Abstract
Many aspects of the biology and epidemiology of influenza B viruses are far less studied than for influenza A viruses, and one of these aspects is effectiveness and resistance to the clinically available antiviral drugs, the neuraminidase (NA) inhibitors (NAIs). Acute respiratory infections are one of the leading causes of death in children and adults, and influenza is among the few respiratory infections that can be prevented and treated by vaccination and antiviral treatment. Recent data has suggested that influenza B virus infections are of specific concern to pediatric patients because of the increased risk of severe disease. Treatment of influenza B is a challenging task for the following reasons:
NAIs (e.g., oseltamivir and zanamivir) are the only FDA-approved class of antivirals available for treatment;
the data suggest that oseltamivir is less effective than zanamivir in pediatric patients;
zanamivir is not prescribed to patients younger than 7;
influenza B viruses are less susceptible than influenza A viruses to NAIs in vitro;
although the level of resistance to NAIs is low, the number of different molecular markers of resistance is higher than for influenza A viruses, and they are not well defined;
the relationship between levels of NAI phenotypic resistance and known molecular markers, frequency of emergence, transmissibility, and fitness of NAI-resistant variants are not well established.
This review presents current knowledge of the effectiveness of NAIs for influenza B virus and antiviral resistance in clinical, surveillance, and experimental studies.
Keywords: influenza B virus, neuraminidase inhibitors, antiviral drug resistance, oseltamivir, zanamivir, peramivir
1. Introduction
Each year, thousands of people worldwide contract influenza virus and develop an acute respiratory infection. The causative agents of these infections are influenza A and B viruses, which lead to annual epidemics with significant morbidity and mortality. Although influenza B has been long overshadowed by seasonal epidemics and pandemic outbreaks of influenza A, recent surveillance and epidemiological data indicate that it contributes to a higher global burden of circulation and disease than traditionally thought. Since 2004, seasonal influenza B epidemics have contributed between 1% and 56% of all circulating influenza virus strains and 1% to 52% of influenza-associated pediatric mortality. Two lineages of influenza B virus, named Victoria and Yamagata after their progenitor strains, co-circulate globally and present challenges for vaccine strain selection. As the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir are currently the only two therapeutic options for the control of influenza B, the establishment and spread of NAI-resistant influenza B viruses would be a public health concern. A clear understanding of the frequency and mechanisms of NAI resistance is therefore needed, so that public health professionals can best diagnose and treat influenza B virus and implement control measures to mitigate emergence and spread of resistant viruses.
This review article consists of six sections in which we present current knowledge of influenza B virus, focusing on treatment options and the emergence of NAI resistance. The first section contains an overview of the historical and biological features of influenza B viruses. Part two discusses epidemiologic data specific to influenza B virus, including symptoms, infection rates, and potential complications. Section three reviews treatment regimens for the control of influenza B virus infection and clinical efficacy of NAIs in adult and pediatric patients. In section four, we provide an in-depth review of phenotypic levels and NA molecular markers associated with reduced susceptibility or resistance to NAIs. We review the contribution of catalytic and framework NA residues to levels of NAI resistance from influenza B viruses in clinical samples, surveillance isolates, and experimentally generated viruses. For each group, we include and discuss the accompanying patient, surveillance, or experimental data. In part five, we review the relative fitness of susceptible and resistant viruses from experimental data. We conclude by reviewing novel anti-influenza drugs in clinical trials as future treatment options.
2. Biology of influenza B viruses
Influenza B virus was first isolated in 1940 from a pediatric patient and designated as influenza B/Lee/40 (distinct from the previously identified influenza A virus) based on the lack of reactivity with post-infection ferret serum to influenza A/Puerto Rico/8/1934 (H1N1) virus (Francis, 1940; Horsfall et al., 1940). Influenza B virus is well documented to cause seasonal influenza outbreaks that coincide with but receive less attention than seasonal outbreaks of influenza A virus. Influenza B viruses belong to the family Orthomyxoviridae and share similar structural features of this family, such as the negative-sense, single-stranded, segmented RNA genome. However, influenza B viruses have features distinct from influenza A viruses that classify them into a different genus.
First, the hemagglutinin (HA) and NA surface proteins are antigenically distinct from those of influenza A viruses. Second, while influenza A and B viruses contain equal numbers of gene segments, the protein products and non-coding regions (NCRs) differ. Influenza B virus encodes fewer viral proteins due to a lack of alternative protein products of the polymerase genes (PB1-F2, N40, PA-X, and PA-M encoded by influenza A virus), but a second protein product (NB) is encoded from the influenza B virus NA gene from a -1 open reading frame. The NB protein is an 11 kDa transmembrane protein with ion-channel activity that is incorporated into virions and required for efficient replication in vivo but is dispensable for virus growth in vitro (Betakova et al., 1996; Hatta and Kawaoka, 2003; Sunstrom et al., 1996). The 5' NCRs are longer for each gene segment in influenza B viruses (Jackson et al., 2011; Stoeckle et al., 1987). Third, the matrix BM2 protein of influenza B viruses, while performing a function similar to the ion channel protein M2 of influenza A viruses, is resistant to the adamantane class of antiviral drugs. Resistance is structurally innate, because adamantanes do not bind to the ion pore of BM2 (Davies et al., 1964). Fourth, as an indication of the persistence of influenza B virus exclusively in humans, the NS1 protein preferentially binds to ISG15 of human and non-human primates (Guan et al., 2011).
Another striking difference is the rate of evolution and ecology of influenza A and B viruses. Influenza A viruses evolve rapidly, are characterized by a broad host range, are maintained in an wild aquatic bird reservoir, and can be isolated from humans, waterfowl, domestic avian species, horses, pigs, seals, dogs, and cats. Influenza B viruses infect humans and evolve at a slower rate, likely due to lack of wild animal reservoir (Chen and Holmes, 2008; Nobusawa and Sato, 2006). Seals were shown to be competent for influenza B virus infection, but their role in transmission or as a source of genetic diversity is unknown (Bodewes et al., 2013; Ohishi et al., 2002; Osterhaus et al., 2000).
Antigenic and genetic variation of the HA protein of influenza B viruses resulted in the emergence of two distinct lineages represented by the prototype viruses B/Victoria/2/87 (Victoria lineage) and B/Yamagata/16/88 (Yamagata lineage) (Shaw et al., 2002). Yamagata was the primary lineage circulating until the 1980s, when Victoria lineage viruses appeared first in China in 1975 then worldwide in 1985; since then, drift variants of both HA lineages have co-circulated globally (Chen et al., 2007; Chen et al., 2008; Matsuzaki et al., 2004; McCullers et al., 2004; Puzelli et al., 2004), with both circulating in recent influenza seasons (Chi et al., 2008; Li et al., 2008; Roy et al., 2011). Importantly, co-circulation of the two lineages results in a different pattern of evolution of influenza B virus and can explain some of the disparate variability of seasonal outbreaks (Yamashita et al., 1988). The same two genetic lineages were identified in the NA genes of influenza B viruses. These two NA lineages have diverged since 1983, and due to the probability of inter-lineage reassortment among influenza B viruses, the viruses carrying mixed HA-NA combinations from the two lineages have been isolated worldwide (Hay et al., 2001; Rota et al., 1992). Though all combinations of HA and NA result in viable virus (McCullers et al., 2004), current strains contain NA of Yamagata lineage and HA of either Victoria or Yamagata lineages (WHO, 2013).
3. Epidemiology and clinical manifestation of infection caused by influenza B viruses
The frequency of laboratory-confirmed cases, clinical burden in different population groups, associated complications, and rates of hospitalizations have been less studied in patients infected with influenza B virus than with influenza A virus. As a result, its epidemiology and impact on public health are less understood and often underestimated. The frequency of laboratory-confirmed influenza B cases in the Northern Hemisphere from 2004 to 2013, based on data reported by the Centers for Disease Control and Prevention for the United States (CDC, 2013a), the National Institute for Medical Research for Europe (NIMR, 2013), and the National Institute of Infectious Diseases for Japan (NIID, 2013), is highly variable, depending on the influenza season (Figure 1A). The average global percentage of circulating influenza B viruses from 2004-13 was 21.4%, ranging in the United States from 1.4% in 2009-10 to 35.7% in 2012-2013, in Europe from 2.5% in 2006-7 to 46% in 2012-2013, and in Japan from 0.9% in 2009-10 to 55.3% in 2004-2005. In the Southern Hemisphere (Australia) the average percentage of circulating influenza B viruses since 2000 was 22.2%, ranging from 0.8% in 2003 to 63.3% in 2008 (Barr and Jelley, 2012). These data suggest that influenza B viruses circulate at lower frequency than influenza A viruses but can vary significantly among different epidemic seasons.
Figure 1.
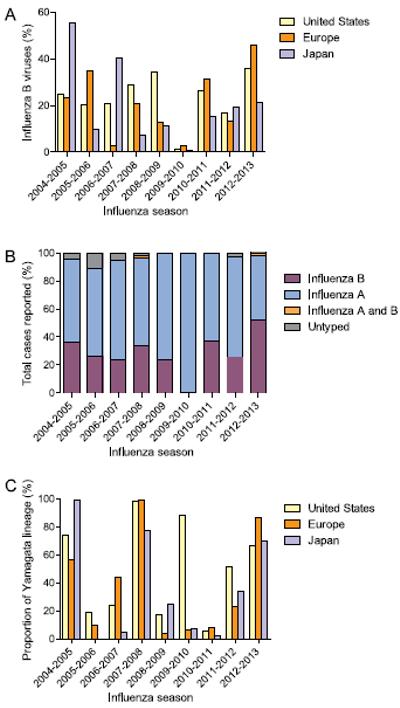
Frequency of laboratory-confirmed cases, influenza B virus-associated pediatric mortality, and proportion of Yamagata lineage across the Northern Hemisphere. A) Frequency of circulation of influenza B viruses during 2004-2013. Data from Centers for Disease Control and Prevention (CDC, USA), National Institute for Medical Research (NIMR, Europe), and National Institute of Infectious Diseases (NIID, Japan). B) Influenza-associated pediatric mortality in the United States (2004-2013) delineated by influenza subtype. Influenza A and B indicate co-infections. Pediatric ages: < 18 years old, data from CDC, USA. C) Proportion of influenza B strains subtyped as Yamagata lineage in the United States, Europe, and Japan. Years Yamagata lineage included in seasonal influenza vaccine: 2004-5, 2005-6, 2008-9, and 2012-13. Data from CDC, WHO, and NIID.
The percentage of circulating strains identified as Yamagata lineage is moderately consistent across the Northern Hemisphere (the United States, Europe, and Japan) during an influenza season (Figure 1C) (CDC, 2013a; EuroFlu; NIID, 2013). However, the low percentage of influenza B strains during the 2009-2010 H1N1pdm season significantly increased the variability. During the 2009-2010 influenza season the percentage of Yamagata-lineage strains was 84% in the United States, but only 6% in Europe and 6.7% in Japan. Excluding the 2009-2010 influenza season, the average standard deviation of Yamagata lineage proportion per year across the three regions was only 12.8%, ranging from 3.1% in 2010-2011 to 21.5% in 2004-2005. There was no significant difference in the variation of the Yamagata lineage during matched or mismatched vaccine years (p = 0.7), nor between different regions (United States versus Europe versus Japan), even excluding the 2009-2010 season.
Surveillance studies suggest higher attack rates among children and young adults infected with influenza B virus than among older individuals, and those infections are more severe (NIID, 2013; WHO, 2011). Whether such a pattern is related to the relatively slow antigenic changes of influenza B viruses is not known. CDC reports of specific influenza-associated pediatric deaths since 2004 (Figure 1B) indicate that during the last nine influenza seasons in the United States, up to 52.3 % (2012-2013) of influenza-associated pediatric deaths were attributed to influenza B (CDC, 2013b). In fact, 27% of all influenza-associated pediatric deaths in the United States during the 2011-2012 epidemic season were attributed to influenza B viruses, even though they comprised only 18% of all circulating viruses (McCullers and Hayden, 2012).
To our knowledge, randomized studies comparing clinical features of influenza A and B virus infections have not been conducted. A systematic clinical and literature review suggested that myalgia, sore throat, and hoarseness are more often associated with influenza B than influenza A, but they are otherwise clinically indistinguishable (Glezen et al., 2013). Some reports suggest that influenza B was more common than influenza A in children with malignancies or other immunosuppression conditions (Carr et al., 2011; Peltola et al., 2003; Peltola et al., 2011). Other studies have underlined the significance of gastrointestinal symptoms in influenza B (Peltola et al., 2003; Wright et al., 2006). It is not clear, however, whether these symptoms are attributed to the pathogenicity of influenza B virus itself or whether they are simply more common in children and young adults in whom this infection is predominant.
The primary hospitalization rate associated with influenza B cases (81.4 per 100,000) is similar to that of seasonal influenza A (H1N1) and A (H3N2) viruses (55.9 and 99.0 per 100,000, respectively) (Thompson et al., 2003). However, a retrospective study of 45 fatal influenza B virus infections in the United Sates observed rapid clinical progression, with an average time from illness onset to death of 3 days. This is shorter than the average time to death for influenza A during the 1918, 1957, 1968, and 2009 pandemics and for seasonal H3N2 virus infections (Paddock et al., 2012).
