism. Alleles at two ADH genes that encode enzymes with higher Vmax values—namely, ADH1B*47His (previously called “ADH2*2”), at the Arg47His (exon 3) SNP, and ADH1C*349Ile (previously called “ADH3*1”), at the Ile349Val (exon 8) SNP—have consistently been found at significantly lower frequencies in alcoholic individuals than in nonalcoholic controls in eastern-Asian samples (Thomasson et al. 1991; Chen et al. 1996; Shen et al. 1997; Tanaka et al. 1997; Osier et al. 1999; Li et al. 2001). Also, in a genomewide linkage study in families mostly of European ancestry, the Collaborative Studies on Genetics of Alcoholism (COGA) group (Reich et al. 1998) found evidence that supports the genetic linkage between alcoholism and the region of chromosome 4 that includes the ADH genes. Long et al. (1998) also found evidence, in a sample of an Amerindian population, that supports the genetic linkage between alcohol dependence and a nearby region on chromosome 4.
The two functional variants are ∼21 kb apart and are in strong linkage disequilibrium. Earlier, we demonstrated, through analysis of haplotype frequencies, that the association with alcoholism is caused by the allelic variation at the ADH1B Arg47His site and that the decreased frequency of the ADH1C*349Ile allele in samples of Taiwanese Chinese alcoholic individuals, relative to the frequency in control subjects, is most likely due to linkage disequilibrium with the ADH1B*47His allele (Osier et al. 1999). Similar haplotype-based analyses will be important for future work that seeks to resolve the role that polymorphisms at the various ADH genes play in protection against alcoholism. Such haplotype analyses must be preceded by the discovery and characterization of new polymorphisms.
The frequencies of ADH alleles are distinctly different in various populations. ADH1B*47His is present at frequencies >0.33 in eastern-Asian populations and <0.25 in populations from other regions (Goedde et al. 1992; Thomasson et al. 1994; Neumark et al. 1998). The low frequency, outside eastern Asia, of the commonly studied ADH1B*47His allele makes it an unlikely contributor, in those regions, to protection against alcoholism, but other polymorphisms in the same or other ADH genes may be important. For example, the ADH1C*349Ile allele (previously called “ADH3*2”) is found in Europeans (e.g., see Edman and Maret 1992; Poupon et al. 1992), and an expressed variant in exon 9 of ADH1B (Cys for Arg at codon 369; previously called “ADH2*3”) is found in African American samples (Bosron et al. 1979; 1983; Carr et al. 1989).
Linkage disequilibrium in the ADH gene family has also been shown to exist for other sites. Murray et al. (1987) found evidence of linkage disequilibrium between StuI and XbaI polymorphic sites in ADH1C in a sample of European Americans. Edman and Maret (1992) examined linkage disequilibrium, in samples of Swedes and mixed European Americans, between sites in ADH1B, ADH1C, ADH4, and ADH5 by use of the measure Δ. They found significant linkage disequilibrium between sites in the two Class I genes and between ADH4 and ADH5. They did not, however, find significant evidence of linkage disequilibrium between the Class I sites and either the ADH4 site or the ADH5 site, possibly because of the small size of the samples that they analyzed. Through analysis of Swedish nuclear families, Edman and Maret (1992) estimated haplotype frequencies and found that 10 of 16 possible haplotypes were present in a sample of 33 chromosomes.
To increase our understanding of both genetic diversity and the nature and extent of linkage disequilibrium in the ADH cluster, we have examined a large number of sites and populations. We have genotyped individuals from 40 populations for seven SNPs—including the two commonly studied coding polymorphisms, another previously described coding polymorphism ADH1B Arg369Cys (Carr et al. 1989), a novel ADH1C HaeIII (exon 5) site silent SNP, the ADH1B RsaI (intron 3) site SNP (Smith 1986; Osier et al. 1999), a novel ADH1C EcoRI (intron 2) site SNP, and a novel ADH7 StyI (intron 5) site SNP. All sites were chosen because they can be typed by PCR and digestion with inexpensive, readily available, restriction enzymes. We have estimated the haplotype frequencies for all six Class I ADH sites, and we have examined linkage disequilibrium among the sets of five and six Class I ADH sites and across the segment between the ADH7 (intron 5) SNP and all six Class I ADH sites. Patterns of linkage disequilibrium are strong within the Class I cluster, but linkage disequilibrium between the ADH7 site and the Class I sites varies among populations. This suggests that studies of ADH and alcoholism should evaluate the effect linkage disequilibrium has in the larger ADH gene family on both a site-by-site and population-by-population basis.
Material and Methods
Samples
The populations studied at Yale are those previously studied for the ADH1B RsaI (intron 3) site (Osier et al. 1999) plus samples of several new populations, noted briefly here. Additional cell lines from Druze and Ethiopian Jews were obtained from the National Laboratory for the Genetics of Israeli Populations (Tel Aviv University, Israel). The Nigerian samples were collected by Prof. Friday Okonofua, Dr. Frank Oronsaye, and Dr. Adekunle Odunsi: most of the Yoruba samples were from urban health care workers from Benin City; the Hausa samples were from Zaria, in north-central Nigeria; and the Ibo samples were from Enugu, in eastern Nigeria. The Kachari sample was collected by Dr. Ranjan Deka from the Assam province, in India. The Irish sample primarily consists of individuals from Roscommon County, in Ireland, and was collected by Prof. Kenneth Kendler and his colleagues in Ireland. The African American sample consists of cell lines that were purchased from the National Institute of General Medical Sciences (NIGMS) Human Genetic Cell Repository (Corielle Institute for Medical Research). The African American cell lines are among those listed by the NIGMS as being from the African American panel and, as such, may represent a broad cross section of African Americans. The samples of southeastern Bantu-speakers and !Kung San were provided by H.S. Both samples consist of unrelated individuals; the southeastern Bantu-speakers are urban South Africans with ancestry involving various native groups, primarily groups that speak different languages from the Bantu family. The Northern Moroccan, Central Moroccan, Saharan, Catalan, and Basque samples were collected by J.B. and D.C. (Comas et al. 2000; Bosch et al. 2001).
All individuals in all population samples were normal, apparently healthy volunteers, and no diagnoses of alcoholism or related disorders were performed, except in the Taiwanese Han, Ami, and Atayal, as described by Osier et al. (1999). These samples were enriched, relative to population frequencies, for individuals with alcohol dependence. All samples were collected with both the approval of the appropriate institutional review boards and informed consent from the participants.
The majority of the samples were typed in the laboratory at Yale. The !Kung San and southeastern-Bantu-speaker samples were typed in the laboratory of H.S. by M.O. while a visitor at the South African Institute for Medical Research (Johannesburg). The Northern Moroccan, Central Moroccan, Saharan, Catalan, and Basque samples were typed in Barcelona by D.C.
Sequencing, Polymorphism Detection, and Ancestral-State Determination
Sequencing of the clone pADH73 (Smith 1986) and PCR products of genomic DNA was performed by cycle sequencing with ABI PRISM Dye Terminator Cycle Sequencing Core Kits on an ABI 373S Automatic DNA Sequencer. The new polymorphisms—ADH1C EcoRI, ADH1C HaeIII, and ADH7 StyI—were ascertained in different ways, as described in the dbSNP records. Following the logic of Iyengar et al. (1998), we determined ancestral states for all ADH sites by direct sequencing of PCR products from genomic DNA from nonhuman primates. The PCR products were generated using the same primers as those in the protocols given for human typing with genomic DNA from at least one chimpanzee (Cheetah, Dodo, and/or Colin), at least one gorilla (M'kubwa, Oko, Machi, and/or Abe), and at least one orangutan (Puti, Tupa, and/or CP81).
Typing Protocols
The ADH1B RsaI (intron 3) site was typed as described elsewhere (Osier et al. 1999). The ADH1B Arg47His and ADH1C Ile349Val polymorphisms were typed as described elsewhere (Osier et al. 1999) for the Taiwanese Chinese, Ami, Atayal, and Maya samples. All other typings, including the ADH1B Arg47His and ADH1C Ile349Val polymorphisms for other populations, were performed as PCR-RFLPs. Details of the individual typing protocols are given in table 1.
Table 1.
