Oncogenic Ras generates building blocks for growth, fuels metabolic pathways, and bolsters metabolism. In this review, White discusses advances that shed light on new opportunities with which to cripple the critical metabolic effector functions of Ras.
Keywords: Ras, macropinocytosis, autophagy, cancer metabolism, glutamine, scavenging
Abstract
Oncogenic Ras promotes glucose fermentation and glutamine use to supply central carbon metabolism, but how and why have only emerged recently. Ras-mediated metabolic reprogramming generates building blocks for growth and promotes antioxidant defense. To fuel metabolic pathways, Ras scavenges extracellular proteins and lipids. To bolster metabolism and mitigate stress, Ras activates cellular self-cannibalization and recycling of proteins and organelles by autophagy. Targeting these distinct features of Ras-driven cancers provides novel approaches to cancer therapy.
A hallmark of cancer long known to distinguish normal tissue and cells from that of tumors is altered metabolism. Tumor-specific metabolic reprograming results from the activation of oncogenes and the inactivation of tumor suppressor genes, a subset of which are themselves metabolic enzymes. The functional importance of altered metabolism in cancer derives from the requirement of tumor cells to produce the energy and building blocks required for cell growth and the generation of new cells (Vander Heiden et al. 2009; Dang 2012). In doing so, they must also maintain redox balance, alter gene expression to sustain tumor growth, and withstand stress. Recent findings are revealing how tumor cells accomplish this, which has stimulated interest in targeting metabolic pathways in cancer to improve therapeutic outcome by starving them of what they need to grow and survive.
Sustained tumor growth requires the up-regulation of nutrient acquisition mechanisms, redirection of nutrient utilization to biosynthetic pathways, and adaptation to metabolic stress. Indeed, nutrient demand is higher in tumor cells than most normal cells because of these requirements imposed by growth and proliferation. While it is known that cancers often up-regulate glucose uptake to fuel aerobic glycolysis and require glutamine to fuel central carbon metabolism, this is just the beginning of our understanding of altered tumor metabolism and its functional role in supporting tumor growth. Knowing the details and function of metabolism is important to determine how to interfere with tumor metabolism to block tumorigenesis.
The concept of starving tumor cells by cutting off their blood supply by inhibiting angiogenesis, which is now in the clinic to treat some cancers, was an early, general approach to depriving tumor cells of what they need. To effectively implement the concept of targeting metabolism, we clearly need a more comprehensive understanding of what the “tumor fuel” is, where it comes from, how it is obtained, and how and where it is used. It will also be important to determine how these processes are altered by different oncogenic events, whether they are distinct from those in normal cells, and whether they can be targeted effectively to improve cancer therapy.
Among the most problematic oncogenic events to target directly are those with activation of Ras oncogenes, the presence of which is common and associated with a poor prognosis in the most deadly cancers. Activation of members of the Ras family of oncogenes up-regulates growth-promoting pathways controlled by PI3 kinase/mTOR and MAP kinases, among others, while they simultaneously shut off the brakes that suppress cell growth. Recent advances have shed light on exciting new opportunities with which to cripple the critical metabolic effector functions of Ras.
Hungry, hungry tumor cells
Metabolic demand is greatly increased by cell growth and proliferation, and this contributes to the elevated nutrient requirements of tumor cells (Rabinowitz and White 2010). To meet this elevated demand imposed by cell growth, tumor cells recruit blood vessels through angiogenesis to increase the delivery of growth factors, nutrients, and oxygen to the tumor microenvironment. Tumor vasculature is often abnormal, and tumor growth may outpace vascularization; as a result, tumors often possess hypoxic regions deprived of oxygen, growth factors, and nutrients (Carmeliet and Jain 2011). As a result, tumor cells activate processes that allow them to adapt, tolerate, and grow despite these circumstances. These metabolic changes include induction of the hypoxia-inducible transcription factors (HIF) and up-regulation of glucose uptake, glycolysis, glutamine dependence, and autophagy (DeBerardinis et al. 2007; Yuneva et al. 2007; Semenza 2012; White 2012).