Complications of influenza B are understudied, and most reports available to date are on single patients with multiple complications; most are from children, often with immunocompromised status (Aebi et al., 2010; Chen et al., 2011; Namendys-Silva et al., 2011). Pulmonary complications such as primary viral pneumonia, secondary bacterial or fungal pneumonia, combined viral-bacterial pneumonia, and exacerbations of chronic pulmonary diseases have been described (Aebi et al., 2010; Chen et al., 2011; Gutierrez-Pizarraya et al., 2012; Krell et al., 2003; Paddock et al., 2012; Wang et al., 2009; Yusuf et al., 2007). Importantly, histological evidence of interstitial pneumonia and immunohistochemical features of alveoli involvements during influenza B virus infections was provided for fatal cases (Paddock et al., 2012). This study also found that bacterial superinfections were more common in patients over 18 (Paddock et al, 2012). Several single-patient reports of severe influenza B suggested complication with acute respiratory distress syndrome (Cardoso et al., 2012; Lu et al., 2004; Namendys-Silva et al., 2011; Wang et al., 2009).
The important extrapulmonary complication of influenza B virus infections is myocardial injury, which is observed in almost 70% of fatal cases (Paddock et al., 2012). Other extrapulmonary complications include central nervous system involvement, such as viral encephalitis and encephalopathy, gastrointestinal symptoms, or myositis (Craver et al., 1997; Frank et al., 2010; Muneuchi et al., 2009; Newland et al., 2003; Tabbutt et al., 2004; Wright, 2006). Additional studies designed specifically for populations with influenza B virus infections are needed to properly estimate the burden of complications.
4. NAIs for the control of influenza B
Vaccination remains the primary means for the prevention of influenza and an essential public health tool for mitigating the impact of epidemics and pandemics. Selection of the influenza B virus strain for the annual trivalent vaccine is challenging; it was matched with the circulating influenza B virus in the Northern Hemisphere only 5 times in 10 seasons between 2000 and 2010 (Belshe, 2010). However, with the approval in the United States of both live attenuated (FluMist Quadrivalent, MedImmune) and inactivated (Fluarix Quadrivalent, GlaxoSmithKline and Fluzone, Sanofi Pasteur) quadrivalent influenza vaccines, the CDC estimates that between 40,000 and 275,000 fewer illnesses per year could be prevented due to reduce influenza B virus infections (FDA, 2013a, b, c; Reed et al., 2012).
Treatment and prophylaxis with antiviral drugs are another option for the control of influenza. There are currently two FDA-approved drugs effective for influenza B virus infections: oseltamivir (Tamiflu®, Roche) and zanamivir (Relenza®, GlaxoSmithKline) that target the viral NA surface protein. The function of the NA protein is to cleave terminal sialic acids from the cell surface, promoting release of newly formed virions. The NAIs were designed to bind the NA enzyme active site of influenza A viruses and thus slow the spread of newly synthesized viral particles (von Itzstein et al., 1993). Another FDA-approved class of anti-influenza drugs, the adamantanes (amantadine and rimantadine), are not effective against influenza B viruses (Davies et al., 1964).
Oseltamivir, the most widely prescribed NAI, is administered orally as a prodrug (oseltamivir phosphate) (Genentech USA, 2012; Hoffman-La Roche, 2007). After processing by liver enzymes into the active form, oseltamivir carboxylate, it is distributed systemically to all potential sites of infection with a maximum plasma concentration of 350 μg/L (He et al., 1999). Oseltamivir is approved for the treatment of acute influenza in patients 2 weeks of age and older. In the United States it is administered orally to adults at 75 mg twice daily for 5 days, to children from 1 to 12 years old on a tiered weight-based schedule, and to infants 2 weeks to 1 year old at 3 mg/kg twice daily for 5 days (Genentech USA, 2012). For adult patients with renal impairment, the recommended dose is reduced to 75 mg once daily. Oseltamivir is also approved for prophylaxis of influenza at a dosage of 75 mg per day for up to 6 weeks for adults and based on body weight for children older than one year old (Harper et al., 2009).
Zanamivir, the other FDA-approved NAI, is administered by inhalation via a disk inhaler and the active compound it deposited directly in the respiratory tract, with a maximum plasma concentration of 17-142 ng/ml (GlaxoSmithKline, 2013). Zanamivir is approved for the treatment of acute influenza in adults and in children 7 years or older with a recommended dosage of 10 mg twice daily for 5 days by inhalation (Harper et al., 2009). Zanamivir is approved for prophylaxis in adults and in children 7 years or older, using a single daily 10 mg dose for 10 days for household prophylaxis and for 28 days for seasonal prophylaxis (Harper et al., 2009). Both oseltamivir and zanamivir are beneficial when administered 48 hours after infection by reducing secondary complication and decreasing mortality.
Soon after the development of oseltamivir and zanamivir multiple studies were conducted to establish their clinical efficacy against influenza in adults and children (Cooper et al., 2003; Hayden et al., 1997; Hayden et al., 1999a; Hayden et al., 1999b; Monto et al., 1999; Nicholson et al., 2000; Treanor et al., 2000; Whitley et al., 2001). None was conducted exclusively for influenza B virus-infected patients, but the presence of laboratory-confirmed influenza B virus was noted in all studies. Because these studies used different criteria and primary efficacy endpoints for analysis, and in conjunction with low numbers of laboratory-confirmed influenza B, specific conclusions regarding efficacy against influenza B virus infections could not be determined, but in aggregate the data suggest that oseltamivir is less effective at treating influenza B infections.
In a randomized, double-blind, placebo-controlled study among children 1-12 years old, oseltamivir (2 mg/kg/dose, twice daily for 5 days) given within 48 hours of the onset of illness was shown to significantly reduce the duration of fever and other symptoms compared with placebo and was a well tolerated therapy for acute influenza B in previously healthy children (Whitley et al., 2001). Three randomized, double-blind, placebo-controlled, parallel group studies evaluated oral oseltamivir for early treatment or prevention of experimental influenza B in healthy susceptible adults (Hayden et al., 2000). Two treatment studies (n=60 and n=117) similarly found that 75 mg doses of oseltamivir administered 24 hours after virus inoculation reduced virus titers and the duration of virus shedding (p = 0.0005). In the prevention study (n=58), oseltamivir did not reduce infection rates but did significantly reduce virus titers (p = 0.03) and the duration of shedding (36 versus 84 hours; p = 0.03) compared to placebo (Hayden et al., 2000). Overall these results indicate that for influenza B virus infection, oseltamivir can reduce the duration of illness during treatment, but cannot prevent infection during prophylaxis.
Recent clinical studies conducted primarily in Japan, the country with the highest per capita NAI use, have shown that oseltamivir is less effective at treating influenza B than influenza A, primarily in children (Hoffman-La Roche, 2007; Kawai et al., 2006; Sato et al., 2008; Sugaya et al., 2007). In a prospective, multicenter study of the 2003-2004 and 2004-2005 influenza seasons, oseltamivir was administered to 1485 patients with influenza B virus infection and 1818 patients with influenza A virus infection (Kawai et al., 2006). The patients were also divided into 4 subgroups on the basis of age (0-6, 7-15, 16-64, and >64 years). After 4-6 days of oseltamivir therapy, the influenza B virus re-isolation rate (51.6%) was significantly higher than the influenza A re-isolation rate (15.9%) (p < 0.001). It was concluded that oseltamivir was less effective for influenza B than for influenza A, with regard to the duration of fever and virus persistence, irrespective of patient age or the timing of administration of the first dose (Kawai et al., 2006).
A study conducted in a Japanese pediatric clinic during four influenza seasons (2001-2005) showed that, although oseltamivir was effective in reducing the duration of fever for both influenza B and A, it was less effective for influenza B which lasted longer than influenza A under treatment (Sato et al., 2008). Notably, this study was conducted over multiple years, and sufficient numbers of patients were enrolled (209 influenza B virus-infected patients were given oseltamivir, and 66 were not treated). Another Japanese group reported the effectiveness of oseltamivir against influenza B and A (H3N2) infections in children on the basis of the duration of fever in which oseltamivir was administered to 362 children with influenza B (mean age, 5.16 years) and 127 children with influenza A (H3N2; mean age, 6.97 years) in outpatient clinics (Sugaya et al., 2007). The authors conclude that fever responded more slowly to oseltamivir in children with influenza B infections than influenza A (H3N2) infections.
In a later study, the same group found that resolution of fever in children treated with oseltamivir was similar between influenza B and seasonal influenza A (H1N1) virus infections but slightly faster for influenza A (H3N2) (Sugaya et al., 2008). In Finland, a randomized, double-blind, placebo-controlled trial addressed the efficacy of oseltamivir given to children 1-3 years old within 24 hours of symptom onset of laboratory-confirmed influenza during the 2007-8 and 2008-9 seasons (Heinonen et al., 2010). The authors did not observe any efficacy of oseltamivir in children with influenza B, although the numbers of children were too small to draw significant conclusions (8 patients in oseltamivir treatment and 11 in the untreated control group).
Zanamivir was shown to be equally effective for influenza A and B infections, in terms of the percentage of patients afebrile after 24 and 48 hours, in a study with a small number of patients (Kawai et al., 2007). Another study of 380 influenza B patients, in which 177 received zanamivir, 171 oseltamivir and 32 no treatment, reported marginally significant differences in the duration of fever after the first dose of zanamivir (31.8 ± 18.4 hours) and oseltamivir (35.5 ± 23.9 hours) for influenza A (p < 0.05) (Kawai et al., 2008). For influenza B, the duration of fever after starting zanamivir (35.8 ± 22.4 hours) was significantly shorter than for oseltamivir (52.7 ± 31.3 hours; p < 0.001). There were no significant differences between influenza A and B in the percentage of patients afebrile 24 or 48 hours after the first inhalation of zanamivir. The re-isolation rate after zanamivir therapy showed marginally significant differences between influenza A and B (p < 0.05). Overall, zanamivir therapy was more effective than oseltamivir for the treatment of influenza B (Kawai et al., 2008).
Meta-analysis has been used to identify significance and draw conclusions from the numerous and disparate reports of oseltamivir efficacy (Ebell et al., 2013; Jefferson et al., 2012; Michiels et al., 2013; Wang et al., 2012). Unfortunately, these meta-analyses have not delineated NAI treatment for influenza B, but have suggested that oseltamivir is marginally effective in reducing the duration of illness for all influenza infections.
5. NAI resistance among influenza B viruses
5.1. Methods of detection of NAI resistance
The emergence of NAI-resistant influenza B viruses is of great concern, as much less is known regarding the significance of molecular markers of NAI resistance than for influenza A viruses. Here we describe the phenotypic susceptibility of influenza B viruses with NAI-resistant mutations, arranged into three groups:
clinical samples isolated before or after NAI treatment with accompanying patient data,
surveillance isolates identified from retrospective studies; and
resistant viruses generated by reverse genetics or passaging under drug selection in vitro. Each group separately provides data important for evaluating the significance of NAI resistance among influenza B viruses, including patient history, the use of NAIs, prevalence of resistance in a given season or geographic region, and characterization of markers in known genetic backgrounds.
Evaluation of influenza B virus susceptibility to NAIs is similar to the approaches used for influenza A, and includes phenotypic and genotypic methods. Phenotypic functional assays are based on biochemical NA inhibition using small synthetic sialic acid conjugates as substrates that produce either a fluorescent or luminescent signal upon cleavage by the viral NA surface protein. As substrates, the fluorescence assay uses 2′-O-(4-methylumbelliferyl)-N-acetylneuraminic acid (MUNANA) and the luminescence assay uses a 2-dioxetane derivative of neuraminic acid (Buxton et al., 2000; Potier et al., 1979). Phenotypic analysis typically determines the concentration of NAI required to inhibit NA activity by 50% relative to a mixture containing virus but no inhibitor (IC50 value). These phenotypic assays utilize cultured influenza viral isolates, and Madin-Darby canine kidney (MDCK) cells are the most commonly used cell line for propagation of influenza B viruses prior to testing in the functional assay. MDCK cells may provide a growth advantage to particular influenza virus variants, and therefore analysis of the NA sequence of virus isolates and their matching original clinical specimens is required to confirm known and novel markers associated with reduced NAI susceptibility. Genotypic methods are variable and utilize different techniques, such as real-time polymerase chain reaction (RT-PCR) and single-nucleotide polymorphism assays or sequencing using Sanger, pyrosequencing, or deep sequencing methods (Deyde et al., 2010; Sheu et al., 2010).
The NAs of influenza A and B viruses are structurally similar, but their overall amino acid homology is only ~30% (Colman, 1994; Russell et al., 2006). However, the 19 amino acids that make up the catalytic site are highly conserved among all known influenza A and B NAs (Figure 2) (Colman et al., 1993). These include catalytic residues (R118, D151, R152, R224, E276, R292, R371, and Y406; N2 numbering here and throughout the text) that have a direct interaction with the substrate (sialic acid) and framework residues (E119, R156, W178, S179, D/N198, I222, E227, H274, E277, N294, and E425) that support the catalytic residues for functional binding and catalysis (Burmeister et al., 1992; Varghese et al., 1992). Genotypic methods can identify the presence of known molecular markers of NAI resistance, and although they can also identify potential mutations involved in the resistant phenotype, all novel mutations must be further evaluated in phenotypic assays.
Figure 2.