PCR Reagents and Conditions[Note]
|
Fragment Size(s)(bp) |
|||||
| Polymorphic Site (Location) | Forward and Reverse Primers | Cycling Conditionsa[No. of Cycles] | Enzymeb | Site Absent | Site Present |
| ADH7 StyI (intron 5) | A7IN5DW2 (5′-TAT TAA ATT ATT GCT TAA TAA CTG G-3′), A7IN5UP1 (5′-TTC CTG TGT CTC TTA CAG TG-3′) | 95°C (15 s), 54°C (15 s), 72°C (60 s) [40] | StyI | 477 | 263 + 214 |
| ADH1C: | |||||
| EcoRI (intron 2)c | A3EX2DW (5′-TTG CAC CTC CTA AGG CTC-3′), A3EcoUP2 (5′-TCT AAT GCA AAT TGA TTG TGA AC-3′) | 95°C (15 s), 51°C (15 s), 72°C (75 s) [40] | EcoRI | 323 | 242 + 81 |
| HaeIII (exon 5) | A3EX5FOR2 (5′-TGA GTT TGC ACA TTA GTT ATG G-3′), A3EX5REV1 (5′-TGC TCT CAG TTC TTT CTG GG-3′) | 95°C (30 s), 56°C (30 s), 72°C (60 s) [35] | HaeIII | 435 | 193 + 242 |
| Ile349Valc | A3FXNFOR1 (5′-TTG TTT ATC TGT GAT TTT TTT TGT-3′), A3FXNREV3 (5′-CGT TAC TGT AGA ATA CAA AGC-3′) | 95°C (15 s), 51°C (15 s), 72°C (75 s) [40] | SspI | 378 | 274 + 104 |
| ADH1B: | |||||
| Arg47His | A2FXNFOR (5′-ATT CTA AAT TGT TTA ATT CAA GAA G-3′), A2FXNREV (5′-ACT AAC ACA GAA TTA CTG GAC-3′) | 95°C (30 s), 56°C (30 s), 72°C (60 s) [35] | MslI | 685 | 443 + 242 |
| Arg369Cysc | HE39d (5′-TGG ACT TCA CAA CAA GCA TGT-3′), HE40d (5′-TTG ATA ACA TCT CTG AAG AGC TGA-3′) | 95°C (15 s), 58°C (15 s), 72°C (60 s) [40] | AlwNI | 201 | 130 + 71 |
Note.— All PCR was performed using 100 ng genomic template, 100 ng each primer, 200 μM dNTP, 2.0 mM MgCl2, 50 mM KCl, 10 mM Tris HCl (pH 8.4), and 0.5 U AmpliTaq DNA polymerase (Perkin Elmer) in a total volume of 25 μl.
All cycling protocols were performed on a Perkin Elmer 9600, with an initial hold at 95°C, for 5 min, and a final hold at 72°C, for 10 min.
PCR products were digested with 5 U of the appropriate restriction enzyme by use of the buffer that was recommended by the manufacturer.
Dimethyl sulfoxide was added to a final concentration of 5% by volume.
Xu et al. (1988).
Statistical Analyses
Genotype and allele frequencies for the separate polymorphisms were calculated by direct counting under the assumption that each was a biallelic codominant system; binomial SEs were calculated separately, for each allele-frequency estimate, as  . Fst values were calculated by the program DISTANCE (cf. Kidd and Cavalli-Sforza 1974), which also calculates Wright's Fst as
. Fst values were calculated by the program DISTANCE (cf. Kidd and Cavalli-Sforza 1974), which also calculates Wright's Fst as  (Wright 1969). Maximum-likelihood estimates of haplotype frequencies were calculated by HAPLO (Hawley and Kidd 1995) from the individual multisite phenotypes of individuals in each population. HAPLO output includes a jackknife estimate of SEs for the frequency estimates. Overall and pairwise measures of linkage disequilibrium were evaluated using the ξ coefficient by the HAPLO program (Zhao et al. 1999; Kidd et al. 2000). Linkage disequilibrium measured by D′ was calculated by the LINKD program (Kidd et al. 2000), which calculates D′ as described by Lewontin (1964).
(Wright 1969). Maximum-likelihood estimates of haplotype frequencies were calculated by HAPLO (Hawley and Kidd 1995) from the individual multisite phenotypes of individuals in each population. HAPLO output includes a jackknife estimate of SEs for the frequency estimates. Overall and pairwise measures of linkage disequilibrium were evaluated using the ξ coefficient by the HAPLO program (Zhao et al. 1999; Kidd et al. 2000). Linkage disequilibrium measured by D′ was calculated by the LINKD program (Kidd et al. 2000), which calculates D′ as described by Lewontin (1964).
Results
Site Descriptions
The three polymorphisms that result in amino acid changes (ADH1C Ile349Val [exon 8], ADH1B Arg47His [exon 3], and ADH1B Arg369Cys [exon 9]) have been documented by Smith (1986) and Carr et al. (1989), and the ADH1B RsaI (intron 3) site by Smith (1986) and Osier et al. (1999). The molecular spacing of the sites on chromosome 4, as well as the Allele Frequency Database (ALFRED) site unique identification numbers (UIDs) and dbSNP submitted-SNP accession numbers (assay IDs), are listed in table 2. Ancestral states for the Class I ADH sites were determined, thereby allowing the ancestral haplotype to be inferred, and are listed in table 3.
Table 2.
SNPs in the ADH Cluster[Note]
|
Unique Identifier |
||||
| Polymorphic Site (Location) | Distance fromADH1C EcoRI Sitea(kb) | ALFRED UIDb | dbSNP Assay IDc | Reference |
| ADH7 StyI (intron 5) | −71.7 | SI000231G | ss2978365 | Present study |
| ADH1C: | ||||
| EcoRI (intron 2) | .0 | SI000226K | ss2978361 | Present study |
| HaeIII (exon 5) | 2.7 | SI000227L | ss2978362 | Present study |
| Ile349Val (exon 8) | 8.0 | SI000228M | ss2978363 | Smith (1986) |
| ADH1B: | ||||
| Arg47His (exon 3) | 29.5 | SI000229N | ss2978360 | Smith (1986) |
| RsaI (intron 3) | 30.4 | SI000002C | ss2978359 | Smith (1986) |
| Arg369Cys (exon 9) | 39.8 | SI000230F | ss2978364 | Carr et al. (1989) |
| Class I SNPs: | ||||
| Five-site haplotype | NA | SI000659U | NA | Present study |
| Six-site haplotype | NA | SI000259C | NA | Present study |
Note.— NA = not applicable.
Negative value indicates distance upstream in the cluster; positive values indicate distance downstream from the ADH1C EcoRI SNP.
“UIDs” are unique identifiers in the ALFRED database for locating allele frequencies and typing protocols.
“Assay IDs” are unique identifiers in the dbSNP database for definition of the molecular position of each SNP.
Table 3.
Sequences and Ancestral States[Note]
|
Sequencea |
|||||
| Polymorphic Site (Location) | Human | Chimpanzee | Gorilla | Orangutan | Consensus AncestralState (Symbol) |
| ADH7 StyI (intron 5) |
G/C CATGG |
C CATGG |
C CATGG |
Unknown | Site present (2) |
| ADH1C: | |||||
| EcoRI (intron 2) | GAAT T/G C |
GAAT T C |
GAAT T C |
AAACT C |
Site presentb (2) |
| HaeIII (exon 5) |
A/G GCC |
A GCC |
A GCC |
A GCC |
Site absent (1) |
| Ile349Val | AAT A/G TT |
AAT G TT |
AAT G TT |
AAT G TT |
349Val (2) |
| ADH1B: | |||||
| Arg47His | C A/G CACAGATG |
C G CACAGATG |
C G CACAGATG |
C G CACAGATG |
47Arg (1) |
| RsaI (intron 3) | G T/C AC |
G C AC |
G C AC |
G C AC |
Site absent (1) |
| Arg369Cys | CAGTATC C/T G |
CAGTATC C G |
CAGTATC C G |
CAGTATC C G |
369Arg (1) |
Note.— SNP are indicated in boldface, and the sequence differences are italicized and underlined.
Oriented in the 5′→3′ direction in the gene cluster.
With the exception of orangutan, in which two different base changes result in the restriction site being absent.
Individual-Site Frequencies
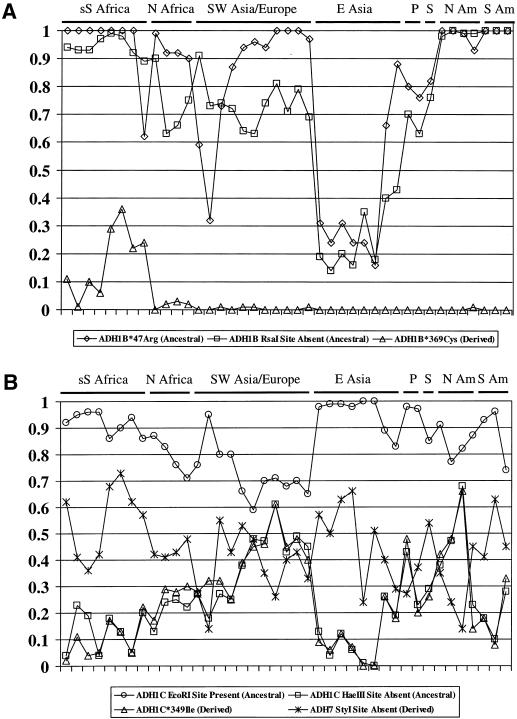
All individual-site allele-frequency data and the five- and six-site haplotype frequency estimates are in ALFRED (see Osier et al. 2001) (table 2). Future updates to the frequency data will be placed in the database. Individual-site frequencies are presented graphically in figure 2. The mean number of typed chromosomes per population, 2N, across all sites is 111.8; the sample size of each population is given in table 4.
Figure 2.
ADH SNP allele frequencies. In each panel, the populations are in the same order, from left to right (as in tables 4, 6, and 7 and from top to bottom). Geographic regions are indicated, across the top of each panel, by the abbreviations “sS Africa,” for “sub-Saharan Africa”; “N Africa,” for “northern Africa”; “SW Asia,” for “southwestern Asia”; “E Asia,” for “eastern Asia”; “P,” for “Pacific” (Micronesians and Nasioi); “S,” for “Siberia” (Yakut); “N Am,” for “North America”; and “S Am,” for “South America.” The allele graphed for each SNP is indicated. A, Allele frequencies at the three ADH1B SNPs. B, Allele frequencies at the three ADH1C SNPs and the ADH7 SNP.
