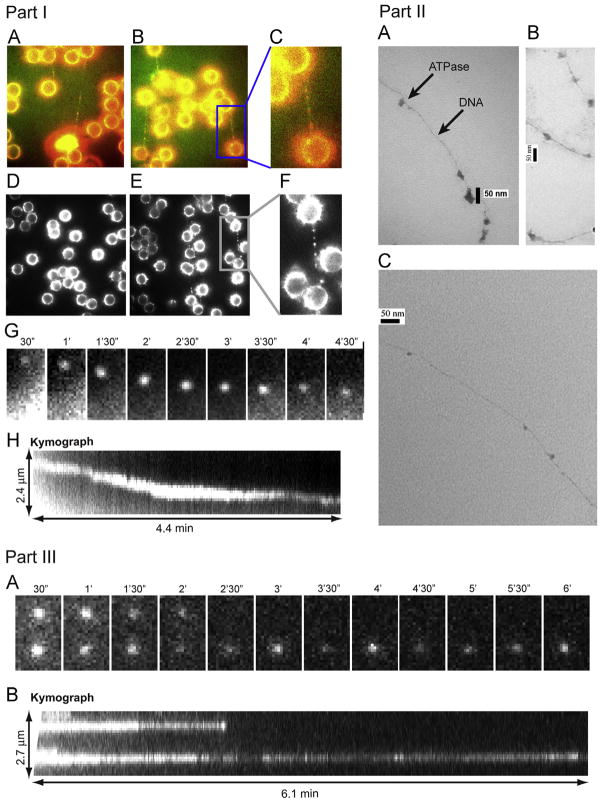
Fig. 8.
Part I. Direct observation of ATPase complex queued and moving along dsDNA. Cy3 conjugated gp16 was incubated with (A, B, E) and without (D) dsDNA, tethered between two polylysine beads where (C, F) are magnified images of the framed regions of (B, E), respectively. (A–C) are overlapped pseudocolor images indicating the binding of Cy3-labeled gp16 along the To-Pro-3 stained dsDNA chain (Red: Cy3-gp16; Green: To-Pro-3 DNA). (G, H) The motion of the Cy3-gp16 spot was analyzed and a kymograph was produced to characterize the ATPase walking. (Actual motion videos can be found in the supplementary information and at http://nanobio.uky.edu/movie.html). Part II. Negatively stained transmission electron microscopy images of ATPase queued along dsDNA. gp16 was bound to non-specific dsDNA in queue. Part III. Recording of two Cy3-gp16/dsDNA complexes showing motionless gp16 spots in a buffer containing no ATP. (A) Sequential images of the recording. (B) Kymograph of the two spots.