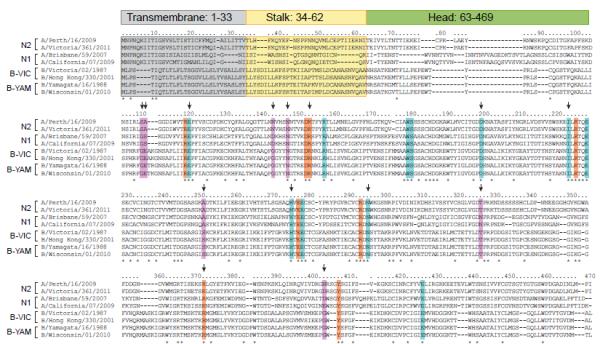
Alignment of NA proteins of N1 and N2 subtypes of influenza A viruses and influenza B viruses. Transmembrane, stalk, and head regions of NA protein are indicated by gray, yellow, or green shading, respectively. Catalytic residues are highlighted in orange (R118, D151, R152, R224, E276, R292, R371, and Y406), framework residues in teal (E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425). Residues associated with NAI resistance in influenza B viruses from clinical, surveillance, or experimental studies are designated with an arrow, with other supporting residues (not catalytic or framework) shown in lavender (G109, E110, S250, T325, G402, G142+N146). Strain subtype and years of use in vaccine composition (shown in parentheses): A/Perth/16/2009 (H3N2, 2010-2012); A/Victoria/361/2011 (H3N2); A/Brisbane/59/2007 (H1N1, 2008-2010); A/California/07/2009 (H1N1pdm, 2010-2013); B/Victoria/02/1987 (HA and NA genes from Victoria lineage); B/Hong Kong/330/2001 (HA and NA genes from Victoria lineage, 2002-2004); B/Yamagata/16/1988 (HA and NA genes from Yamagata lineage, 1989-1992); B/Wisconsin/01/2010 (HA and NA genes from Yamagata lineage, 2012-2013).
One subtle, but clinically important difference between influenza A and B viruses is the consistent observation that influenza B viruses lacking known molecular markers of NAI resistance have up to 10 times higher baseline IC50 values (McKimm-Breschkin et al., 2003; Monto et al., 2006). Importantly, this observation is often specific to the fluorescent phenotypic assay: IC50 values for oseltamivir of susceptible H1N1pdm and H3N2 influenza A viruses range from 0.1 to 0.5 nM, while influenza B viruses exhibit elevated IC50 values ranging from 5.19 to 38.9 nM. Conversely, influenza B viruses exhibit moderately elevated IC50 values for oseltamivir with the chemiluminescent substrate (2.07 to 5.21 nM) (Escuret et al., 2008; Kawai et al., 2007; McKimm-Breschkin et al., 2003; Okomo-Adhiambo et al., 2010; Sheu et al., 2008; Tashiro et al., 2009; Wetherall et al., 2003). These values are still lower than the maximum plasma concentrations for oseltamivir (He et al., 1999).
The basis for the elevated IC50 values for oseltamivir of influenza B viruses is structural: NA proteins appear to be less flexible than that of influenza A viruses as incomplete rotation of the E276 side chain of the active site prevents complete binding to the hydrophobic pocket of oseltamivir but not zanamivir or peramivir (Oakley et al., 2010; Taylor et al., 1998). The criteria recommended by the World Health Organization (WHO) Antiviral Working Group for data interpretation for the resistant phenotype are based on fold change in IC50 values compared with susceptible viruses and these criteria are different for influenza B viruses. The criteria are the following: normal inhibition (NAI-susceptible influenza B viruses) < 5-fold; reduced inhibition, 5- to 50-fold; highly reduced inhibition, > 50-fold increase in IC50 value over susceptible viruses (Pozo et al., 2013; WHO, 2012a). The molecular markers associated with NAI-resistant influenza A viruses have been reviewed elsewhere (Nguyen et al., 2012a; Samson et al., 2013).
5.2. NAI-resistant NA mutations in clinical studies
Clinical isolates identified from patients undergoing NAI treatment provide direct evidence of the development of NAI resistance and thus are of particular importance from a public health perspective. Information regarding patient age, immunocompetent status, initiation and duration of antiviral treatment, and the presence of infected close household contacts are important parameters for understanding the frequency of generation and transmissibility of influenza B viruses with reduced susceptibility to NAIs.
To date, only four reports have described the isolation of influenza B viruses with reduced susceptibility to NAIs after treatment with either oseltamivir (Bastien et al., 2011; Hatakeyama et al., 2007; Ison et al., 2006) or zanamivir (Gubareva et al., 1998). Each of them identified individual NA mutations (Table 1) located either in the catalytic [R152K, (Gubareva et al., 1998)] or framework NA residues [D198N, (Ison et al., 2006)] or near the sialidase enzymatic center [G109E and G402S, (Bastien et al., 2011; Hatakeyama et al., 2007)]. Importantly, two of the four reports identified drug-resistant influenza B viruses from immunocompromised patients (Gubareva et al., 1998; Ison et al., 2006). Influenza B/Memphis/20/1996 virus with an R152K catalytic NA mutation was identified from an 18-month-old immunocompromised patient treated with zanamivir (Gubareva et al., 1998). The R152K NA mutation was first identified after 12 days of zanamivir treatment in virus passaged in MDCK cells and was later confirmed in a clinical sample after 14 days of zanamivir treatment by RNA isolation directly from patient aspirates. This R152K NA mutation caused cross-resistance to all NAIs tested and resulted in the reduction of susceptibility to oseltamivir (100-fold) and zanamivir (70-fold) (Mishin et al., 2005). The D198N NA framework mutation was identified from a 2-year-old immunocompromised patient after 4 days of prophylactic oseltamivir treatment (10 mg twice daily) and 23 days of therapeutic oseltamivir treatment (30 mg twice daily for 14 days, followed by 20 mg twice daily for 9 days). The MDCK-passaged clinical isolate contained a quasi-species mixture of virus with D198N/D NA mutations, but analysis of individual viral clones revealed that the D198N mutation conferred 8- and 11-fold decrease in susceptibility to oseltamivir and zanamivir, respectively (Ison et al., 2006).
Table 1.
Clinical isolates of influenza B virus with NA mutations resulting in decreased susceptibility to FDA-approved NAIs.
| NA mutation and function a |
Method of testing b |
Susceptibility to NAIs (range fold difference) c |
Patient information |
||||
|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Age (years) |
Immune status |
NAI treatment (days of treatment prior to resistance) |
Reference | ||
| Catalytic mutation | |||||||
| R152K | FL | 100-187 | 28-70 | 1.5 | IC | zanamivir (14) | Gubareva et al., 1998, 2001 |
| Framework mutation | |||||||
| D198N | FL | 9.2 | 9.4 | 2 | IC | oseltamivir (27) | Ison et al., 2006 |
| D198N d | FL | 2.8 - 3.5 | 7.2 - 9.3 | 1, 6, 8 | -- | oseltamivir (0) | Hatakeyama et al., 2007 |
| I222T d | FL | 6.1 - 7.1 | 3.4 - 4.5 | 3, 5, 6 | -- | oseltamivir (0) | Hatakeyama et al., 2007 |
| H274Y | CL | 11.4 | 1 | 33 | -- | no treatment (0) | Higgins et al., 2012 |
| N294S | FL | 23 | S | 7 | IC | oseltamivir (0) | Carr et al., 2011 |
| Other mutation | |||||||
| G109E | CL | 5.5 | 3.6 | 87 | -- | oseltamivir (3) | Bastien et al., 2011 |
| E105K | CL | 4.2 | 14.6 | 35 | -- | no treatment (0) | Fujisaki et al., 2012 |
| S250G | FL | 0.7 | 29 | 22 | -- | no treatment (0) | Hatakeyama et al., 2007 |
| G402S | FL | 3.9 | 7.1 | 7 | -- | oseltamivir (3) | Hatakeyama et al., 2007 |
The function of NA mutation is based on Colman et al. (1989), N2 numbering.
Fold difference in mean IC50 values derived from FL, fluorescence-based or CL, chemiluminescence-based assay.
Fold difference as compared to mean IC50 value of susceptible isolates.
Cluster of 3 patients.
Abbreviations: S, susceptible; IC, immunocompromised patient (leukemia); -- immunocompetent patient.
Patients who are immunocompromised and those who are very young or elderly have delayed influenza virus clearance (Aoki et al., 2007; Ison et al., 2006; Klimov et al., 1995). For patients receiving therapeutic NAIs, replicating virus will be exposed to the NAI for a greater length of time, increasing the opportunity for the acquisition and propagation of NAI-resistant mutations. Importantly, mutations that reduce NAI susceptibility and viral fitness can proliferate in this host environment, as the viral population is under less pressure from the immune system. As with influenza A viruses, this suggests that immunocompromised patients may increase the chances for emergence of NAI-resistant influenza B viruses, and this should be considered when developing infection control measures.
The other two reports described the isolation of influenza B viruses with reduced susceptibility to NAIs in immunocompetent patients (Bastien et al., 2011; Hatakeyama et al., 2007). Influenza B/Ontario/RV75-11/2010 virus was isolated from a nasopharyngeal swab of an 87-year-old patient after 3 days of oseltamivir treatment (75 mg twice daily) and had a G109E NA mutation (Bastien et al., 2011). The mechanism by which this change leads to reduced susceptibility to NAIs is unknown, but it is possible that residue G109 affects interactions between residue R118, the sialic acid, and the conserved E119 residue that provides a structural framework for the NA active site (Burmeister et al., 1992). In the fourth report from Japan during the 2004-2005 influenza season, an influenza B virus with reduced NAI susceptibility and a G402S NA mutation was isolated in 1 (1.4%) of the 74 children who had received oseltamivir treatment (Hatakeyama et al., 2007). This G402S NA mutation resulted in 3- and 7-fold reductions in susceptibility to oseltamivir and zanamivir, respectively.
Influenza B viruses with reduced susceptibility to NAIs have been identified in patients before initiation of antiviral therapy and without any known exposure to the drug (Carr et al., 2011; Fujisaki et al., 2012; Hatakeyama et al., 2007; Higgins et al., 2012) (Table 1). To date, clinical studies have identified influenza B viruses carrying NA mutations either in the framework (D198N, I222T, H274Y, and N294S) (Carr et al., 2011; Hatakeyama et al., 2007; Higgins et al., 2012) or in close proximity to the enzyme active site (E105K and S250G) (Fujisaki et al., 2012; Hatakeyama et al., 2007). One common feature of these reports has been the isolation of viruses with NAI resistance-associated mutations from other close household contacts infected with influenza B and treated with antiviral drugs.
Table 2.
Surveillance samples of influenza B virus with NA mutations resulting in decreased susceptibility to FDA-approved NAIs.
| NA mutation and function a |
Method of testing b |
Susceptibility to NAIs (range of fold-difference) c |
Reference | |
|---|---|---|---|---|
| Oseltamivir | Zanamivir | |||
| Catalytic mutation | ||||
| R371K | CL | 407 | 29 | Sheu et al., 2008 |
| Framework mutation | ||||
| D198E | on FL | 8.4 - 14 | 6.4 - 9 | Hurt et al., 2004b, 2006 |
| CL | 14.2 | 2.2 | ||
| D198E | CL | 4.0 | 3.1 | Sheu et al., 2010, Okomo-Adhiambo et al., 2010 |
| D198N | CL | 5 | 7 | Wang et al., 2013 |
| D198N | CL | > 10 | > 10 | WHO, 2007, Tashiro et al., 2009 |
| D198Y | FL | 14.5 | 14.31 | Escuret et al., 2008 |
| I222T | CL | 3.2 | 2.1 | Sheu et al., 2008 |
| I222T d | FL | 5 - 8 | S | Wang et al., 2013 |
| CL | 9 | S | ||
| I222T | CL | 7.3 - 13.4 | 2.3 - 6.5 | Monto et al., 2006 |
| I222V e | FL | 5.5 - 6.9 | 2.5 - 3.3 | Sleeman et al., 2011 |
| CL | 2.6 - 4.8 | 1.2 - 2.2 | ||
| H274Y | CL | 3.3 | 0.4 | Sheu et al., 2008 |
| Other mutation | ||||
| T325I | CL | 2.7 | 0.9 | Sheu et al., 2008 |
| G142R+N146K | FL | 6 | 10 | Okomo-Adhiambo et al., 2013 |
The function of NA mutation is based on Colman et al. (1989), N2 numbering.
Fold difference in mean IC50 values derived from FL, fluorescence-based or CL, chemiluminescence-based assay.
Fold difference as compared to mean IC50 value of susceptible isolates.
Cluster of 4 isolates (3 from Chongqing, China and 1 from Shanghai, China).
Cluster of 14 isolates (North Carolina, United States).
Abbreviations: S, susceptible
Influenza B viruses containing either the D198N or I222T NA mutations were isolated from young children prior to oseltamivir treatment in Japan in 2005 (Hatakeyama et al., 2007). In both cases of patients with D198N or I222T mutations had contact with infected family members on therapeutic oseltamivir treatment. Phenotypic testing of the cell-passaged isolate containing either the D198N or I222T NA mutation resulted in reductions in susceptibility to oseltamivir and zanamivir (Hatakeyama et al., 2007). Influenza B virus with an S250G NA mutation was isolated from a patient prior to oseltamivir treatment (Hatakeyama et al., 2007). This NA mutation resulted in a 29-fold decrease in susceptibility to zanamivir, but no change in susceptibility to oseltamivir. The specific role of this residue in NA enzymatic activity is not known, but it is internal to the NA protein and may play a role in NA stability (Burmeister et al., 1992).