Table 4.
Class I ADH Six-Site Haplotype Frequencies[Note]
|
Estimated Frequency of Haplotypec |
||||||||||||||||||
| Population | 2N | E(Het)a | Residualb | 111111 | 112111d | 112121 | 121111 | 2 11111d | 212111d,e | 212121 | 212211f | 221111d | 221112d,g | 221121d | 221211d,f | 221221d,f | 222111 | 222211f |
| Sub-Saharan African: | ||||||||||||||||||
| Southeastern Bantu-speakers | 96 | 407 | 15 | 14 | 28 | 0 | 42 | 0 | 0 | 0 | 0 | 761 | 92 | 49 | 0 | 0 | 0 | 0 |
| !Kung San | 82 | 524 | 4 | 23 | 0 | 0 | 26 | 86 | 106 | 3 | 0 | 672 | 12 | 68 | 0 | 0 | 0 | 0 |
| Biaka | 134 | 497 | 48 | 0 | 24 | 0 | 0 | 108 | 0 | 0 | 0 | 694 | 59 | 67 | 0 | 0 | 0 | 0 |
| Mbuti | 74 | 230 | 0 | 0 | 27 | 13 | 0 | 0 | 0 | 0 | 0 | 875 | 57 | 14 | 0 | 0 | 14 | 0 |
| Yoruba | 152 | 612 | 0 | 0 | 147 | 13 | 0 | 0 | 16 | 0 | 0 | 528 | 294 | 0 | 0 | 0 | 0 | 0 |
| Ibo | 94 | 616 | 0 | 0 | 96 | 0 | 0 | 0 | 31 | 0 | 0 | 501 | 350 | 21 | 0 | 0 | 0 | 0 |
| Hausa | 78 | 543 | 0 | 0 | 51 | 0 | 13 | 0 | 0 | 0 | 0 | 630 | 225 | 81 | 0 | 0 | 0 | 0 |
| African Americans | 176 | 684 | 47 | 0 | 136 | 3 | 0 | 12 | 28 | 0 | 0 | 503 | 198 | 67 | 0 | 6 | 0 | 0 |
| Northern African: | ||||||||||||||||||
| Ethiopians | 60 | 720 | 0 | 0 | 131 | 0 | 0 | 0 | 19 | 0 | 0 | 374 | 0 | 68 | 340 | 35 | 33 | 0 |
| Northern Moroccans | 186 | 802 | 95 | 22 | 107 | 0 | 6 | 0 | 61 | 19 | 0 | 357 | 19 | 221 | 0 | 37 | 55 | 0 |
| Central Moroccans | 174 | 782 | 49 | 0 | 172 | 14 | 15 | 0 | 53 | 6 | 0 | 358 | 28 | 227 | 0 | 69 | 10 | 0 |
| Saharans | 118 | 771 | 66 | 0 | 131 | 30 | 89 | 0 | 47 | 7 | 0 | 431 | 12 | 92 | 0 | 71 | 23 | 0 |
| European/Middle Eastern: | ||||||||||||||||||
| Yemenite Jews | 76 | 764 | 41 | 0 | 195 | 5 | 0 | 0 | 50 | 0 | 0 | 269 | 0 | 78 | 340 | 22 | 0 | 0 |
| Samaritans | 80 | 716 | 13 | 0 | 50 | 0 | 0 | 0 | 7 | 0 | 131 | 19 | 0 | 250 | 439 | 0 | 0 | 92 |
| Druze | 140 | 821 | 37 | 0 | 176 | 16 | 0 | 0 | 73 | 0 | 0 | 191 | 0 | 230 | 228 | 0 | 21 | 29 |
| Adygei | 106 | 746 | 0 | 0 | 198 | 0 | 0 | 0 | 57 | 0 | 0 | 390 | 0 | 223 | 78 | 54 | 0 | 0 |
| Catalans | 176 | 770 | 34 | 0 | 293 | 19 | 7 | 0 | 52 | 0 | 0 | 256 | 11 | 270 | 0 | 46 | 6 | 6 |
| Basque | 190 | 797 | 57 | 16 | 308 | 54 | 16 | 0 | 70 | 0 | 0 | 206 | 1 | 235 | 0 | 37 | 5 | 0 |
| Russians | 92 | 767 | 0 | 0 | 304 | 0 | 0 | 0 | 148 | 0 | 4 | 245 | 0 | 237 | 38 | 24 | 0 | 0 |
| Finns | 70 | 745 | 14 | 0 | 293 | 0 | 0 | 0 | 307 | 0 | 0 | 209 | 0 | 177 | 0 | 0 | 0 | 0 |
| Danes | 96 | 759 | 34 | 0 | 290 | 12 | 0 | 10 | 125 | 0 | 0 | 256 | 0 | 275 | 0 | 0 | 0 | 0 |
| Irish | 136 | 741 | 0 | 0 | 294 | 0 | 0 | 8 | 155 | 0 | 0 | 317 | 0 | 219 | 0 | 0 | 7 | 0 |
| European North Americans | 176 | 778 | 35 | 0 | 292 | 33 | 0 | 7 | 74 | 4 | 0 | 272 | 6 | 232 | 46 | 0 | 0 | 0 |
| Eastern Asian: | ||||||||||||||||||
| San Francisco Chinese | 96 | 503 | 21 | 0 | 21 | 0 | 0 | 10 | 63 | 0 | 0 | 94 | 0 | 104 | 0 | 688 | 0 | 0 |
| Taiwanese Chinese | 94 | 454 | 22 | 0 | 0 | 0 | 0 | 0 | 32 | 0 | 0 | 106 | 0 | 107 | 11 | 723 | 0 | 0 |
| Hakka | 82 | 505 | 0 | 0 | 0 | 12 | 0 | 0 | 98 | 12 | 0 | 98 | 0 | 98 | 0 | 683 | 0 | 0 |
| Japanese | 84 | 422 | 14 | 0 | 24 | 0 | 0 | 0 | 34 | 0 | 0 | 97 | 0 | 83 | 0 | 748 | 0 | 0 |
| Ami | 80 | 566 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 196 | 0 | 29 | 154 | 609 | 0 | 0 |
| Atayal | 82 | 314 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 133 | 0 | 25 | 25 | 816 | 0 | 0 |
| Cambodians | 46 | 790 | 65 | 0 | 140 | 0 | 0 | 0 | 56 | 0 | 0 | 205 | 0 | 251 | 0 | 283 | 0 | 0 |
| Kachari | 30 | 595 | 33 | 0 | 133 | 0 | 0 | 0 | 0 | 0 | 0 | 174 | 0 | 592 | 67 | 0 | 0 | 0 |
| Pacific: | ||||||||||||||||||
| Nasioi | 44 | 756 | 23 | 0 | 0 | 0 | 0 | 0 | 380 | 74 | 0 | 234 | 0 | 62 | 0 | 182 | 45 | 0 |
| Micronesians | 66 | 736 | 47 | 0 | 0 | 0 | 0 | 30 | 146 | 20 | 0 | 423 | 0 | 110 | 0 | 222 | 0 | 0 |
| Yakut | 98 | 748 | 13 | 0 | 140 | 0 | 0 | 13 | 131 | 0 | 0 | 424 | 0 | 97 | 24 | 158 | 0 | 0 |
| North American: | ||||||||||||||||||
| Cheyenne | 110 | 587 | 0 | 0 | 91 | 0 | 0 | 0 | 291 | 0 | 0 | 564 | 0 | 9 | 0 | 0 | 45 | 0 |
| Arizona Pima | 88 | 634 | 0 | 11 | 210 | 0 | 0 | 0 | 246 | 0 | 0 | 511 | 0 | 0 | 0 | 0 | 23 | 0 |
| Mexican Pima | 106 | 658 | 9 | 0 | 166 | 13 | 0 | 0 | 481 | 10 | 0 | 286 | 0 | 35 | 0 | 0 | 0 | 0 |
| Maya | 94 | 508 | 13 | 92 | 23 | 0 | 0 | 0 | 117 | 0 | 0 | 683 | 11 | 11 | 51 | 0 | 0 | 0 |
| South American: | ||||||||||||||||||
| Ticuna | 130 | 306 | 0 | 0 | 69 | 0 | 0 | 0 | 108 | 0 | 0 | 823 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rondonian Surui | 76 | 197 | 0 | 0 | 26 | 0 | 15 | 0 | 66 | 0 | 0 | 893 | 0 | 0 | 0 | 0 | 0 | 0 |
| Karitiana | 100 | 523 | 12 | 11 | 227 | 0 | 11 | 0 | 32 | 0 | 0 | 649 | 0 | 0 | 0 | 0 | 58 | 0 |
Note.— All values of frequencies and estimated heterozygosities are multiplied by 1,000.
Expected haplotype heterozygosity.
Haplotypes with a frequency <5% in all populations.
Haplotype names in the present article are derived from the alleles of the six sites in order (5′→3′), from top to bottom, on the chromosome (cf. table 3): ADH1C EcoRI, ADH1C HaeIII, ADH1C Ile349Val, ADH1B Arg47His, ADH1B RsaI, and ADH1B Arg369Cys.
Haplotype in the schema in figure 3.
The 212111 haplotype is ancestral (i.e., each of the six sites is represented by the ancestral allele).