The autophagy starvation response (Kuma et al. 2004; Mizushima et al. 2004) is induced in hypoxic tumor regions, where it is required for tumor cell survival (Degenhardt et al. 2006). Autophagy is also induced in many Ras-driven cancers and is critical for their survival (Guo et al. 2011; Lock et al. 2011; Yang et al. 2011). This suggests that both the environment and activation of oncogenes such as Ras create metabolic stress. Thus, some tumors have deregulated growth but are living at the limits of the availability of nutrients to sustain that growth.
Ras's sweet tooth
A critical fuel for cancers is glucose. Oncogenic Ras promotes glucose uptake (Flier et al. 1987; Ying et al. 2012) and its utilization in glycolysis, which ultimately converts glucose to pyruvate (Racker et al. 1985; Yun et al. 2009). This occurs in the presence of oxygen and is an example of the Warburg effect, long known to be a prominent characteristic of many tumors (Warburg 1956). It is now becoming clear that this plays a causal role in tumorigenesis by facilitating the diversion of glycolytic intermediates into biosynthetic pathways (Vander Heiden et al. 2009; Dang 2012). Examples include shunting glucose carbon through the pentose phosphate pathway (PPP) to increase ribose production for nucleic acid biosynthesis and NADPH production for antioxidant defense (Fig. 1). Glycolysis also generates glycerol for lipid synthesis and can promote hexosamine biosynthesis for protein glycosylation and nutrient uptake as well as serine biosynthesis for α-ketoglutarate production and tricarboxylic acid (TCA) cycle anaplerosis (Wellen et al. 2010; Possemato et al. 2011; Dang 2012). With glucose uptake and activity of glycolysis increased, flux can be slowed at the end to supply biosynthetic pathways by suppression of pyruvate kinase M2 (Christofk et al. 2008a,b). This suggests that interfering with glucose uptake, glycolysis, and the interrelated biosynthetic pathways is a potential approach to impair the growth and survival of Ras-driven and other cancers.
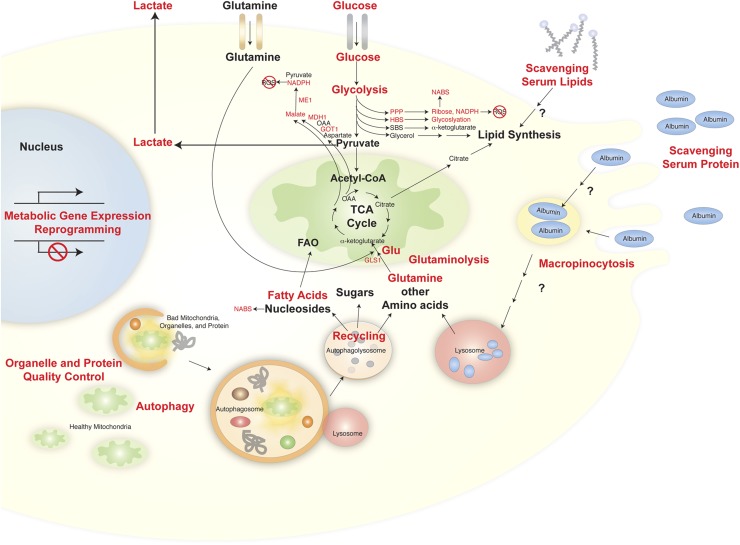
Figure 1.
Fueling Ras. Ras-driven cancer cells alter glucose and glutamine metabolism and activate scavenging and self-cannibalization to supply metabolic pathways. Red indicates processes modulated by or required for Ras. (HBS) Hexosamine biosynthesis pathway; (ME1) malic enzyme 1; (MDH1) malate dehydrogenase; (GOT1) aspartate/glutamate–oxaloacetate transaminase 1; (OAA) oxaloacetate; (GLS1) glutaminase; (GLU) glutamate; (SBS) serine biosynthesis; (ROS) reactive oxygen species; (NABS) nucleic acid biosynthesis; (FAO) fatty acid oxidation.