An H274Y NA framework mutation was identified from a patient with no known previous antiviral treatment (Higgins et al., 2012). This mutation conferred 11- and 35-fold reductions in susceptibility to oseltamivir and peramivir, respectively, but the virus retained susceptibility to zanamivir. The novel E105K NA mutation was identified from MDCK cell-passaged isolates of a Japanese patient in 2011, but as pyrosequencing was unable to detection the mutation in the original clinical sample, the significance is unknown (Fujisaki et al., 2012; WHO, 2012b). Finally, an N294S NA framework mutation was identified in a virus from a young immunocompromised patient prior to treatment with oseltamivir (Carr et al., 2011). The mutation was present in isolates collected over 25 days of virus shedding, 15 of which included oseltamivir treatment. This mutation conferred a 23-fold reduction in susceptibility to oseltamivir, but the virus remained susceptible to zanamivir. It was suggested the virus containing this N294S NA mutation was likely transmitted from another person undergoing NAI treatment, but patient or family contact information was not available to support this hypothesis.
5.3. NAI-resistant NA mutations in surveillance studies
An important part of surveillance studies is the determination of antiviral susceptibility which allows detection of the emergence and spread of drug-resistant influenza variants. Subtle changes in susceptibility can also be identified when compared with viruses from previous season or from different geographic areas. Although retrospective in most cases, antiviral surveillance studies are based on statistical analysis of data obtained from a large number of samples and can thus provide guidance for the antiviral management of influenza infection. A limitation of antiviral surveillance studies is the lack of information about NAI treatments that complicate our understanding of the sources of emergence and clinical relevance of the identified drug-resistant variants.
Before the introduction of the clinical use of NAIs, no drug-resistant human influenza A or B viruses were detected (McKimm-Breschkin et al., 2003; Zambon and Hayden, 2001). The Neuraminidase Inhibitor Susceptibility Network evaluated 2,287 viruses isolated during the first 3 years of NAI use (1999 to 2002) for susceptibility to oseltamivir and zanamivir and determined that 8 (0.3%) viruses had a >10-fold decrease in susceptibility to oseltamivir, 2 of which were influenza B viruses (Monto et al., 2006).
Antiviral surveillance studies conducted during different seasons and in different geographic areas have shown activity of NAIs against the NA of influenza B viruses in vitro and indicated low frequency of circulation of drug-resistant variants (Escuret et al., 2008; Hurt et al., 2004a; Monto et al., 2006; Okomo-Adhiambo et al., 2010; Sheu et al., 2008; Tashiro et al., 2009; WHO, 2007). Several molecular markers of drug resistance were identified, located either at the conserved NA residues (E119A, D198N, D198E, I222T, I222V, H274Y, and R371K) or in surrounding locations (T325I and G142R+N146K) (Table 2). The growing number of NA mutations in influenza B viruses acquired as a result of drug selection pressure or natural drift presents a challenge for NAI susceptibility testing by surveillance laboratories and requires validated knowledge of the role of specific mutations on the reduction in NAI susceptibility (Nguyen et al., 2012b).
Hurt and colleagues reported detection of an influenza B/Perth/211/2001 virus among 126 influenza B viruses (0.8%) isolated between 1998 and 2002 from Australasia and the Asia-Pacific region (Hurt et al., 2004b). The virus was isolated from an 8-month-old girl with acute respiratory illness and no history of treatment or close contact with patients under treatment with either zanamivir or oseltamivir (Hurt et al., 2006). This virus contained a mixture of NAI-sensitive and -resistant viral clones, and the D198E NA mutation was responsible for the drug-resistant phenotype. A second B/Texas/38/2008 virus containing a D198E mutation with slightly reduced susceptibility was reported by Sheu and colleagues (Sheu et al., 2010) and fold difference was reported by Okomo-Adhiambo and colleagues (Okomo-Adhiambo et al., 2010). A variation of this mutation was found in one other surveillance sample, D198Y in B/Lyon CHU/8.614/2006 (Escuret et al., 2008). The D198Y NA mutation resulted in 14-fold reduction in susceptibility to both oseltamivir and zanamivir while the D198E NA mutation of B/Texas/38/2008 conferred only slight 4-14- and 2-3-fold reductions in susceptibility to oseltamivir and zanamivir, respectively (Escuret et al., 2008; Hurt et al., 2004b; Okomo-Adhiambo et al., 2010).
A large-scale antiviral surveillance study among influenza B viruses circulating worldwide from 2004 to 2008 identified one extreme outlier and three mild outliers among 1070 viruses tested (0.4%) (Sheu et al., 2008). Influenza B/Hong Kong/36/2005 with a single novel amino acid change, R371K, at the highly conserved catalytic position resulted in 407-fold reduction in susceptibility to oseltamivir and 29 fold reduction to zanamivir. This virus had not been selected in the presence of any NAI, either in vitro or in vivo. Three mild outliers detected among influenza B viruses had a change at positions I222T, H274Y, and T325I (Sheu et al., 2008). A later study identified influenza B/Illinois/03/2008 with an E119A NA mutation that had greatly reduced susceptibility to oseltamivir and zanamivir, and was the only virus described with a mutation at residue E119 from a patient sample (Sheu et al., 2010). Another study among 346 influenza B viruses isolated worldwide in April-September 2011 identified two viruses (0.6%) with reduced susceptibility to NAIs (Okomo-Adhiambo et al., 2013). Full NA sequence analysis showed that B/Ontario/1256/2011 had two previously unreported G142R and N146K NA mutations. It is unknown whether substitutions G142R and N146K were present in the virus before culturing because clinical specimens were unavailable for testing (Okomo-Adhiambo et al., 2013). Interestingly, the N146 residue is a predicted site of N-linked glycosylation in N1-N9 subtypes (Air, 2012; Chen et al., 2012), of which alteration can affect NA activity and affinity in N1 and N8 subtypes (Li et al., 1993; Saito and Kawano, 1997).
Three studies have examined changes in susceptibility of influenza B viruses isolated over time. In one study, IC50 values among viruses isolates from 1994-2005 were not significantly different between the Yamagata and Victoria HA lineages (Sugaya et al., 2007). A second study tested 500 isolates from 2001-6 and found a small decrease in IC50 values to oseltamivir from 2001-2 and 2002-3 seasons to the 2003-4 and 2004-5 (mean IC50 34 versus 25.75 nM, p < 0.05) influenza seasons (Tashiro et al., 2009). However, this change was independent of influenza B lineage variation. More recent analysis found no significant difference in NAI susceptibility by HA lineage of influenza B viruses collected in the Southern Hemisphere in 2011 (Okomo-Adhiambo et al., 2013).
Antiviral surveillance of influenza B viruses collected in mainland China during 2010-2011 identified 5 viruses among 680 tested (0.7%) with reduced susceptibility to NAIs (Wang et al., 2013). A single virus with reduced susceptibility to both oseltamivir and zanamivir contained an amino acid substitution at the conserved framework residue D198N, while a cluster of three and one other virus resistant to oseltamivir but not zanamivir and shared the amino acid framework substitution I222T (Wang et al., 2013). In the United States (North Carolina) during the 2010-11 influenza season surveillance testing identified a cluster of 14 influenza B isolates with an I222V framework mutation in mixed populations varying from 62% to 95% in the clinical specimen; two contained double I222V and K360E or S288N NA mutations, but did not exhibit significantly altered susceptibility. Patients had not received antiviral treatment prior to sample collection, although some family contacts had received oseltamivir (Garg et al., 2013). Interestingly, oseltamivir treatment lengthened the duration of illness in patients with the NAI-resistant I222V influenza B virus compared with infection with the susceptible WT (5 versus 3 days, I222T and WT, p = 0.02), while non-treated patients positive for the I222V mutant influenza B virus had a similar duration of illness (3 versus 3.5 days, I222V and WT) (Garg et al., 2013). This suggests that, in the absence of oseltamivir, the mutation confers a slight loss of fitness, but with the addition of oseltamivir, the influenza B virus with an I222V NAI-resistant mutation can cause disease longer than WT. Both influenza B virus NA mutations I222T and I222V resulted in 5- to 8-fold reductions in susceptibility to oseltamivir. In conjunction with the clinical report from Japan describing 3 patients with I222T mutations (Hatakeyama et al., 2007), these clusters of I222T and I222V isolates from China and the United States provide strong evidence that mutations conferring small reductions in susceptibility can be transmitted. Viruses with I222T and I222V mutations have also been identified from individual isolates without evidence of clustering or transmission (Monto et al., 2006; Sheu et al., 2008; Wang et al., 2013).
Most systematic antiviral surveillance studies have evaluated NAI susceptibility of influenza viruses collected with imperfect knowledge of antiviral treatment history. In contrast, the Influenza Resistance Information Study (IRIS) is a 5-year, prospective, multicenter, observational study (Whitley et al., 2013). This report summarized results from 79 centers (64 primary care centers and 15 hospitals) in the United States, China, France, Germany, Poland, Norway, and Australia from December 2008 to March 2011, comprising three Northern and two Southern Hemisphere seasons and including the 2009-10 pandemic. It is one of the largest surveys to specifically examine the prevalence and development of influenza antiviral resistance during the course of disease and its clinical correlates. Of 518 influenza B virus positive samples, 256 patients received antiviral treatment, and no emergent resistance was found (Whitley et al., 2013).
5.4. NAI-resistant NA mutations in experimental studies
The existing experimental approaches used to generate NAI-resistant influenza viruses in vitro are limited to multiple virus passages under drug selection pressure or reverse genetics technology (Table 3). Notably, selection of NAI-resistant influenza B variants in normal or immunodeficient animal models has not been reported. Most of the studies that used drug selection pressure in cell culture for the generation of NAI-resistant viruses were conducted either during preclinical drug development or shortly after introduction into clinical practice (Barnett et al., 1999; Cheam et al., 2004; Colacino et al., 1997; Staschke et al., 1995).
Table 3.
Experimental studies of influenza B virus with NA mutations resulting in decreased susceptibility to FDA-approved NAIs.
| NA mutation and function a |
Method of testing b |
Susceptibility to NAIs (range fold difference) c |
Method of virus generation (drug concentration) d |
Reference | |
|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | ||||
| Catalytic mutation | |||||
| R152K | CL | 252 | 4.7 | RG | Jackson et al., 2005 |
| R292K | CL | >300 | 28.5 | RG | Jackson et al., 2005 |
| Framework mutation | |||||
| E119A | CL | >300 | >560 | RG | Jackson et al., 2005 |
| E119D | FL | 117 | 32667 | Zanamivir (10 μM) | Cheam et al., 2004 |
| E119D | CL | >300 | >560 | RG | Jackson et al., 2005 |
| E119G | P | nd | 86 | Zanamivir (60 μM) | Staschke et al., 1995 Colacino et al., 1997 |
| E119G | FL | nd | 1243 - 1586 | Zanamivir (300 μM) | Barnett et al., 1999 |
| E119G | CL | 31.1 | >560 | RG | Jackson et al., 2005 |
| E119V | CL | >300 | 1.9 | RG | Jackson et al., 2005 |
| D198N | FL | 3.0 - 3.2 | 4.9 - 5.8 | Zanamivir (1μM) | Hatakeyama et al., 2011 |
| I222T | FL | 7.2 | 2.1 | Oseltamivir (1μM) | Hatakeyama et al., 2011 |
The function of NA mutation is based on Colman et al. (1989), N2 numbering.
Fold difference in mean IC50 values derived from FL, fluorescence-based or CL, chemiluminescence-based assay or P, plaque reduction assay in MDCK cells.
Fold difference as compared to mean IC50 value of susceptible isolates.
Concentration of drug during passaging which lead to resistant virus.
Abbreviations: nd, no data; RG, reverse genetics.
Passaging of influenza B viruses in MDCK cells in the presence of zanamivir resulted in resistant variants with NA amino acid substitutions E119G (Barnett et al., 1999; Colacino et al., 1997; Staschke et al., 1995), E119D (Cheam et al., 2004), or D198N (Hatakeyama et al., 2011) (Table 3). These experimentally generated resistant viruses had much higher levels of resistance to both oseltamivir and zanamivir than are typically observed in clinical or surveillance studies. Passaging of influenza B virus in MDCK cells with oseltamivir identified a virus with an I222T NA mutation, but it resulted in only a small reduction of viral susceptibility to oseltamivir (Hatakeyama et al., 2011). This experimental evidence lends strong support to the clinical evidence of transmission of influenza B viruses with similar mutations from patients receiving oseltamivir in Japan and the United States (North Carolina) (Garg et al., 2013; Hatakeyama et al., 2007; Sleeman et al., 2011). The cell culture experiments required multiple passages for the generation of viruses resistant to NAIs, which suggests resistance to these agents arises infrequently. Some experimentally derived NAI-resistant influenza B viruses had NA mutations similar to those identified in clinical and surveillance studies, and, to some degree, validated the two most common NA mutations identified, D198N and I222T.