Haplotypes with the high-efficiency ADH1B allele are indicated in boldface italic.
Haplotype with the ADH1B*369Cys allele.
Fst Values
For some sites, allele frequencies in eastern-Asian populations seem markedly different from those in non–eastern-Asian populations. To examine the magnitude of these differences, we calculated Fst values for a set of 33 populations that includes the 8 eastern-Asian populations and compared them to Fst values that were calculated for a subset of 25 non–eastern-Asian populations (table 5). For a comparison with other loci, we calculated Fst values, in the same populations, for a total of 86 single sites that were not linked to the ADH cluster (Pakstis et al. 2002). For the full set of 33 populations, these 86 sites had a mean ± SD Fst value of 0.15±0.08. The Fst value for the ADH1B Arg47His polymorphism (Fst=0.48) is the highest, for the full set of 33 populations, that we have seen among the 86 single sites that were not linked to the ADH cluster; the value for this polymorphism is followed closely by that for the ADH1B RsaI polymorphism (Fst=0.41). The ADH1B RsaI and ADH1B Arg47His polymorphisms, respectively, are 3.3 and 4.3 SDs from the mean Fst value. All Fst values for the ADH sites decrease when the eastern-Asian populations are removed from the calculation, but the change is most extreme for the ADH1B Arg47His and ADH1B RsaI (intron 3) polymorphisms. When the eastern-Asian populations are removed from the Fst calculations, the 86 sites had a mean ± SD Fst value of 0.14±0.07. The ADH1B Arg47His site is still 2.8 SDs from the mean Fst value, but the ADH1B RsaI site is at the mean. The higher heterozygosity for the ADH1B Arg47His site in southwestern-Asian populations relative to other populations probably accounts for the high Fst value even when the eastern-Asian populations are removed. The difference between Fst values with and without the sample data from the eastern-Asian populations that we report does not appear to be due to the informativeness of these two sites, since the expected “global” heterozygosity, Ht (mean of the estimated heterozygosity for all populations) is comparable for the ADH1C exon 5 HaeIII and ADH1C Ile349Val sites when the eastern-Asian populations are included (for ADH1C HaeIII [exon 5], Fst=0.323; for ADH1C Ile349Val, Fst=0.317; for ADH1B Arg47His, Fst=0.166; and for ADH1B RsaI [intron 3], Fst=0.256).
Table 5.
Fst Values in Analyses with and without Populations from Eastern Asia and Siberia
|
Fst Value for |
||||||
|
ADH1C |
ADH1B |
|||||
| Populations Testeda | EcoRI (Intron 2) | HaeIII (Exon 5 ) | Ile349Val | Arg47His | RsaI (Intron 3) | Arg369Cys |
| With eastern Asians (n=33) | .10 | .16 | .17 | .48 | .41 | .22 |
| Without eastern Asians (n=25) | .08 | .13 | .14 | .32 | .13 | …b |
The eastern Asian populations include the San Francisco Chinese, Taiwanese Chinese, Hakka, Japanese, Ami, Atayal, Cambodian, and Yakut; n refers to the number of populations in the test.
Not calculated, because the ADH1B Arg369Cys site is not polymorphic in eastern Asia.
Haplotype Frequencies
Table 4 lists all estimated frequencies of six-site Class I haplotypes that were observed at a frequency of at least 5% in some population that we have studied. The “Residual” column is the total frequency, in the specific population, of all haplotypes that never exceeded a frequency, in any population studied, of 5%. In all but five populations, the haplotypes listed account for >95% of all chromosomes in the sample. Most of the eastern-Asian samples (San Francisco Han, Taiwanese Han, Hakka, Japanese, Ami, and Atayal) have high frequencies (>0.60) of the 221221 haplotype. This haplotype consists of the ADH1C EcoRI site-present allele, the ADH1C HaeIII (exon 5) site-present allele, the ADH1C*349Ile allele (high-activity enzyme), the ADH1B*47His allele (high-activity enzyme), and the ADH1B RsaI site-present allele. This haplotype was not observed in most African or in any Amerindian populations that we studied and was observed at low frequencies in some European/Middle Eastern samples, the Nasioi, the Micronesians, and the Yakut.
The haplotype composed of the ancestral alleles at all Class I sites—that is, 212111, the ancestral haplotype—is infrequent to absent in Africa, except in the !Kung San sample, in which it is estimated as having a frequency of 10.6%. This haplotype is much more frequent in some European (e.g., Finns, with a frequency of 0.31) and Native American (e.g., Mexican Pima, with a frequency of 0.49) samples. It is also present in most eastern-Asian populations.
Linkage Disequilibrium
When the calculation is possible, pairwise linkage disequilibrium measured by D′ is generally strong and significant within the Class I ADH cluster. There are sporadic combinations of sites in some populations for which pairwise linkage disequilibrium is not significant or strong, but this can almost always be accounted for by small sample size or low heterozygosity for one or both sites.
The ξ coefficient measures overall nonrandomness of alleles on chromosomes (i.e., of alleles constituting haplotypes), for a series of polymorphic sites. The ξ coefficient is based on a comparison of the likelihood based on estimated haplotype frequencies (in table 4) with the distribution of likelihoods under the assumption of no linkage disequilibrium (Zhao et al. 1999). Values of the ξ coefficient for the Class I ADH sites are given in table 6. For the Class I ADH sites, we have examined five sites—specifically, the ADH1C EcoRI, ADH1C HaeIII (exon 5), ADH1C Ile349Val, ADH1B Arg47His, and ADH1B RsaI polymorphic sites, spanning 39.8 kb—that are variable in a wide range of global population samples. We have calculated separate ξ values for the above five Class I ADH sites and the “African-specific” ADH1B Arg369Cys polymorphism in the African populations.
Table 6.
Linkage Disequilibrium for Five Class I ADH Sites, for Six Class I ADH Sites, and for the Segment between the Six Class I ADH Sites and the ADH7 StyI Site
|
Five Sitesa |
Six Sitesb |
ADH7 Class I Segment |
||||
| Population | ξ | P | ξ | P | ξ | P |
| African: | ||||||
| Southeastern Bantu-speakers | .68 | <.001 | .62 | .004 | −.04 | .649 |
| !Kung San | .45 | .006 | .50 | .018 | −.15 | .951 |
| Biaka | 1.00 | <.001 | 1.15 | <.001 | .03 | .285 |
| Mbuti | 1.63 | <.001 | 1.72 | <.001 | .04 | .299 |
| Yoruba | 3.72 | <.001 | 3.75 | <.001 | .06 | .574 |
| Ibo | 2.32 | <.001 | 2.54 | <.001 | −.03 | .574 |
| Hausa | 1.67 | <.001 | 1.66 | <.001 | −.02 | .527 |
| African Americans | 3.29 | <.001 | 3.32 | <.001 | .09 | .072 |
| Ethiopians | 2.22 | <.001 | 2.20 | <.001 | .58 | .005 |
| Northern Moroccans | .98 | <.001 | 1.11 | <.001 | .01 | .359 |
| Central Moroccans | 2.47 | <.001 | 2.98 | <.001 | .22 | .005 |
| Saharans | 1.71 | <.001 | 2.20 | <.001 | .09 | .167 |
| Non-African:c | ||||||
| Yemenites | 2.62 | <.001 | … | … | .08 | .279 |
| Samaritans | 3.24 | <.001 | … | … | .64 | <.001 |
| Druze | 2.50 | <.001 | … | … | .07 | .176 |
| Adygei | 3.68 | <.001 | … | … | .05 | .231 |
| Catalans | 2.82 | <.001 | … | … | .03 | .250 |
| Basque | 3.16 | <.001 | … | … | .02 | .330 |
| Russians | 3.72 | <.001 | … | … | .02 | .343 |
| Finns | 2.46 | <.001 | … | … | .23 | .027 |
| Danes | 2.69 | <.001 | … | … | .15 | .059 |
| Irish | 2.46 | <.001 | … | … | −.01 | .438 |
| European North Americans | 3.48 | <.001 | … | … | −.03 | .625 |
| San Francisco Chinese | 3.01 | <.001 | … | … | .00 | .466 |
| Taiwanese Chinesed | 2.36 | <.001 | … | … | −.05 | .757 |
| Hakka | 3.32 | <.001 | … | … | .08 | .154 |
| Japanese | 2.50 | <.001 | … | … | .14 | .088 |
| Amid | .52 | .001 | … | … | .13 | .068 |
| Atayald | .90 | <.001 | … | … | .09 | .086 |
| Cambodians | 2.86 | <.001 | … | … | −.03 | .525 |
| Kachari | 3.01 | <.001 | … | … | −.04 | .491 |
| Nasioi | 1.97 | <.001 | … | … | .35 | .032 |
| Micronesians | 2.60 | <.001 | … | … | .63 | .001 |
| Yakut | 3.30 | <.001 | … | … | .01 | .416 |
| Cheyenne | 2.28 | <.001 | … | … | .18 | .007 |
| Arizona Pima | 1.71 | <.001 | … | … | .06 | .195 |
| Mexican Pima | 3.34 | <.001 | … | … | −.04 | .619 |
| Maya | 2.36 | <.001 | … | … | −.02 | .508 |
| Ticuna | 2.20 | <.001 | … | … | .20 | .002 |
| Rondonian Surui | 1.52 | <.001 | … | … | .19 | .026 |
| Karitiana | 2.22 | <.001 | … | … | .21 | .010 |
ADH1C EcoRI, ADH1C HaeIII, ADH1C Ile349Val, ADH1B Arg47His, and ADH1B RsaI.
ADH1C EcoRI, ADH1C HaeIII, ADH1C Ile349Val, ADH1B Arg47His, ADH1B RsaI, and ADH1B Arg369Cys.
The ADH2 Arg369Cys site generally does not vary in non-African populations; therefore, the ξ values will usually not vary between the five- and six-site polymorphisms in non-African populations.
Alcoholic individuals comprise ∼50% of these samples.
Pairwise linkage disequilibrium between the ADH7 StyI site and each of the five Class I sites was examined for all 41 samples (table 7). The permutation test was significant at P⩽.01 for 10.2% of all tests and at P⩽.05 for 24.4% of all tests, suggesting that most of the D′ values with P values .05 are likely to be meaningful, even though many (197) tests were performed. However, the individual significance levels are probably not reliable above the P⩽.01 level. Significant (P⩽.030) and moderate-to-strong D′ values between the ADH1B Arg369Cys and ADH7 StyI sites (∼112 kb) are observed in the !Kung San (D′=-1.00), Biaka (D′=-0.88), Yoruba (D′=-0.54), and Ibo (D′=-0.75) samples, but values were not significant for the other sub-Saharan African samples for any other combination of Class I sites with the ADH7 StyI site (excepting the Ethiopians in northeastern Africa). Interestingly, in the European samples, where several of the Class I sites were initially identified (ADH1C EcoRI and HaeIII) or have high individual-site heterozygosities (ADH1B RsaI; ADH1C Ile349Val) linkage disequilibrium with the ADH7 StyI site was generally not significant. The few exceptions are one Class I site (ADH1C EcoRI) in the Finns (D′=0.38; P⩽.05) and Irish (D′<0.51; P⩽.05), one site in the Catalans (ADH1B Arg47His [D′=1.00; P⩽.01]), and one site in the Basque (ADH1B Arg369Cys [D′=1.00; P⩽.01]). In the Japanese sample, there was significant (P⩽.05) evidence of strong linkage disequilibrium (|D′|>0.80) between the ADH7 StyI site and four of five polymorphic Class I sites (ADH1C EcoRI, ADH1C HaeIII, ADH1B Arg47His, and ADH1B RsaI). There was not significant evidence of linkage disequilibrium between the ADH7 StyI site and any Class I site for the other eastern-Asian samples, except for a few specific pairwise combinations for the San Francisco Chinese (ADH1B Arg47His), Hakka (ADH1C Ile349Val), Atayal (ADH1B RsaI), and Cambodians (ADH1C Ile349Val). In the South American Amerindian samples, there was significant evidence of strong linkage disequilibrium between the ADH7 and ADH1C sites except for one pairwise combination (Surui, ADH1C EcoRI). Otherwise, the sporadic occurrences of significant P values for linkage disequilibrium showed no geographic pattern.
Table 7.
Pairwise Linkage Disequilibrium, |D′|, between the ADH7 StyI Site and Each of the Class I Sites[Note]
|
|D′| between ADH7 StyI and |
||||||
| Population | ADH1C EcoRI | ADH1C HaeIII | ADH1C Ile349Val | ADH1B Arg47His | ADH1B RsaI | ADH1B Arg369Cys |
| Southeastern Bantu-speakers | .37 | .13 | .24 | … | .74 | .28 |
| !Kung San | .56 | .14 | .35 | … | .47 | 1.00** |
| Biaka | .20 | .14 | .33 | … | .50 | .88* |
| Mbuti | .23 | .22 | .42 | … | .13 | 1.00 |
| Yoruba | .26 | .27 | .27 | … | .24 | .54* |
| Ibo | .55 | .31 | .31 | … | 1.00 | .75* |
| Hausa | .04 | .33 | .33 | … | .17 | .36 |
| African Americans | .08 | .33 | .25 | 1.00 | .40 | .21 |
| Ethiopians | .41 | .43 | .62* | .83** | .62 | … |
| Northern Moroccans | .21 | .05 | .32 | .79* | .05 | 1.00* |
| Central Moroccans | .37* | .13 | .22 | 1.00** | .34* | 1.00* |
| Saharans | .14 | .17 | .10 | .67 | .56* | 1.00** |
| Yemenites | .85* | .86* | .86 | .59 | .36 | … |
| Samaritans | 1.00 | 1.00 | 1.00* | 1.00** | 1.00** | … |
| Druze | .02 | .10 | .07 | .47* | .43 | 1.00 |
| Adygei | .14 | .20 | .20 | .37 | .26 | … |
| Catalans | .07 | .07 | .00 | 1.00** | .14 | 1.00 |
| Basque | .04 | .05 | .13 | .16 | .09 | 1.00** |
| Russians | .27 | .12 | .12 | .14 | .53 | … |
| Finns | .38* | .06 | .08 | … | .36 | … |
| Danes | .14 | .09 | .02 | … | .41 | … |
| Irish | .51* | .25 | .26 | … | .20 | … |
| European North Americans | .12 | .16 | .04 | 1.00 | .03 | 1.00 |
| San Francisco Chinese | .11 | .51 | .78 | .52* | .64 | … |
| Taiwanese Chinese | 1.00 | .77 | .77 | .01 | .24 | … |
| Hakka | 1.00 | .77 | .77* | .32 | .58 | … |
| Japanese | 1.00* | 1.00* | 1.00** | .32 | .80** | … |
| Ami | … | 1.00 | … | .22 | .08 | … |
| Atayal | … | … | … | .51 | .71* | … |
| Cambodians | 1.00 | .78 | .79* | .38 | .44 | … |
| Kachari | .50 | .50 | .50 | 1.00 | .40 | … |
| Nasioi | 1.00** | .54 | .83 | .54 | .15 | … |
| Micronesians | .23 | .40 | .45* | 1.00** | .80** | … |
| Yakut | .29 | .42* | .47 | .53 | .36 | … |
| Cheyenne | .48 | .87** | .76** | … | 1.00 | … |
| Arizona Pima | 1.00* | .36 | .26 | … | … | … |
| Mexican Pima | .68 | .25 | .25 | 1.00 | .20 | … |
| Maya | .60 | .50 | .52 | 1.00 | 1.00 | 1.00 |
| Ticuna | 1.00* | 1.00** | 1.00** | … | … | … |
| Rondonian Surui | 1.00 | 1.00** | 1.00* | … | … | … |
| Karitiana | .86** | .87** | .57* | … | … | … |
Note.— Ellipses indicate when comparison was not possible.
P⩽.05.
P⩽.01.
Using a segment test for evidence of linkage disequilibrium between the six Class I ADH sites considered as a group and the ADH7 site should clarify linkage disequilibrium between the ADH7 StyI site and the Class I cluster as a whole by effectively using the Class I haplotypes. This integrates into one test the information from all five polymorphic sites (six sites in the African populations). At the .01 level of significance, we observe (table 6) evidence of linkage disequilibrium in only seven populations from different geographic regions. These are primarily populations that have (or had) undergone a population bottleneck (e.g., Ethiopian Jews, Samaritans, Micronesians, Ticuna, and Karitiana), but, for other populations (Central Moroccans and Cheyenne), that simple explanation for the significant linkage disequilibrium does not seem likely. In all populations, ξ values for the specified segment are moderate or weak (0<|ξ|<0.64).
Discussion
Global Patterns of Allele-Frequency Variation
As shown in figure 2, most of the SNPs that we studied were segregating in all major geographic regions of the world, and allele frequencies for the individual sites tend to be similar within each geographic region. The most notable patterns among geographic regions occur for two of the expressed sites, the ADH1B Arg47His and Arg369Cys sites.
These data confirm the well known high frequency of the ADH1B*47His (high-activity) allele in eastern-Asian populations and its presence in some European and southwestern-Asian populations. We now document the presence of the ADH1B*47His allele in northern-African populations for the first time. In southwestern-Asian samples, the frequency of the ADH1B*47His allele is greater than that previously reported in other populations from the region. We found frequencies as high as 0.68 (in Samaritans), 0.27 (in Druze), 0.41 (in Yemenite Jews), and 0.38 (in Ethiopian Jews). Previous reports of this allele in populations from this general region showed much lower frequencies: 0.20 (in Israeli Jews) (Neumark et al. 1998) and 0.125 (in Turks) (Goedde et al. 1992). In all other populations studied from Europe, the Middle East, and Africa, the ADH1B*47His allele was observed at frequencies no higher than 0.13 (in Adygei) and was usually either much lower or absent. These are similar to frequencies that have previously been reported for populations in these regions (e.g., frequencies of 1%–5%, in central and northern Europe; see Goedde et al. 1992). We find the site to be monomorphic for ADH1B*47Arg in sub-Saharan African populations except for Ethiopians and to be rarely heterozygous in Native American populations.