Reverse genetics technology allows the generation of influenza viruses with mutations of choice, in particular with different mutations in the NA enzyme active site, and the determination of the role of single or multiple mutations against a homogeneous virus genetic background. Further analysis of resistant phenotypes will reveal potential resistance mutations that should be monitored during antiviral treatment. The contributions of the framework NA residue E119 and two catalytic NA residues (R152 and R292) to the reduction of susceptibility to NAIs were addressed by the generation of recombinant influenza B/Beijing/1/1987-like viruses by reverse genetics (Jackson et al., 2005). Introduction of either the E119A or the E119D NA mutation resulted in high resistance to oseltamivir (> 300-fold reduction) and zanamivir (> 560-fold reduction). The E119G NA mutation resulted in high resistance to zanamivir (> 560-fold reduction), but only medium reduction in sensitivity to oseltamivir (31.1-fold reduction), while the E119V mutation resulted in high resistance to oseltamivir (> 300-fold reduction), but no reduction to zanamivir (1.9-fold reduction). Both R152K and R292K NA mutations conferred high resistance to oseltamivir (252- and > 300-fold reductions) but differential resistance to zanamivir (4.7- and 28.5-fold reductions).
Overall, mutations of the catalytic residues (R152K, R292K, R371K) confer the highest levels of resistance, more pronounced to oseltamivir than to zanamivir. Mutations H274Y and N294S, which are commonly associated with NAI resistance in N1 and N2 subtypes, confer moderate resistance to oseltamivir, but remain susceptible to zanamivir in influenza B viruses. The most common mutations are at the framework positions D198 and I222. These residues are permissible for different amino acid substitutions (D198E, D198N, D198Y and I222T and I222V) that confer a similar reduction in susceptibility. Of significant concern are substitutions of the E119 residue, as all mutations (E119A, E119D, E119G or E119V) conferred decreased susceptibility to oseltamivir or zanamivir.
6. Fitness of influenza B viruses with NAI-resistant mutations
The fitness of NAI-resistant influenza B viruses in comparison with their drug-susceptible counterparts remains largely uncharacterized, leaving open questions of their clinical significance. Although potential detrimental effects of NAI-resistant mutations on viral fitness can be assessed by multiple approaches, the most clinically relevant models utilize competitive co-infections (WT and mutant viruses) in vitro and in vivo and virus transmission in a ferret model. The critical limitation of these studies for influenza B virus is its low pathogenicity in mice and ferrets, the commonly used animal models for influenza research (Kim et al., 2009; McCullers et al., 2005). However, a guinea pig model was recently established and can be used as an important tool to study the fitness of NAI-resistant influenza B viruses (Pica et al., 2012).
The fitness of 6 experimentally generated NAI-resistant viruses was tested by examination of their growth kinetics in MDCK cells. It was found that recombinant influenza B/Beijing/1/1987-like viruses with substitutions at residue E119 had different patterns of growth kinetics (Jackson et al., 2005). Compared with the WT, a virus with an E119G NA mutation had no reduction in virus yield, while viruses with E119A or E119V NA mutations had up to a 1 log reduction, and the virus with E119D NA mutation had the yield reduced by almost 2 logs. These differences in growth kinetics correlated with a decrease in NA activity and stability of viruses with different NA mutations. These findings are also in agreement with a previous characterization of differences in growth kinetics between WT B/Hong Kong/8/1973 and the zanamivir-resistant virus with an E119G mutation (Staschke et al., 1995). Two catalytic-site NA mutations, R152K and R292K, introduced into the NA of an influenza B/Beijing/1/1987-like virus also significantly reduced its growth kinetics. The reduction in growth as a result of these two mutations was correlated with a reduction in the stability (R152K) or activity (R292K) of the NA enzyme, suggesting that these two factors contribute significantly to decreased replication of NAI-resistant viruses in cell culture.
To date, two reports have assessed the contribution of the NA catalytic R152K (Gubareva et al., 1998) and framework D198N mutations (Mishin et al., 2005) to the fitness of influenza B viruses in a ferret model. In a competitive co-infection model, when ferrets were inoculated with a 1:60 ratio of WT and R152K viruses, 2 of 3 animals resolved to equal WT/R152K mixed populations after 3 days and entirely to WT after 6 days. In 1 of 3 animals, the resolution was delayed as the mixed population was identified after 6 days. However, when ferrets co-infected with WT and R152K viruses were treated with zanamivir, in all 3 animals only the R152K NA mutant virus was identified after 3 and 6 days. Thus, an influenza B virus with an R152K NA mutation was less virulent, but it had a growth advantage over the WT virus under drug pressure (Gubareva et al., 1998). Another study investigated the role of the D198N NA mutation on virus fitness in the competitive co-infection ferret model (Mishin et al., 2005). When ferrets were inoculated with a 50:50 mixture of the WT virus and the D198N NA mutant, the WT virus was unable to outcompete the D198N NA mutant, as determined by genotyping virus collected from the nasal washes. After inoculation of ferrets with a mixture of viruses, treatment with oseltamivir did not significantly alter the ratio of WT and D198N NA viruses, but did reduce the number of influenza-positive animals.
Although the role of NAI resistance-associated mutations in the transmissibility of influenza B viruses has not been studied in animal models, epidemiological data suggest that drug-resistant mutants can be transmitted to close contacts. Clusters of influenza B viruses with D198N, I222T, and I222V NA mutations have been identified in Japan, China, and the United States, supporting the hypothesis that viruses containing these mutations are transmissible, and therefore were acquired from family or community contacts (Garg et al., 2013; Hatakeyama et al., 2007; Sleeman et al., 2011; Wang et al., 2013). It is also possible that drug-resistant variant viruses can outgrow drug-sensitive variants from mixed populations in the presence of NAIs. A virus with an R152K NA mutation was identified in isolates from an immunocompromised patient after 14 days of zanamivir treatment, indicating virus replication in the presence of the drug (Gubareva et al., 1998). A virus containing the D198N NA mutation isolated from an immunocompromised patient was also identified in a mixed viral population after 27 days of oseltamivir treatment (Ison et al., 2006).
7. Investigational NAIs
Oral oseltamivir and inhaled zanamivir are currently available worldwide for clinical use, and parenteral formulations of these drugs are in phase III clinical trials in the United States. They are being considered for severely ill hospitalized patients who are unable to tolerate oral or inhaled administration of NAIs, such as those with extracorporeal membrane oxygenation, gastric stasis, or bleeding. Although the levels of NAI resistance among seasonal influenza A and B viruses are currently low, the possible emergence and spread of NAI-resistant variants cannot be excluded, emphasizing the need for continued development of novel NAIs and other anti-influenza drugs.
Peramivir is an investigational NAI that was developed using a structure-based design (Babu et al., 2000). Like the other NAIs, peramivir mimics the sialic acid residues of the cell membrane and binds to the NA active site, competing with neuraminic acid, thereby preventing the release of progeny virions from infected host cells. In addition to the carboxyl group that is present in oseltamivir and zanamivir, which binds to amino acids R118, R292, and R317, peramivir has two side chains. One chain is formed by a hydrophobic acetamido group, which binds to the hydrophobic pocket formed by I222 and W178, similar to that of oseltamivir, and the other is a guanidino group which binds to E119, D151, E227, and E277, similar to that of zanamivir. These unique structural properties allow peramivir to bind tightly the N9 NA site for more than 24 hours and slowly dissociate (Bantia et al., 2006; Castillo et al., 2010).
Peramivir is active against influenza B viruses in vitro and in mice and ferrets (Kitano et al., 2011; Sidwell et al., 2001), with a single intravenous administration giving mice 100% protection (Kitano et al., 2011). Peramivir was safe and well tolerated in clinical trials and was approved in Japan and South Korea in 2011 (Shetty and Peek, 2012; Sugaya et al., 2012). A post-marketing study from Japan assessed the efficacy of intravenous peramivir in influenza B cases and showed that it was superior to oseltamivir in reducing the duration of fever (1 vs. 3 days) in children 5-18 years old. However, there was no difference between oseltamivir and peramivir treatment in children less than 9 years old with influenza B (Hikita et al., 2012). The mechanism and molecular markers of influenza B virus resistance to peramivir have not been fully elucidated. The emergence of peramivir-resistant variants was detected after 15 passages in MDCK cells under drug pressure, and the mutant virus had an H274Y NA mutation that conferred a 714-fold reduction in susceptibility to peramivir (Baum et al., 2003).
Laninamivir octanoate was discovered in 2004 and approved for the treatment of influenza in adults and children in Japan in 2010 (Yamashita et al., 2009). It is a pro-drug (CS-8958, previously known as R-118958) that after inhalation is converted in the lungs to its active form, laninamivir (R-125489). It was shown to inhibit the NA activity of the N1-N9 subtypes of influenza A and influenza B viruses in vitro (Yamashita et al., 2009). The metabolized plasma drug concentration in vivo is reported to be 1100 nM after 144 hours (Ishizuka et al., 2010), which is approximately 50 times higher than the level necessary to inhibit influenza B virus replication (Kubo et al., 2012). Based on this pharmacokinetic characteristic, influenza can be treated with a single administration of laninamivir octanoate by oral inhalation. A single dose prolonged the survival of mice infected with a mouse-adapted A/Puerto Rico/8/34 (H1N1) virus and was effective in vitro against influenza A viruses with the H274Y and R292K NA mutations (Yamashita et al., 2009).
Laninamivir was effective effect against influenza B viruses for both Victoria and Yamagata lineages in vitro with similar mean IC50 values of 19.8 ± 8.2 and 18.1 ± 6.3 nM, respectively (Yamashita et al., 2009). There was no difference in the IC50 values between laninamivir and oseltamivir for influenza A virus with a N294S mutation. The IC50 values for laninamivir among influenza B viruses with the D198N and I222T mutations were comparable to those for oseltamivir and zanamivir, suggesting similar effectiveness in vitro (Yamashita et al., 2009). In two prospective, multicenter, randomized, double-blind control studies in adults and children, the clinical efficacy of laninamivir octanoate was comparable to that of oseltamivir against influenza A viruses (Sugaya and Ohashi, 2010; Watanabe et al., 2010). However, the number of patients with influenza B was too small (10 children and 2 adults) to allow a solid conclusion regarding the effectiveness of laninamivir against influenza B viruses. Further clinical studies are needed to confirm the clinical efficacy of laninamivir against susceptible and resistant influenza B virus infections.
8. Concluding remarks
Our knowledge about the efficacy of NAIs against influenza B virus is based on observational studies, which report lower clinical efficacy of oseltamivir against influenza B than influenza A, and in particular, lower efficacy against influenza B in young children. Specific studies must be done to determine whether higher dosages of oseltamivir or the use of other approved antivirals, such as zanamivir in the United States or laninamivir and peramivir in Japan and South Korea, can overcome the reduced effectiveness of oseltamivir for the control of influenza B, with a specific focus on children. Recent developments in rapid antigen detection tests have made it possible to differentiate between influenza A and B viruses, allowing physicians to make informed decisions regarding drug and treatment regimens, based on accurate determination of the virus causing the disease.
The symptoms of patients infected with influenza B viruses with reduced susceptibility to NAIs were similar to those infected with susceptible viruses, indicating that these mutant viruses, at least those with mutations in framework residues, have no significant loss of virulence, even though they have developed drug resistance. The defects and deficiencies in NA enzymatic functions can vary by NA mutations located in different functional regions. Continued evaluation of the biological properties of NAI-resistant influenza B viruses, with an emphasis on specific NA mutations, is needed to fully assess their fitness and transmissibility in humans. Further studies are needed to evaluate the NAI susceptibility of influenza B viruses of different NA lineages, as a majority of antiviral surveillance studies are limited to evaluation of only HA lineages. With the advent of molecular diagnostic tools and rapid genotyping techniques, this information can be readily obtained, and we encourage the reporting of both HA and NA lineages for influenza B viruses.
Several questions regarding NAI efficacy and resistance in influenza B viruses remain unanswered. One of them is why the pattern of NA mutations observed in clinical and surveillance studies for influenza B viruses is different from that of human influenza A viruses of the N1 and N2 subtypes. The most common NA mutations are limited to H274Y and N294S for the N1 subtype, and E119V and R292K for N2 subtype viruses. For NAI-resistant influenza B viruses, multiple mutations are found at the two most common residues, D198 and I222, which lead to emergence of D198N, D198E, D198Y and I222T and I222V NAI-resistant viruses. Another question is why the overall frequency of NAI-resistant influenza B viruses is lower than that observed for influenza A. Does this relate to elevated baseline IC50 values and the lower frequency of circulation of influenza B viruses? Finally, what are the specific clinical features of influenza B virus infections? We know very little regarding the influence of viral load and virulence factors on disease severity and viral persistence, nor do we understand the relationship between pre-existing immunity and the co-circulation of different lineages.
With new NAIs advancing into clinical practice, more studies are needed to define the mechanisms of influenza B virus resistance and elucidate the importance of the resistant genotypes and phenotypes in clinical practice. Continued antiviral surveillance studies are essential to monitor naturally occurring istrains that are resistant to NAIs and to establish molecular markers of resistance.
Highlights.
The role of influenza B viruses in morbidity and mortality is often underestimated. Pediatric patients are at risk for developing severe influenza B virus infections.
Neuraminidase inhibitor-resistant influenza B viruses harbor a spectrum of neuraminidase mutations.
Molecular markers and fitness of neuraminidase inhibitor-resistant influenza B viruses are not well defined.
Acknowledgments
We gratefully acknowledge the editorial assistance of David Galloway and Betsy Williford for help with illustrations. This work was funded, in part, by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under contract number HHSN266200700005C, and by ALSAC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest:
The authors have no personal or financial affiliation with a commercial entity that might pose a conflict of interest. EAG reports receiving research funding from Hoffmann-La Roche and BioCryst Pharmaceuticals.