Few populations have previously been found to have the ADH1B*369Cys allele; our results are similar to those that have previously been reported, but we now show that the allele is present in all the African samples that we examined except the Ethiopians. Indeed, this allele is essentially an African-specific allele. Bosron et al. (1983) first reported the observation, on the basis of protein electrophoresis, of the ADH1B*369Cys allele (then called “ADH2*3”) in a sample of African Americans from Indianapolis. Wall et al. (1997) did report a very low frequency (0.063) of this allele, in a sample of Amerindians from southern California. It has not generally been tested for in other samples. We observed the ADH1B*369Cys allele not only in the African American sample that we studied but also in all but one of the African samples that we studied (mean frequency of the ADH1B*369Cys allele is 0.11 in the 11 African populations). There were also individual occurrences of that allele in five non-African samples—the European American, Druze, Catalan, Basque, and Mayan samples—but not elsewhere. Its presence in both the Amerindian sample that Wall et al. (1997) studied and the Mayan sample that we studied can be explained by recent migration and/or admixture. In the southwestern-Asian and European populations, the allele may have persisted at a low frequency (estimated at 0–3%) since modern humans arrived but may be only rarely observed, as in our samples of the Druze and European Americans, by happenstance. Alternatively, occurrence of the allele in these few sampled individuals may reflect more recent gene flow from Africa (e.g., by northern Africans migrating into the Iberian peninsula).
At the ADH1C Ile349Val site, the ancestral, lower-activity allele (ADH1C*349Val or ADH1C*2) is the more common allele in all but two populations (Finns and Mexican Pima), and the site has heterozygosities >0.2 in all northern-African, European, Pacific, and Amerindian (except for the Surui) populations. Heterozygosities are generally lower in the sub-Saharan African populations and the eastern-Asian populations.
The allele-frequency distribution of the noncoding ADH1B RsaI (intron 3) site shows close parallels with the nearby ADH1B Arg47His site in eastern-Asian, Pacific, and Native American populations but not in African, southwestern-Asian, and European populations. The RsaI site is fixed (or nearly fixed) only for the Native American populations. The allele-frequency distribution for the noncoding ADH1C HaeIII site very closely tracks that for the ADH1C Ile349Val site, which is 5.3 kb away. The pattern for the ADH1C EcoRI site is similar to—but not as highly correlated as—the other two ADH1C sites, even though it is closer to the HaeIII site (2.7 kb) than the HaeIII site is to the Ile349Val site. These allele-frequency correlations among the diverse population samples are a reflection of—indeed, a measure of—linkage disequilibrium.
Population frequencies of the other Class I ADH sites (ADH1B RsaI, ADH1C EcoRI, ADH1C HaeIII, and ADH1C Ile349Val) generally cluster by region. The exceptions to strong geographic patterning primarily appear to be populations that have probably undergone strong founder effects. For example, some of the southwestern-Asian samples (Samaritans, Yemenite Jews, and Ethiopian Jews) have high frequencies of the ADH1B*47His allele. The Samaritans have a noticeably lower frequency of the ADH1C EcoRI site-present allele than do other southwestern-Asian populations. The Finns, the Nasioi, and the Mexican Pima have distinctly higher frequencies of the ADH1C HaeIII site-absent and ADH1C*349Ile alleles than do their neighboring populations. The unusually high frequency of the ADH1B*47His allele in the African Americans (graphed as a low frequency of the ADH1B*47Arg allele in fig. 2) is not explainable by either founder effect or European admixture, since it is higher than that seen in Europe. There may be other African populations with high frequencies of this high-activity allele that have not yet been identified.
Fst Values
The high Fst values for the ADH1B Arg47His and RsaI (intron 3) site polymorphisms clearly place these two sites as outliers in the distribution of global variation that can be attributed to random genetic drift among populations. The high Fst values reflect the large frequency differences, for these sites, between eastern-Asian populations and most other populations. With the eastern-Asian allele frequencies at these two sites included in the calculation, the Fst value for the ADH1B RsaI site is three times as high as it is when the frequencies for these populations are omitted; the Fst value for the ADH1B Arg47His polymorphism is also much higher when calculated including, rather than when calculated excluding, the eastern-Asian populations. We have not observed such a large difference—nor such high Fst values across the set of 33 populations—for any of the 86 other non-ADH sites that our laboratory has studied (Pakstis et al. 2002) and are not familiar with such high values for studies in the literature of samples from a range of global populations. Interestingly, the changes in Fst value for the three ADH1C polymorphisms when the eastern-Asian populations are removed are not very large—even though the Ile349Val polymorphism, like the ADH1B Arg47His polymorphism, encodes proteins with different metabolic efficiency for the breakdown of ethanol, and the other sites are generally in disequilibrium with it, as suggested in figure 2.
Haplotype Distribution and Evolution
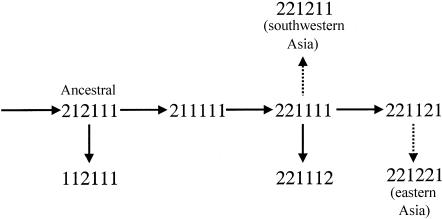
The ancestral haplotype must be the one with ancestral alleles at all sites, although not all occurrences of this haplotype are necessarily identical by descent with the ancestral haplotype. This haplotype is named “212111,” on the basis of a binary system of allele names (cf. tables 3 and 4). Random genetic drift could explain this pattern of ancestral haplotype frequencies. A possible concomitant factor would be regeneration by recombination of the ancestral combination of alleles at the individual sites. There is only one common haplotype with the ADH1C HaeIII site-absent allele, 112111, and it differs from the ancestral haplotype only at the ADH1C EcoRI site. The common haplotypes with the ADH1C EcoRI site-present allele differ from the ancestral haplotype at two or three other sites, including the ADH1C HaeIII site. Thus, only two heterozygotes that are likely to occur commonly, 112111/221111 or 112111/221121, would be able to regenerate the ancestral haplotype by recombination, and the crossovers would have to be between the two ADH1C sites, only 2.7 kb apart. This seems unlikely to be a systematic “force” but could have regenerated a few instances that random genetic drift raised to modest frequencies. It seems more parsimonious, pending possible clarification from data on additional sites, especially sites closely flanking these Class I sites, to assume that this distribution of the ancestral haplotype is the consequence of random genetic drift.
The haplotypes that are common (i.e., those with a frequency ⩾10% of the chromosomes) in any African population and in most non-African populations can be accounted for by a simple tree of mutations starting from the ancestral haplotype (fig. 3). These haplotypes account for between 76.5% (Samaritans) and 100% (Adygei and Ticuna) of the chromosomes in these 41 populations. The other haplotypes in table 4, none of which exceeds 10% anywhere, require recombination if recurrent mutation is assumed not to have occurred. From the ancestral haplotype, two independent derivatives seem likely. One involves mutation of the ADH1C EcoRI site to generate the 112111 haplotype now common in Europe and seen in most populations around the world. An independent mutation of the ADH1C Ile349Val site on the ancestral haplotype generates the 211111 haplotype. This haplotype is rarely seen today but is present in the !Kung San, Biaka Pygmies, and African Americans. The single haplotype that is most common around the world involves a mutation that starts from this currently uncommon haplotype of the ADH1C HaeIII site to generate the 221111 haplotype. The ADH1B*369Cys allele, found only in African populations, is commonly seen on a single haplotype and, thus, likely represents a single mutation on this common haplotype to generate the 221112 haplotype. The rare occurrences of this allele on other haplotypes indicate that recombination events have occurred.
Figure 3.
Class I ADH–haplotype evolutionary tree. Haplotype names are listed in table 4. All haplotypes in the figure are observed at frequencies >5% in one or more African samples. Each solid arrow represents a single base mutation. The two dashed arrows cannot both be single mutations if recurrent mutation to ADH1B*47His is excluded; one of the two presumably arose by recombination. The 221221 haplotype is the one most common in eastern Asia and associated with the protective effect against alcoholism. The 221112 haplotype is the only one with the African-specific ADH1B*369Cys allele.
At this level of analysis, gene conversion cannot be distinguished from ordinary crossing-over, and, with one exception, the other haplotypes can be explained by a single crossover between two of the common noncrossover haplotypes in figure 3. The exception is the 222211 haplotype, which can arise from a crossover between 221111 and 212211, which is itself a crossover product. Collectively, the haplotypes in table 4 require inference of crossovers in every interval except the last. As noted above, the African-specific ADH1B*369Cys allele does exist on other haplotypes in the residual class. The 221122 haplotype (with frequencies of 1.5% among southeastern Bantu-speakers and 0.8% among African Americans) could be the result of a crossover in the last interval between the common 221121 and 221112 haplotypes, and the 211112 haplotype (with a frequency of 2.6% among Biaka) could be the result of a crossover between the haplotypes 211111 and 221122 that are common in the Biaka.
In eastern Asia, the 221221 haplotype (ADH1C EcoRI site present, ADH1C HaeIII site present, ADH1C*349Ile, ADH1B*47His, ADH1B RsaI site present, and ADH1B*Arg369) is estimated at frequencies >60%—as high as 82% in the Atayal. There may be a unique shared evolutionary history of the ADH1B*47His-containing haplotypes in the populations of eastern Asia, north Africa, Iberia, and southwestern Asia, especially in light of the fact that the ADH1B*47His allele in the Middle Eastern samples occurs primarily in a haplotype observed in northern Africa and Iberia and was only slightly different from the haplotype common in eastern Asia. These two common haplotypes (indicated by the dotted arrows in fig. 3) cannot be jointly explained without requiring recurrent mutation or recombination.
Because it occurs in essentially all parts of the world on a specific haplotype, the derived allele at the ADH1B RsaI site seems likely to have arisen on the common 221111 haplotype to generate the now reasonably common 221121 haplotype. The ADH1B*47His allele does not fit simply into such a scheme since it occurs as part of two different common haplotypes in Asia (221221 and 221211) that differ only at the RsaI site, 906 bp downstream from the ADH1B Arg47His site. If we exclude recurrent mutation as being very unlikely, then two schemes, both involving recombination, could explain the origins. Ethiopian Jews have the ADH1B*47His allele, suggesting that it may have arisen in northeastern Africa before the expansion out of Africa. This would be analogous to the large-normal (CTG)n alleles, at the locus causing myotonic dystrophy (DM), that exist in northeastern African and non-African populations but not in sub-Saharan populations (Tishkoff et al. 1998). If the haplotype common in Ethiopians and southwestern-Asian populations, 221211, is the original haplotype that carries the ADH1B*47His allele, then the haplotype that is common in eastern Asia must have arisen by recombination with the common 221121 haplotype, the only common haplotype for the RsaI site-present allele.
This recombination event in southwestern Asia or northeastern Africa with a subsequent increase in frequency of the 221221 haplotype in eastern Asia seems most compatible with the spread of modern humans out of Africa and then, from west to east, across Asia. The alternative—that the ADH1B*47His allele arose on the haplotype with the RsaI site-present allele (i.e., 221121) and that the 221211 haplotype arose by recombination—seems less likely but certainly cannot be excluded. The occurrence of the 221221 haplotype in northwestern Africa could be considered an argument for this scenario. A proper statistical evaluation of these alternatives would require historical demographic and frequency data, which are not available. Additional polymorphic sites downstream from the existing six Class I markers may help clarify this aspect of haplotype evolution.
Selection on the ADH1B Arg47His Polymorphism?
The common eastern-Asian haplotype, which has the ADH1B*47His allele, could have attained such a high frequency through genetic drift or the effects of selection. Drift seems to be an insufficient explanation, for several reasons. The high Fst values for this and the adjacent RsaI site are clearly outliers, showing more genetic variation among populations than is seen for any other markers. It would take a strong population bottleneck and/or strong subsequent random genetic drift within eastern Asia for this rare allele to become frequent. The aldehyde dehydrogenase 2 (ALDH2) locus provides additional evidence that drift may not be the sole factor determining the frequency of this haplotype. ALDH2 is functionally monomorphic in most populations, but not in those of eastern-Asian descent (Goedde et al. 1992; Peterson et al. 1999). The protein produced by this eastern-Asian allele acts as a dominant null allele that dramatically reduces the ability to breakdown acetaldehyde, the by-product of ADH metabolism of ethanol. In heterozygous individuals, only ∼1/16 of the ALDH2 enzyme tetramers would be functionally active. Heterozygosity and homozygosity for the null ALDH2 allele combined with alleles for high-efficiency ADH enzymes should result in a high level of acetaldehyde, a toxin, in the body following ethanol ingestion. It seems unlikely that alleles of unlinked genes in two gene families in the same metabolic pathway that both increase acetaldehyde levels would both have high frequencies in the same populations by random genetic drift alone. Again, although it cannot be proven that the high frequency of this ADH haplotype is primarily due to selection, it seems unlikely that random genetic drift is solely responsible. We hope that future studies will help our understanding of the causes behind the high frequency of this haplotype in only one region of the world.
Extent of Linkage Disequilibrium
In all population samples that we studied, there was significant evidence of linkage disequilibrium across the five Class I ADH sites. This is expected in the non-African samples on the basis of our other studies of multiple populations (Tishkoff et al. 1996; Kidd et al. 1998, 2000), since the five sites polymorphic outside of Africa span a small region that is slightly larger than 30 kb. It also agrees with other empiric studies on more restricted sets of populations (Reich et al. 2001). It does not agree with expectations for some theoretical models (Kruglyak et al. 1999) nor some results for individual loci, such as lipoprotein lipase (LPL), where a recombination hotspot disrupts linkage disequilibrium across a short segment (Templeton et al. 2000; Jeffreys et al. 2001). Also, in contrast to data at several loci we have studied in the same population samples (Pakstis et al. 2000), in the African samples linkage disequilibrium at this locus has a wide range of ξ values (0.45<ξ<3.72) and, in several cases, is average or above average in comparison to non-African samples. One African population, the Yoruba, has the same ξ value as the maximum observed in all other samples. As a rule, based on analyses of data from multiple loci (Pakstis et al. 2000 and unpublished data from Kidd Lab), values of ξ and other measures of linkage disequilibrium are generally low in African populations and less than those observed in samples from other parts of the world.
Interestingly, pairwise linkage disequilibrium, as measured by D′, between the ADH7 StyI and some Class I sites is observed at moderate values and is significant in some samples. For the segment test of linkage disequilibrium between the ADH7 site and the six Class I ADH sites, very few populations had significant evidence of linkage disequilibrium, although some did in certain pairwise combinations of sites. It may be that some pairwise tests are significant while the segment test is insignificant for these samples when some specific pairwise site combinations tested are nonrandom (linkage disequilibrium) but the other specific pairwise site combinations are random (linkage equilibrium) and “outweigh” the nonrandom combinations. Because there are no clear overall patterns of variation or linkage disequilibrium between the non–Class I ADH7 site and the Class I sites, studies of the role that the Class I ADH genes play in alcoholism may need to examine the degree of linkage disequilibrium across large regions on a population-by-population and site-by-site basis to eliminate any effects of linkage disequilibrium with a nonmember of the Class I ADH cluster.
Since linkage disequilibrium between causative and noncausative sites has resulted in positive association studies of alcoholism in samples from eastern Asia for those sites that appear not to be causative (Osier et al. 1999) and since linkage disequilibrium may extend over large regions in this gene cluster, the range of possible locations of sites that could be truly causative is much larger than had previously been anticipated. The range of sequence that could be suspected encompasses at least the ADH1C and ADH1B genes and all intergenic sequence, a distance of >40 kb. It is possible that even though the ADH1B Arg47His polymorphism incorrectly appears to be causative by association studies because of linkage disequilibrium with another, truly functional variant, possibly in the promoter region. Also, if other functional sites show positive associations in populations outside eastern Asia, then it will be important to ensure that such results cannot be accounted for by linkage disequilibrium with yet other, possibly undiscovered, sites, perhaps in other members of the ADH cluster.
The need to screen for effects of non–Class I ADH polymorphisms is especially true for the Japanese, for whom samples have been extensively studied for association between Class I ADH sites and alcoholism. We observed significant pairwise linkage disequilibrium between the ADH7 StyI site and four of five Class I sites polymorphic in the Japanese, even though the segment test between ADH7 and the six Class I sites was not significant. The cause for this pattern of linkage disequilibrium in eastern Asia and any effect on association studies in the Japanese will need to be pursued. The contrasting pattern of linkage disequilibrium between the Japanese and other eastern-Asian samples is not due to the informativeness of individual markers, since the heterozygosities for individual sites in non-Japanese eastern-Asian samples are generally comparable to or higher than those observed in the Japanese sample. This pattern of linkage disequilibrium also means that it is not valid to assume that all eastern-Asian populations are homogeneous in genetic studies of the ADH genes and possibly in studies of other genes for which the assumption of homogeneity has not been tested.
To date, almost all case-control association studies of alcoholism in populations of European ancestry have used only the ADH1C Ile349Val polymorphism because of its higher heterozygosity. Those studies have had largely negative results but may have had low power to detect an indirect effect on the ADH1C Ile349Val site through linkage disequilibrium with some other, as-yet-undetected, polymorphism in the cluster. Use of haplotypes in such a study would help overcome this problem (Kidd et al. 1996) and may resolve the conflict between positive linkage studies to the region and negative association studies with known variants in European samples.
A significant effort is under way to design generalized linkage disequilibrium maps that are based on “haplotype blocks” punctuated by recombination hotspots (Daly et al. 2001; Jeffreys et al. 2001; Johnson et al. 2001), so that only the sites distinguishing common haplotypes need be studied. Most of these studies focus on samples of Europeans and only one or two non-European samples. In the global samples that we studied, we found strong, but incomplete, linkage disequilibrium across the six Class I sites. In any one region of the world, the most common haplotypes (i.e., those that account for a minimum of 80% of the chromosomes in the region) can be distinguished by two or three of the sites. However, different regions of the world require different combinations of sites. Globally, five of the six sites are essential.
The ADH genes are an interesting exception to the patterns of variation and linkage disequilibrium observed at other loci in the human genome. Untangling the forces at play in the evolutionary history of the ADH genes in humans presents a unique challenge. The resulting knowledge will provide insight into how to refine the design and interpretation of studies of the role that the ADH genes play in the protection against alcoholism in different populations.