References
- Aebi T, Weisser M, Bucher E, Hirsch HH, Marsch S, Siegemund M. Co-infection of Influenza B and Streptococci causing severe pneumonia and septic shock in healthy women. BMC infectious diseases. 2010;10:308. doi: 10.1186/1471-2334-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Air GM. Influenza neuraminidase. Influenza and other respiratory viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki FY, Boivin G, Roberts N. Influenza virus susceptibility and resistance to oseltamivir. Antivir Ther. 2007;12:603–616. [PubMed] [Google Scholar]
- Babu YS, Chand P, Bantia S, Kotian P, Dehghani A, El-Kattan Y, Lin TH, Hutchison TL, Elliott AJ, Parker CD, Ananth SL, Horn LL, Laver GW, Montgomery JA. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 2000;43:3482–3486. doi: 10.1021/jm0002679. [DOI] [PubMed] [Google Scholar]
- Bantia S, Arnold CS, Parker CD, Upshaw R, Chand P. Anti-influenza virus activity of peramivir in mice with single intramuscular injection. Antiviral Res. 2006;69:39–45. doi: 10.1016/j.antiviral.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Barnett JM, Cadman A, Burrell FM, Madar SH, Lewis AP, Tisdale M, Bethell R. In vitro selection and characterisation of influenza B/Beijing/1/87 isolates with altered susceptibility to zanamivir. Virology. 1999;265:286–295. doi: 10.1006/viro.1999.0058. [DOI] [PubMed] [Google Scholar]
- Barr IG, Jelley LL. The coming era of quadrivalent human influenza vaccines: who will benefit? Drugs. 2012;72:2177–2185. doi: 10.2165/11641110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Bastien N, Gubbay JB, Richardson D, Sleeman K, Gubareva L, Li Y. Detection of an influenza B virus strain with reduced susceptibility to neuraminidase inhibitor drugs. J. Clin. Microbiol. 2011;49:4020–4021. doi: 10.1128/JCM.05069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum EZ, Wagaman PC, Ly L, Turchi I, Le J, Bucher D, Bush K. A point mutation in influenza B neuraminidase confers resistance to peramivir and loss of slow binding. Antiviral Res. 2003;59:13–22. doi: 10.1016/s0166-3542(03)00011-1. [DOI] [PubMed] [Google Scholar]
- Belshe RB. The need for quadrivalent vaccine against seasonal influenza. Vaccine. 2010;28(Suppl 4):D45–53. doi: 10.1016/j.vaccine.2010.08.028. [DOI] [PubMed] [Google Scholar]
- Betakova T, Nermut MV, Hay AJ. The NB protein is an integral component of the membrane of influenza B virus. J. Gen. Virol. 1996;77(Pt 11):2689–2694. doi: 10.1099/0022-1317-77-11-2689. [DOI] [PubMed] [Google Scholar]
- Bodewes R, Morick D, de Mutsert, Osinga N, Bestebroer T, van der Vliet S, Smits SL, Kuiken T, Rimmelzwaan GF, Fouchier RA, Osterhaus AD. Recurring influenza B virus infections in seals. Emerg. Infect. Dis. 2013;19:511–512. doi: 10.3201/eid1903.120965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister WP, Ruigrok RW, Cusack S. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 1992;11:49–56. doi: 10.1002/j.1460-2075.1992.tb05026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton RC, Edwards B, Juo RR, Voyta JC, Tisdale M, Bethell RC. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal. Biochem. 2000;280:291–300. doi: 10.1006/abio.2000.4517. [DOI] [PubMed] [Google Scholar]
- Cardoso Y, Oliveira E, Vasconcelos J, Cohen AL, Francisco M. Characteristics of patients with influenza-like illness, severe acture respiratory illness, and laboratory-confirmed influenza at a major children's hospital in Angola, 2009-2011. J. Infect. Dis. 2012;206(Suppl 1):S136–139. doi: 10.1093/infdis/jis534. [DOI] [PubMed] [Google Scholar]
- Carr S, Ilyushina NA, Franks J, Adderson EE, Caniza M, Govorkova EA, Webster RG. Oseltamivir-resistant influenza A and B viruses pre- and postantiviral therapy in children and young adults with cancer. Pediatr. Infect. Dis. J. 2011;30:284–288. doi: 10.1097/INF.0b013e3181ff863b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo R, Holland LE, Boltz DA. Peramivir and its use in H1N1 influenza. Drugs Today (Barc) 2010;46:399–408. doi: 10.1358/dot.2010.46.6.1459659. [DOI] [PubMed] [Google Scholar]
- CDC Influenza National and Regional Level Graphs and Data. 2013a http://gis.cdc.gov/grasp/fluview/fluportaldashboard.html.
- CDC Influenza-Associated Pediatric Mortality. 2013b http://gis.cdc.gov/GRASP/Fluview/PedFluDeath.html.
- Cheam AL, Barr IG, Hampson AW, Mosse J, Hurt AC. In vitro generation and characterisation of an influenza B variant with reduced sensitivity to neuraminidase inhibitors. Antiviral Res. 2004;63:177–181. doi: 10.1016/j.antiviral.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Chen JM, Guo YJ, Wu KY, Guo JF, Wang M, Dong J, Zhang Y, Li Z, Shu YL. Exploration of the emergence of the Victoria lineage of influenza B virus. Arch. Virol. 2007;152:415–422. doi: 10.1007/s00705-006-0852-6. [DOI] [PubMed] [Google Scholar]
- Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008;66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Huang IA, Wu CT, Hsia SH, Hung PC, Chiu CH. Complicated features in a young child with influenza B virus pneumonia and co-infection with Stenotrophomonas maltophilia. Ann. Trop. Paediatr. 2011;31:159–162. doi: 10.1179/1465328111Y.0000000012. [DOI] [PubMed] [Google Scholar]
- Chen W, Zhong Y, Qin Y, Sun S, Li Z. The evolutionary pattern of glycosylation sites in influenza virus (H5N1) hemagglutinin and neuraminidase. PloS one. 2012;7:e49224. doi: 10.1371/journal.pone.0049224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi CY, Wang SM, Lin CC, Wang HC, Wang JR, Su IJ, Liu CC. Clinical features of children infected with different strains of influenza B in southern Taiwan. Pediatr. Infect. Dis. J. 2008;27:640–645. doi: 10.1097/INF.0b013e31816be008. [DOI] [PubMed] [Google Scholar]
- Colacino JM, Laver WG, Air GM. Selection of influenza A and B viruses for resistance to 4-guanidino-Neu5Ac2en in cell culture. J. Infect. Dis. 1997;176(Suppl 1):S66–68. doi: 10.1086/514179. [DOI] [PubMed] [Google Scholar]
- Colman PM, Hoyne PA, Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J. Virol. 1993;67:2972–2980. doi: 10.1128/jvi.67.6.2972-2980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman PM. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci. 1994;3:1687–1696. doi: 10.1002/pro.5560031007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper NJ, Sutton AJ, Abrams KR, Wailoo A, Turner D, Nicholson KG. Effectiveness of neuraminidase inhibitors in treatment and prevention of influenza A and B: systematic review and meta-analyses of randomised controlled trials. BMJ. 2003;326:1235. doi: 10.1136/bmj.326.7401.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craver RD, Sorrells K, Gohd R. Myocarditis with influenza B infection. The Pediatric Infectious Disease Journal. 1997;16:629–630. doi: 10.1097/00006454-199706000-00018. [DOI] [PubMed] [Google Scholar]
- Davies WL, Grunert RR, Haff RF, McGahen JW, Neumayer EM, Paulshock M, Watts JC, Wood TR, Hermann EC, Hoffmann CE. Antiviral Activity of 1- Adamantanamine (Amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- Deyde VM, Sheu TG, Trujillo AA, Okomo-Adhiambo M, Garten R, Klimov AI, Gubareva LV. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob. Agents Chemother. 2010;54:1102–1110. doi: 10.1128/AAC.01417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebell MH, Call M, Shinholser J. Effectiveness of oseltamivir in adults: a meta-analysis of published and unpublished clinical trials. Fam. Pract. 2013;30:125–133. doi: 10.1093/fampra/cms059. [DOI] [PubMed] [Google Scholar]
- Escuret V, Frobert E, Bouscambert-Duchamp M, Sabatier M, Grog I, Valette M, Lina B, Morfin F, Ferraris O. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors. J. Clin. Virol. 2008;41:25–28. doi: 10.1016/j.jcv.2007.10.019. [DOI] [PubMed] [Google Scholar]
- EuroFlu EuroFlu Electronic Bulletin. http://www.euroflu.org.
- FDA Approved Products - Fluzone Quadrivalent. 2013a http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm356091.htm.
- FDA Approved Products - FluMist Quadrivalent. 2013b http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm293952.htm.
- FDA Approved Products - Fluarix Quadrivalent. 2013c http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm342391.htm.
- Francis T., Jr. A New Type of Virus from Epidemic Influenza. Science. 1940;92:405–408. doi: 10.1126/science.92.2392.405. [DOI] [PubMed] [Google Scholar]
- Frank H, Wittekind C, Liebert UG, Siekmeyer M, Siekmeyer W, Schuster V, Kiess W. Lethal influenza B myocarditis in a child and review of the literature for pediatric age groups. Infection. 2010;38:231–235. doi: 10.1007/s15010-010-0013-4. [DOI] [PubMed] [Google Scholar]
- Fujisaki S, Takashita E, Yokoyama M, Taniwaki T, Xu H, Kishida N, Sato H, Tashiro M, Imai M, Odagiri T. A single E105K mutation far from the active site of influenza B virus neuraminidase contributes to reduced susceptibility to multiple neuraminidase-inhibitor drugs. Biochem. Biophys. Res. Commun. 2012;429:51–56. doi: 10.1016/j.bbrc.2012.10.095. [DOI] [PubMed] [Google Scholar]
- Garg S, Moore Z, Lee N, McKenna J, Bishop A, Fleischauer A, Springs CB, Nguyen HT, Sheu TG, Sleeman K, Finelli L, Gubareva L, Fry AM. A cluster of patients infected with I221V influenza b virus variants with reduced oseltamivir susceptibility--North Carolina and South Carolina, 2010-2011. J. Infect. Dis. 2013;207:966–973. doi: 10.1093/infdis/jis776. [DOI] [PubMed] [Google Scholar]
- Genentech USA, I. Tamiflu® (oseltamivir phosphate) Prescribing Information. Genentech; South San Francisco, CA: 2012. [Google Scholar]
- GlaxoSmithKline Relenza (Zanamivir for Inhalation) prescribing information. 2013 us.gsk.com/products/assets/us_relenza.pdf.
- Glezen WP, Schmier JK, Kuehn CM, Ryan KJ, Oxford J. The Burden of Influenza B: A Structured Literature Review. Am. J. Public Health. 2013;103:e43–e51. doi: 10.2105/AJPH.2012.301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Ma LC, Leonard PG, Amer BR, Sridharan H, Zhao C, Krug RM, Montelione GT. Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus. Proc. Natl. Acad. Sci. 2011;108:13468–13473. doi: 10.1073/pnas.1107032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 1998;178:1257–1262. doi: 10.1086/314440. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Pizarraya A, Perez-Romero P, Alvarez R, Aydillo TA, Osorio-Gomez G, Milara-Ibanez C, Sanchez M, Pachon J, Cordero E. Unexpected severity of cases of influenza B infection in patients that required hospitalization during the first postpandemic wave. J. Infect. 2012;65:423–430. doi: 10.1016/j.jinf.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Harper SA, Bradley JS, Englund JA, File TM, Gravenstein S, Hayden FG, McGeer AJ, Neuzil KM, Pavia AT, Tapper ML, Uyeki TM, Zimmerman RK. Seasonal influenza in adults and children--diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1003–1032. doi: 10.1086/604670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Sugaya N, Ito M, Yamazaki M, Ichikawa M, Kimura K, Kiso M, Shimizu H, Kawakami C, Koike K, Mitamura K, Kawaoka Y. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA. 2007;297:1435–1442. doi: 10.1001/jama.297.13.1435. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Ozawa M, Kawaoka Y. In vitro selection of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. Clin Microbiol Infect. 2011;17:1332–1335. doi: 10.1111/j.1469-0691.2010.03313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Kawaoka Y. The NB protein of influenza B virus is not necessary for virus replication in vitro. J. Virol. 2003;77:6050–6054. doi: 10.1128/JVI.77.10.6050-6054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay AJ, Gregory V, Douglas AR, Lin YP. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:1861–1870. doi: 10.1098/rstb.2001.0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG, Osterhaus AD, Treanor JJ, Fleming DM, Aoki FY, Nicholson KG, Bohnen AM, Hirst HM, Keene O, Wightman K. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenzavirus infections. GG167 Influenza Study Group. N. Engl. J. Med. 1997;337:874–880. doi: 10.1056/NEJM199709253371302. [DOI] [PubMed] [Google Scholar]
- Hayden FG, Atmar RL, Schilling M, Johnson C, Poretz D, Paar D, Huson L, Ward P, Mills RG. Use of the selective oral neuraminidase inhibitor oseltamivir to prevent influenza. N. Engl. J. Med. 1999a;341:1336–1343. doi: 10.1056/NEJM199910283411802. [DOI] [PubMed] [Google Scholar]
- Hayden FG, Treanor JJ, Fritz RS, Lobo M, Betts RF, Miller M, Kinnersley N, Mills RG, Ward P, Straus SE. Use of the oral neuraminidase inhibitor oseltamivir in experimental human influenza: randomized controlled trials for prevention and treatment. JAMA. 1999b;282:1240–1246. doi: 10.1001/jama.282.13.1240. [DOI] [PubMed] [Google Scholar]
- Hayden FG, Jennings L, Robson R, Schiff G, Jackson H, Rana B, McClelland G, Ipe D, Roberts N, Ward P. Oral oseltamivir in human experimental influenza B infection. Antivir Ther. 2000;5:205–213. [PubMed] [Google Scholar]
- He G, Massarella J, Ward P. Clinical pharmacokinetics of the prodrug oseltamivir and its active metabolite Ro 64-0802. Clin. Pharmacokinet. 1999;37:471–484. doi: 10.2165/00003088-199937060-00003. [DOI] [PubMed] [Google Scholar]
- Heinonen S, Silvennoinen H, Lehtinen P, Vainionpaa R, Vahlberg T, Ziegler T, Ikonen N, Puhakka T, Heikkinen T. Early oseltamivir treatment of influenza in children 1-3 years of age: a randomized controlled trial. Clin. Infect. Dis. 2010;51:887–894. doi: 10.1086/656408. [DOI] [PubMed] [Google Scholar]
- Higgins RR, Beniprashad M, Chong-King E, Li Y, Bastien N, Low DE, Gubbay JB. Recovery of influenza B virus with the H273Y point mutation in the neuraminidase active site from a human patient. J. Clin. Microbiol. 2012;50:2500–2502. doi: 10.1128/JCM.00682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T, Hikita H, Hikita F, Hikita N, Hikita S. Clinical effectiveness of peramivir in comparison with other neuraminidase inhibitors in pediatric influenza patients. Int J Pediatr. 20122012:834181. doi: 10.1155/2012/834181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman-La Roche I. Nutley, New Jersey. Pediatric Advisory Committee Breifing Document for Tamiflu. 2007 http://www.fda.gov/ohrms/dockets/ac/07/briefing/2007-4325b_02_15_Sponsor%20Background%20Package%20Roche.pdf.
- Horsfall FL, Hahn RG, Rickard ER. Four Recent Influenza Epidemics: An Experimental Study. J. Clin. Invest. 1940;19:379–392. doi: 10.1172/JCI101140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Barr IG, Hartel G, Hampson AW. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res. 2004a;62:37–45. doi: 10.1016/j.antiviral.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Hurt AC, McKimm-Breschkin JL, McDonald M, Barr IG, Komadina N, Hampson AW. Identification of a human influenza type B strain with reduced sensitivity to neuraminidase inhibitor drugs. Virus Res. 2004b;103:205–211. doi: 10.1016/j.virusres.2004.02.035. [DOI] [PubMed] [Google Scholar]
- Hurt AC, Iannello P, Jachno K, Komadina N, Hampson AW, Barr IG, McKimm-Breschkin JL. Neuraminidase inhibitor-resistant and -sensitive influenza B viruses isolated from an untreated human patient. Antimicrob. Agents Chemother. 2006;50:1872–1874. doi: 10.1128/AAC.50.5.1872-1874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka H, Yoshiba S, Okabe H, Yoshihara K. Clinical pharmacokinetics of laninamivir, a novel long-acting neuraminidase inhibitor, after single and multiple inhaled doses of its prodrug, CS-8958, in healthy male volunteers. J. Clin. Pharmacol. 2010;50:1319–1329. doi: 10.1177/0091270009356297. [DOI] [PubMed] [Google Scholar]
- Ison MG, Gubareva LV, Atmar RL, Treanor J, Hayden FG. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J. Infect. Dis. 2006;193:760–764. doi: 10.1086/500465. [DOI] [PubMed] [Google Scholar]
- Jackson D, Barclay W, Zurcher T. Characterization of recombinant influenza B viruses with key neuraminidase inhibitor resistance mutations. J. Antimicrob. Chemother. 2005;55:162–169. doi: 10.1093/jac/dkh528. [DOI] [PubMed] [Google Scholar]
- Jackson D, Elderfield RA, Barclay WS. Molecular studies of influenza B virus in the reverse genetics era. J. Gen. Virol. 2011;92:1–17. doi: 10.1099/vir.0.026187-0. [DOI] [PubMed] [Google Scholar]
- Jefferson T, Jones MA, Doshi P, Del Mar CB, Heneghan CJ, Hama R, Thompson MJ. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. The Cochrane database of systematic reviews. 2012;1 doi: 10.1002/14651858.CD008965.pub3. CD008965. [DOI] [PubMed] [Google Scholar]
- Kawai N, Ikematsu H, Iwaki N, Maeda T, Satoh I, Hirotsu N, Kashiwagi S. A comparison of the effectiveness of oseltamivir for the treatment of influenza A and influenza B: a Japanese multicenter study of the 2003-2004 and 2004-2005 influenza seasons. Clin. Infect. Dis. 2006;43:439–444. doi: 10.1086/505868. [DOI] [PubMed] [Google Scholar]
- Kawai N, Ikematsu H, Iwaki N, Kawashima T, Maeda T, Mitsuoka S, Kondou K, Satoh I, Miyachi K, Yamaga S, Shigematsu T, Hirotsu N, Kashiwagi S. Longer virus shedding in influenza B than in influenza A among outpatients treated with oseltamivir. J. Infect. 2007;55:267–272. doi: 10.1016/j.jinf.2007.05.176. [DOI] [PubMed] [Google Scholar]
- Kawai N, Ikematsu H, Iwaki N, Maeda T, Kanazawa H, Kawashima T, Tanaka O, Yamauchi S, Kawamura K, Nagai T, Horii S, Hirotsu N, Kashiwagi S. A comparison of the effectiveness of zanamivir and oseltamivir for the treatment of influenza A and B. J. Infect. 2008;56:51–57. doi: 10.1016/j.jinf.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kim YH, Kim HS, Cho SH, Seo SH. Influenza B virus causes milder pathogenesis and weaker inflammatory responses in ferrets than influenza A virus. Viral Immunol. 2009;22:423–430. doi: 10.1089/vim.2009.0045. [DOI] [PubMed] [Google Scholar]
- Kitano M, Itoh Y, Kodama M, Ishigaki H, Nakayama M, Ishida H, Baba K, Noda T, Sato K, Nihashi Y, Kanazu T, Yoshida R, Torii R, Sato A, Ogasawara K. Efficacy of single intravenous injection of peramivir against influenza B virus infection in ferrets and cynomolgus macaques. Antimicrob. Agents Chemother. 2011;55:4961–4970. doi: 10.1128/AAC.00412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimov AI, Rocha E, Hayden FG, Shult PA, Roumillat LF, Cox NJ. Prolonged shedding of amantadine-resistant influenzae A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J. Infect. Dis. 1995;172:1352–1355. doi: 10.1093/infdis/172.5.1352. [DOI] [PubMed] [Google Scholar]
- Krell S, Adams I, Arnold U, Kalinski T, Aumann V, Konig W, Konig B. Influenza B pneumonia with Staphylococcus aureus superinfection associated with parvovirus B19 and concomitant agranulocytosis. Infection. 2003;31:353–358. doi: 10.1007/s15010-003-3091-8. [DOI] [PubMed] [Google Scholar]
- Kubo S, Tokumitsu A, Tomozawa T, Kakuta M, Yamashita M. High and continuous exposure of laninamivir, an anti-influenza drug, may work suppressively to generate low-susceptibility mutants in animals. J Infect Chemother. 2012;18:69–74. doi: 10.1007/s10156-011-0292-4. [DOI] [PubMed] [Google Scholar]
- Li S, Schulman J, Itamura S, Palese P. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J. Virol. 1993;67:6667–6673. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Shih SR, Huang YC, Chen GW, Chang SC, Hsiao MJ, Tsao KC, Lin TY. Clinical and genetic characterization of severe influenza B-associated diseases during an outbreak in Taiwan. J. Clin. Virol. 2008;42:45–51. doi: 10.1016/j.jcv.2007.11.026. [DOI] [PubMed] [Google Scholar]
- Lu KC, Chen PY, Huang FL, Yu HW, Kao CH, Lau YJ. Influenza B virus associated pneumonia: report of one case. Acta Paediatr Taiwan. 2004;45:242–245. [PubMed] [Google Scholar]
- Matsuzaki Y, Sugawara K, Takashita E, Muraki Y, Hongo S, Katsushima N, Mizuta K, Nishimura H. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J. Med. Virol. 2004;74:132–140. doi: 10.1002/jmv.20156. [DOI] [PubMed] [Google Scholar]
- McCullers JA, Saito T, Iverson AR. Multiple genotypes of influenza B virus circulated between 1979 and 2003. J. Virol. 2004;78:12817–12828. doi: 10.1128/JVI.78.23.12817-12828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Hoffmann E, Huber VC, Nickerson AD. A single amino acid change in the C-terminal domain of the matrix protein M1 of influenza B virus confers mouse adaptation and virulence. Virology. 2005;336:318–326. doi: 10.1016/j.virol.2005.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Hayden FG. Fatal influenza B infections: time to reexamine influenza research priorities. J. Infect. Dis. 2012;205:870–872. doi: 10.1093/infdis/jir865. [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin J, Trivedi T, Hampson A, Hay A, Klimov A, Tashiro M, Hayden F, Zambon M. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 2003;47:2264–2272. doi: 10.1128/AAC.47.7.2264-2272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels B, Van Puyenbroeck K, Verhoeven V, Vermeire E, Coenen S. The value of neuraminidase inhibitors for the prevention and treatment of seasonal influenza: a systematic review of systematic reviews. PloS one. 2013;8:e60348. doi: 10.1371/journal.pone.0060348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob. Agents Chemother. 2005;49:4515–4520. doi: 10.1128/AAC.49.11.4515-4520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto AS, Robinson DP, Herlocher ML, Hinson JM, Jr., Elliott MJ, Crisp A. Zanamivir in the prevention of influenza among healthy adults: a randomized controlled trial. JAMA. 1999;282:31–35. doi: 10.1001/jama.282.1.31. [DOI] [PubMed] [Google Scholar]
- Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG, Zambon M. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 2006;50:2395–2402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneuchi J, Kanaya Y, Takimoto T, Hoshina T, Kusuhara K, Hara T. Myocarditis mimicking acute coronary syndrome following influenza B virus infection: a case report. Cases journal. 2009;2:6809. doi: 10.4076/1757-1626-2-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namendys-Silva SA, Gonzalez-Herrera MO, Texcocano-Becerra J, Herrera-Gomez A. Acute respiratory distress syndrome caused by influenza B virus infection in a patient with diffuse large B-cell lymphoma. Case reports in medicine. 2011;2011:647528. doi: 10.1155/2011/647528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newland JG, Romero JR, Varman M, Drake C, Holst A, Safranek T, Subbarao K. Encephalitis associated with influenza B virus infection in 2 children and a review of the literature. Clin. Infect. Dis. 2003;36:e87–95. doi: 10.1086/368184. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Fry AM, Gubareva LV. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther. 2012a;17:159–173. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Trujillo AA, Sheu TG, Levine M, Mishin VP, Shaw M, Ades EW, Klimov AI, Fry AM, Gubareva LV. Analysis of influenza viruses from patients clinically suspected of infection with an oseltamivir resistant virus during the 2009 pandemic in the United States. Antiviral Res. 2012b;93:381–386. doi: 10.1016/j.antiviral.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Nicholson KG, Aoki FY, Osterhaus AD, Trottier S, Carewicz O, Mercier CH, Rode A, Kinnersley N, Ward P. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomised controlled trial. Neuraminidase Inhibitor Flu Treatment Investigator Group. Lancet. 2000;355:1845–1850. doi: 10.1016/s0140-6736(00)02288-1. [DOI] [PubMed] [Google Scholar]
- NIID Flash report of influenza virus. 2013 http://www.nih.go.jp/niid/en/iasr-inf-e.html.