Acknowledgments
We thank Akashnie Maharaj and Heeran Makkan, from the laboratory of H.S., for their help with this research and Mònica Vallés, from the laboratory of J.B., for her technical assistance. This work was funded, in part, by National Institute of Alcohol Abuse and Alcoholism grant AA09379. Support was also provided by a grant (to K.K.K. and J.R.K.) from the Alfred P. Sloan Foundation for collection of population samples, by a contract (to K.K.K.) from the National Institute of Diabetes and Digestive and Kidney Diseases, and by grants (to R.B.L.) from the National Health Research Institute (Taiwan, ROC; NHRI-EX91-8939SP) and the National Science Council (Taiwan, ROC; NSC 90-2314-B-016-081). We acknowledge and thank the following researchers, who helped in the assembly of the samples from the individual populations: F. L. Black, L. L. Cavalli-Sforza, R. Deka, J. Friedlaender, K. Kendler, W. Knowler, F. Oronsaye, L. Peltonen, and K. Weiss. We also thank the numerous participants who donated the DNA samples used in this study.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- ALFRED, http://alfred.med.yale.edu/alfred/
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for 4q21-23 [accession numbers AP002026, AP002027, AP002028, and AC097530])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ADH1A [MIM 103700], ADH1B [MIM 103720], ADH1C [MIM 103730], ADH7 [MIM 600086], ADH6 [MIM 103735], ADH4 [MIM 103740], and ADH5 [MIM 103710])
References
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern African and the Iberian Peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosron WF, Li TK, Vallee BL (1979) Heterogeneity and new molecular forms of human liver alcohol dehydrogenase. Biochem Biophys Res Commun 91:1549–1555 [DOI] [PubMed] [Google Scholar]
- Bosron WF, Magnes LJ, Li T-K (1983) Human liver alcohol dehydrogenase: ADHIndianapolis results from genetic polymorphism at the ADH2 gene locus. Biochem Genet 21:735–744 [DOI] [PubMed] [Google Scholar]
- Carr LG, Xu Y, Ho W-H, Edenberg HJ (1989) Nucleotide sequence of the ADH23 gene encoding the human alcohol dehydrogenase β3 subunit. Alcohol Clin Exp Res 13:594–586 [DOI] [PubMed] [Google Scholar]
- Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT (1996) Alcohol-metabolizing genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3, and ALDH2. Br J Psychiatry 168:762–767 [DOI] [PubMed] [Google Scholar]
- Comas D, Calafell F, Noufissa B, Helal A, Lefranc G, Stoneking M, Batzer MA, Bertranpetit J, Sajantila A (2000) Alu insertion polymorphisms in NW Africa and the Iberian Peninsula: evidence for a strong genetic boundary through the Gibraltar straits. Hum Genet 107:312–319 [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Edenberg HJ (2000) Regulation of the mammalian alcohol dehydrogenase genes. Prog Nucleic Acid Res Mol Biol 64:295–341 [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Bosron WF (1997) Alcohol dehydrogenases. In: Guengerich FP (ed) Comprehensive toxicology. Vol 3: Biotransformation. Pergamon Press, New York, pp 119–131 [Google Scholar]
- Edman K, Maret W (1992) Alcohol dehydrogenase genes: restriction fragment length polymorphisms for ADH4 (Π-ADH) and ADH5 (χ-ADH) and construction of haplotypes among different ADH classes. Hum Genet 90:395–401 [DOI] [PubMed] [Google Scholar]
- Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckman G, Bhatia K, Chen LZ (1992) Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet 88:344–346 [DOI] [PubMed] [Google Scholar]
- Hawley HE, Kidd KK (1995) HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered 86:409–411 [DOI] [PubMed] [Google Scholar]
- Iyengar S, Seaman M, Deinard AS, Rosenbaum HC, Sirugo G, Castiglione CM, Kidd JR, Kidd KK (1998) Analyses of cross species polymerase chain reaction products to infer the ancestral state of human polymorphisms. DNA Sequence 8:317–327 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the Class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Johnson GCL, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RCJ, Tuomilehto F, Gough SCL, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 [DOI] [PubMed] [Google Scholar]
- Kidd KK, Cavalli-Sforza L (1974) The role of genetic drift in the differentiation of Icelandic and Norwegian cattle. Evolution 28:381–395 [DOI] [PubMed] [Google Scholar]
- Kidd KK, Morar B, Castiglione CM, Zhao H-Y, Pakstis AJ, Speed WC, Bonne-Tamir B, et al (1998) A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet 103:211–227 [DOI] [PubMed] [Google Scholar]
- Kidd KK, Pakstis AJ, Castiglione CM, Kidd JR, Speed WC, Goldman D, Knowler WC, Lu R-B, Bonne-Tamir B (1996) DRD2 haplotypes containing the TaqI A1 allele: implications for alcoholism research. Alcohol Clin Exp Res 20:697–705 [DOI] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao HY, Lu R-B, Okonofua FE, Odunsi A, Grigorenko E, Bonne-Tamir B, Friedlaender J, Schulz LO, Parnas J, Kidd KK (2000) Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 66:1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage. I. General considerations: heterotic models. Genetics 49:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Yin SJ, Crabb DW, O'Connor S, Ramchandani VA (2001) Genetic and environmental influences on alcohol metabolism in humans. Alcohol Clin Exp Res 25:136–144 [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed] [Google Scholar]
- Murray JC, Shiang R, Carlock LR, Smith M, Buetow KH (1987) Rapid RFLP screening procedure identifies new polymorphisms at albumin and alcohol dehydrogenase loci. Hum Genet 76:274–277 [DOI] [PubMed] [Google Scholar]
- Neumark YD, Friedlander Y, Thomasson HR, Li T-K (1998) Association of the ADH2*2 allele with reduced ethanol consumption in Jewish men in Israel: a pilot study. J Stud Alcohol 59:133–139 [DOI] [PubMed] [Google Scholar]
- Osier M, Pakstis AJ, Kidd JR, Lee J-F, Yin S-J, Ko H-C, Edenberg HJ, Lu R-B, Kidd KK (1999) Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 64:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier MV, Cheung K-H, Kidd JR, Pakstis AJ, Miller PL, Kidd KK (2001) ALFRED: an allele frequency database for diverse populations and DNA polymorphisms—an update. Nucleic Acids Res 29:317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakstis AJ, Kidd KK, Kidd JR (2002) A reference distribution of Fst values for SNPs. Am J Phys Anthropol 117 Suppl 34:121 [Google Scholar]
- Pakstis AJ, Zhao H, Kidd JR, Kidd KK (2000) Patterns of linkage disequilibrium for multisite haplotypes at 14 genetic loci in human populations worldwide. Am J Hum Genet 67 Suppl 2:24 [Google Scholar]
- Peterson RJ, Goldman D, Long JC (1999) Effects of worldwide population subdivision on ALDH2 linkage disequilibrium. Genome Res 9:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupon RE, Nalpas B, Coutelle C, Fleury B, Couzigou P, Higueret D, French Group for Research on Alcohol and Liver (1992) Polymorphism of alcohol dehydrogenase, alcohol and aldehyde dehydrogenase activities: implication in alcoholic cirrhosis in white patients. Hepatology 15:1017–1022 [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, et al (1998) Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81:207–215 [PubMed] [Google Scholar]
- Shen Y-C, Fan J-H, Edenberg HJ, Li T-K, Cui Y-H, Wang Y-F, Tian C-H, Zhou CF, Zhou RL, Wang J, Zhao ZL, Xia GY (1997) Polymorphisms of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–1277 [PubMed] [Google Scholar]
- Smith M (1986) Genetics of human alcohol and aldehyde dehydrogenases. Adv Hum Genet 15:249–290 [DOI] [PubMed] [Google Scholar]
- Tanaka FY, Shiratori Y, Yokosuka O, Imazeki F, Tsukada Y, Omata M (1997) Polymorphisms of alcohol-metabolizing genes affects drinking behavior and alcoholic liver disease in Japanese men. Alcohol Clin Exp Res 21:596–601 [PubMed] [Google Scholar]
- Templeton AR, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF (2000) Cladistic structure within the human lipoprotein lipase gene and its implications for phenotypic association studies. Genetics 156:1259–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li T-K, Hwu H-G, Chen C-C, Yeh E-K, Yin S-J (1994) Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res 18:640–643 [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai X-L, Jerome RE, Li T-K, Wang S-P, Lin YT, Lu R-B, Yin S-J (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48:677–681 [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Dietzsch E, Speed W, Pakstis AJ, Kidd JR, Cheung K, Bonne-Tamir B, Santachiara-Benerecetti AS, Moral P, Krings M, Pääbo S, Watson E, Risch N, Jenkins T, Kidd KK (1996) Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 271:1380–1387 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, Kidd JR, Pakstis AJ, Jenkins T, Kidd KK (1998) A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet 62:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall TL, Garcia-Andrade C, Thomasson HR, Carr LG, Ehlers CL (1997) Alcohol dehydrogenase polymorphisms in Native Americans: identification of the ADH2*3 allele. Alcohol Alcohol 32:129–132 [DOI] [PubMed] [Google Scholar]
- Wright S (1969) Evolution and the genetics of populations: the theory of gene frequencies. Vol 2: The theory of gene frequencies. University of Chicago Press, Chicago [Google Scholar]
- Xu Y, Carr LG, Bosron WF, Li T-K, Edenberg HJ (1988) Genotyping of human alcohol dehydrogenases at the ADH2 and ADH3 loci following DNA sequence amplification. Genomics 2:209–214 [DOI] [PubMed] [Google Scholar]
- Zhao H, Pakstis AJ, Kidd JR, Kidd KK (1999) Assessing linkage disequilibrium in a complex genetic system. I. Overall deviation from random association. Ann Hum Genet 63:167–179 [DOI] [PubMed] [Google Scholar]