- NIMR WHO Influenza Centre - Annual and interim reports | MRC National Institute for Medical Research. 2013 London, http://www.nimr.mrc.ac.uk/who-influenza-centre/annual-and-interim-reports/
- Nobusawa E, Sato K. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 2006;80:3675–3678. doi: 10.1128/JVI.80.7.3675-3678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley AJ, Barrett S, Peat TS, Newman J, Streltsov VA, Waddington L, Saito T, Tashiro M, McKimm-Breschkin JL. Structural and functional basis of resistance to neuraminidase inhibitors of influenza B viruses. J. Med. Chem. 2010;53:6421–6431. doi: 10.1021/jm100621s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Ninomiya A, Kida H, Park CH, Maruyama T, Arai T, Katsumata E, Tobayama T, Boltunov AN, Khuraskin LS, Miyazaki N. Serological evidence of transmission of human influenza A and B viruses to Caspian seals (Phoca caspica) Microbiol. Immunol. 2002;46:639–644. doi: 10.1111/j.1348-0421.2002.tb02746.x. [DOI] [PubMed] [Google Scholar]
- Okomo-Adhiambo M, Sleeman K, Ballenger K, Nguyen HT, Mishin VP, Sheu TG, Smagala J, Li Y, Klimov AI, Gubareva LV. Neuraminidase inhibitor susceptibility testing in human influenza viruses: a laboratory surveillance perspective. Viruses. 2010;2:2269–2289. doi: 10.3390/v2102269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okomo-Adhiambo M, Sleeman K, Lysen C, Nguyen HT, Xu X, Li Y, Klimov AI, Gubareva LV. Neuraminidase inhibitor susceptibility surveillance of influenza viruses circulating worldwide during the 2011 Southern Hemisphere season. Influenza and other respiratory viruses. 2013 doi: 10.1111/irv.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. Influenza B virus in seals. Science. 2000;288:1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- Paddock CD, Liu L, Denison AM, Bartlett JH, Holman RC, Deleon-Carnes M, Emery SL, Drew CP, Shieh WJ, Uyeki TM, Zaki SR. Myocardial injury and bacterial pneumonia contribute to the pathogenesis of fatal influenza B virus infection. The Journal of infectious diseases. 2012;205:895–905. doi: 10.1093/infdis/jir861. [DOI] [PubMed] [Google Scholar]
- Peltola V, Ziegler T, Ruuskanen O. Influenza A and B virus infections in children. Clin. Infect. Dis. 2003;36:299–305. doi: 10.1086/345909. [DOI] [PubMed] [Google Scholar]
- Peltola V, Ruuskanen O, Heikkinen T. Targeting influenza vaccinations of children. CMAJ. 2011;183:1464–1465. doi: 10.1503/cmaj.111055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pica N, Chou YY, Bouvier NM, Palese P. Transmission of influenza B viruses in the guinea pig. J. Virol. 2012;86:4279–4287. doi: 10.1128/JVI.06645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal. Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- Pozo F, Lina B, Andrade HR, Enouf V, Kossyvakis A, Broberg E, Daniels R, Lackenby A, Meijer A. Guidance for clinical and public health laboratories testing for influenza virus antiviral drug susceptibility in Europe. J. Clin. Virol. 2013;57:5–12. doi: 10.1016/j.jcv.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Puzelli S, Frezza F, Fabiani C, Ansaldi F, Campitelli L, Lin YP, Gregory V, Bennett M, D'Agaro P, Campello C, Crovari P, Hay A, Donatelli I. Changes in the hemagglutinins and neuraminidases of human influenza B viruses isolated in Italy during the 2001-02, 2002-03, and 2003-04 seasons. J. Med. Virol. 2004;74:629–640. doi: 10.1002/jmv.20225. [DOI] [PubMed] [Google Scholar]
- Reed C, Meltzer MI, Finelli L, Fiore A. Public health impact of including two lineages of influenza B in a quadrivalent seasonal influenza vaccine. Vaccine. 2012;30:1993–1998. doi: 10.1016/j.vaccine.2011.12.098. [DOI] [PubMed] [Google Scholar]
- Rota PA, Hemphill ML, Whistler T, Regnery HL, Kendal AP. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J. Gen. Virol. 1992;73(Pt 10):2737–2742. doi: 10.1099/0022-1317-73-10-2737. [DOI] [PubMed] [Google Scholar]
- Roy T, Agrawal AS, Mukherjee A, Mishra AC, Chadha MS, Kaur H, Chawla-Sarkar M. Surveillance and molecular characterization of human influenza B viruses during 2006-2010 revealed co-circulation of Yamagata-like and Victoria-like strains in eastern India. Infect Genet Evol. 2011;11:1595–1601. doi: 10.1016/j.meegid.2011.05.022. [DOI] [PubMed] [Google Scholar]
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- Saito T, Kawano K. Loss of glycosylation at Asn144 alters the substrate preference of the N8 influenza A virus neuraminidase. J. Vet. Med. Sci. 1997;59:923–926. doi: 10.1292/jvms.59.923. [DOI] [PubMed] [Google Scholar]
- Samson M, Pizzorno A, Abed Y, Boivin G. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res. 2013;98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Sato M, Saito R, Sato I, Tanabe N, Shobugawa Y, Sasaki A, Li D, Suzuki Y, Sakai T, Oguma T, Tsukada H, Gejyo F, Suzuki H. Effectiveness of oseltamivir treatment among children with influenza A or B virus infections during four successive winters in Niigata City, Japan. Tohoku J. Exp. Med. 2008;214:113–120. doi: 10.1620/tjem.214.113. [DOI] [PubMed] [Google Scholar]
- Shaw MW, Xu X, Li Y, Normand S, Ueki RT, Kunimoto GY, Hall H, Klimov A, Cox NJ, Subbarao K. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology. 2002;303:1–8. doi: 10.1006/viro.2002.1719. [DOI] [PubMed] [Google Scholar]
- Shetty AK, Peek LA. Peramivir for the treatment of influenza. Expert Rev Anti Infect Ther. 2012;10:123–143. doi: 10.1586/eri.11.174. [DOI] [PubMed] [Google Scholar]
- Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 2008;52:3284–3292. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu TG, Deyde VM, Garten RJ, Klimov AI, Gubareva LV. Detection of antiviral resistance and genetic lineage markers in influenza B virus neuraminidase using pyrosequencing. Antiviral Res. 2010;85:354–360. doi: 10.1016/j.antiviral.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Sidwell RW, Smee DF, Huffman JH, Barnard DL, Bailey KW, Morrey JD, Babu YS. In vivo influenza virus-inhibitory effects of the cyclopentane neuraminidase inhibitor RJW-270201. Antimicrob. Agents Chemother. 2001;45:749–757. doi: 10.1128/AAC.45.3.749-757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K, Sheu TG, Moore Z, Kilpatrick S, Garg S, Fry AM, Gubareva LV. Influenza B viruses with mutation in the neuraminidase active site, North Carolina, USA, 2010-11. Emerg. Infect. Dis. 2011;17:2043–2046. doi: 10.3201/eid1711.110787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staschke KA, Colacino JM, Baxter AJ, Air GM, Bansal A, Hornback WJ, Munroe JE, Laver WG. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology. 1995;214:642–646. doi: 10.1006/viro.1995.0078. [DOI] [PubMed] [Google Scholar]
- Stoeckle MY, Shaw MW, Choppin PW. Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2703–2707. doi: 10.1073/pnas.84.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya N, Mitamura K, Yamazaki M, Tamura D, Ichikawa M, Kimura K, Kawakami C, Kiso M, Ito M, Hatakeyama S, Kawaoka Y. Lower clinical effectiveness of oseltamivir against influenza B contrasted with influenza A infection in children. Clin. Infect. Dis. 2007;44:197–202. doi: 10.1086/509925. [DOI] [PubMed] [Google Scholar]
- Sugaya N, Tamura D, Yamazaki M, Ichikawa M, Kawakami C, Kawaoka Y, Mitamura K. Comparison of the clinical effectiveness of oseltamivir and zanamivir against influenza virus infection in children. Clin. Infect. Dis. 2008;47:339–345. doi: 10.1086/589748. [DOI] [PubMed] [Google Scholar]
- Sugaya N, Ohashi Y. Long-acting neuraminidase inhibitor laninamivir octanoate (CS-8958) versus oseltamivir as treatment for children with influenza virus infection. Antimicrob. Agents Chemother. 2010;54:2575–2582. doi: 10.1128/AAC.01755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya N, Kohno S, Ishibashi T, Wajima T, Takahashi T. Efficacy, safety, and pharmacokinetics of intravenous peramivir in children with 2009 pandemic H1N1 influenza A virus infection. Antimicrob. Agents Chemother. 2012;56:369–377. doi: 10.1128/AAC.00132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunstrom NA, Premkumar LS, Premkumar A, Ewart G, Cox GB, Gage PW. Ion channels formed by NB, an influenza B virus protein. J. Membr. Biol. 1996;150:127–132. doi: 10.1007/s002329900037. [DOI] [PubMed] [Google Scholar]
- Tabbutt S, Leonard M, Godinez RI, Sebert M, Cullen J, Spray TL, Friedman D. Severe influenza B myocarditis and myositis. Pediatr Crit Care Med. 2004;5:403–406. doi: 10.1097/01.pcc.0000123555.10869.09. [DOI] [PubMed] [Google Scholar]
- Tashiro M, McKimm-Breschkin JL, Saito T, Klimov A, Macken C, Zambon M, Hayden FG. Surveillance for neuraminidase-inhibitor-resistant influenza viruses in Japan, 1996-2007. Antivir Ther. 2009;14:751–761. doi: 10.3851/IMP1194. [DOI] [PubMed] [Google Scholar]
- Taylor NR, Cleasby A, Singh O, Skarzynski T, Wonacott AJ, Smith PW, Sollis SL, Howes PD, Cherry PC, Bethell R, Colman P, Varghese J. Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. J. Med. Chem. 1998;41:798–807. doi: 10.1021/jm9703754. [DOI] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Treanor JJ, Hayden FG, Vrooman PS, Barbarash R, Bettis R, Riff D, Singh S, Kinnersley N, Ward P, Mills RG. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. US Oral Neuraminidase Study Group. JAMA. 2000;283:1016–1024. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- Varghese JN, McKimm-Breschkin JL, Caldwell JB, Kortt AA, Colman PM. The structure of the complex between influenza virus neuraminidase and sialic acid, the viral receptor. Proteins. 1992;14:327–332. doi: 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- Wang CC, Chen PY, Wang JD, Liu FC, Huang FL, Lee CY. Clinical and laboratory analysis of influenza B infection in children in Taichung, Taiwan during the 2006-2007 flu season. Pediatrics and neonatology. 2009;50:54–58. doi: 10.1016/S1875-9572(09)60033-4. [DOI] [PubMed] [Google Scholar]
- Wang D, Sleeman K, Huang W, Nguyen HT, Levine M, Cheng Y, Li X, Tan M, Xing X, Xu X, Klimov AI, Gubareva LV, Shu Y. Neuraminidase inhibitor susceptibility testing of influenza type B viruses in China during 2010 and 2011 identifies viruses with reduced susceptibility to oseltamivir and zanamivir. Antiviral Res. 2013;97:240–244. doi: 10.1016/j.antiviral.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Wang K, Shun-Shin M, Gill P, Perera R, Harnden A. Neuraminidase inhibitors for preventing and treating influenza in children (published trials only) The Cochrane database of systematic reviews. 2012;4 doi: 10.1002/14651858.CD002744.pub4. CD002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Chang SC, Kim MJ, Chu DW, Ohashi Y. Long-acting neuraminidase inhibitor laninamivir octanoate versus oseltamivir for treatment of influenza: A double-blind, randomized, noninferiority clinical trial. Clin. Infect. Dis. 2010;51:1167–1175. doi: 10.1086/656802. [DOI] [PubMed] [Google Scholar]
- Wetherall NT, Trivedi T, Zeller J, Hodges-Savola C, McKimm-Breschkin JL, Zambon M, Hayden FG. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 2003;41:742–750. doi: 10.1128/JCM.41.2.742-750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley RJ, Hayden FG, Reisinger KS, Young N, Dutkowski R, Ipe D, Mills RG, Ward P. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 2001;20:127–133. doi: 10.1097/00006454-200102000-00002. [DOI] [PubMed] [Google Scholar]
- Whitley RJ, Boucher CA, Lina B, Nguyen-Van-Tam JS, Osterhaus A, Schutten M, Monto AS. Global assessment of resistance to neuraminidase inhibitors, 2008-2011: the Influenza Resistance Information Study (IRIS) Clin. Infect. Dis. 2013;56:1197–1205. doi: 10.1093/cid/cis1220. [DOI] [PubMed] [Google Scholar]
- WHO Monitoring of neuraminidase inhibitor resistance among clinical influenza virus isolates in Japan during the 2003-2006 influenza seasons. Wkly. Epidemiol. Rec. 2007;82:149–150. [PubMed] [Google Scholar]
- WHO Review of the 2010-2011 winter influenza season, northern hemisphere. Wkly. Epidemiol. Rec. 2011;86:222–227. [PubMed] [Google Scholar]
- WHO WHO information for molecular diagnosis of influenza virus in humans - update. 2012a http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/
- WHO Antiviral-resistant B viruses with a novel mutation detected by antiviral resistance surveillance in Japan. 2012b http://www.wpro.who.int/emerging_diseases/meetings/docs/AntiviralResistantBVirusesinJapan.pdf.
- WHO Recommended composition of influenza virus vaccines for use in the 2013-2014 northern hemisphere influenza season. Wkly. Epidemiol. Rec. 2013;88:101–114. [PubMed] [Google Scholar]
- Wright PF. The use of inactivated influenza vaccine in children. Semin Pediatr Infect Dis. 2006;17:200–205. doi: 10.1053/j.spid.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Wright PF, Neumann G, Kawaoka Y. In: Fields Virology. 5 David Knipe PH, editor. Lippincott Williams & Wilkins; 2006. pp. 1691–1740. [Google Scholar]
- Yamashita M, Krystal M, Fitch WM, Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology. 1988;163:112–122. doi: 10.1016/0042-6822(88)90238-3. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Tomozawa T, Kakuta M, Tokumitsu A, Nasu H, Kubo S. CS-8958, a prodrug of the new neuraminidase inhibitor R-125489, shows long-acting anti-influenza virus activity. Antimicrob. Agents Chemother. 2009;53:186–192. doi: 10.1128/AAC.00333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf K, Soraisham AS, Fonseca K. Fatal influenza B virus pneumonia in a preterm neonate: case report and review of the literature. J. Perinatol. 2007;27:623–625. doi: 10.1038/sj.jp.7211802. [DOI] [PubMed] [Google Scholar]
- Zambon M, Hayden FG. Position statement: global neuraminidase inhibitor susceptibility network. Antiviral Res. 2001;49:147–156. doi: 10.1016/s0166-3542(01)00124-3. [DOI] [PubMed] [Google Scholar]