To determine which aspects of glucose metabolism were most essential for Ras-driven cancers, examination of an inducible mouse model for oncogenic KrasG12D pancreatic ductal adenocarcinoma (PDAC) was revealing. PDACs induced by oncogenic Ras expression showed dramatic regression upon extinction of Ras expression (Ying et al. 2012). An examination of the changes induced by Ras extinction revealed that Ras promoted the expression of metabolic genes, including those in pathways for sterol biosynthesis, pyrimidine metabolism, O-glycan biosynthesis, and glycan structures biosynthesis (Ying et al. 2012). Assessment of metabolite pool sizes and tracer studies following Ras extinction demonstrated that Ras also promotes glucose uptake, glycolytic flux, and channeling of intermediates into the PPP and hexosamine biosynthesis. Flux of glucose carbon through the nonoxidative arm of the PPP and ribose biogenesis is particularly important, as knocking down expression of enzymes in this pathway suppressed tumor growth (Fig. 1; Ying et al. 2012). These Ras-driven cancers may rely on the nonoxidative arm of the PPP for production of precursors essential for nucleic acid biosynthesis and cell growth. These recent revelations suggest that Ras can promote glucose metabolism by altering gene expression (Yun et al. 2009; Ying et al. 2012); although this is just the beginning of our understanding of this process, transcriptional changes are not likely to be the only mechanism of regulation. Regulation of enzymatic activities, quantitative assessment of metabolic changes, and tissue-specific and Ras oncogene-specific differences remain to be addressed.
Glutamine addiction
In contrast to most normal cells, cancers syphon off glucose carbon, and much of the pyruvate that is eventually produced is converted to lactate, which is excreted. This regenerates NAD+ and alters the tumor microenvironment, the consequences of which are poorly understood. The increased conversion of pyruvate to lactate in tumors also potentially limits the availability of pyruvate for conversion to acetyl-CoA to drive the TCA cycle. TCA cycle intermediates may need to be replenished (anaplerosis) by substrates other than glucose. The TCA cycle is important for production of ATP for energy homeostasis, citrate for lipid synthesis, and amino acids and NADH for protein synthesis and metabolism. Glutamine serves as a major anaplerotic substrate for the TCA cycle and also supplies nitrogen for nucleotide, nonessential amino acid, and hexosamine biosynthesis (Wise et al. 2011). Ras- as well as Myc-transformed cells have elevated dependence on glutamine for growth and survival (Wu et al. 1978; Gaglio et al. 2009; Gao et al. 2009), which has attracted interest in determining what the glutamine is used for in cancer cell metabolism and how it is obtained. Indeed, tracer studies during Ras extinction revealed that glutamine is the major carbon source for the TCA cycle when Ras is activated (Ying et al. 2012).
Glutamine enters the TCA cycle downstream from isocitrate following conversion to glutamate by glutaminase and then to α-ketoglutarate by glutamate dehydrogenase (Fig. 1). The TCA cycle can then be completed in the forward direction by converting α-ketoglutarate to succinate, fumarate, malate, and then oxaloacetate to provide ATP and precursors. In the absence of oxygen (hypoxia), glutamine-produced α-ketoglutarate can go in the reverse direction in the TCA cycle by undergoing reductive carboxylation to citrate. This citrate can then serve as a substrate for lipid synthesis in the absence of oxygen (Wise et al. 2011; Metallo et al. 2012; Mullen et al. 2012). This has suggested that inhibiting glutamine metabolism by blocking glutaminase, for example, will compromise the growth of Ras and other glutamine-dependent cancers. In support of this, interfering with glutamine metabolism compromises Ras-driven cancer growth, identifying a metabolic vulnerability (Son et al. 2013). However, this is not the sole, essential use of glutamine in these cancers.
In contrast to normal cells, in pancreatic cancers, where >90% have activating mutations in Kras, glutamine is converted to glutamate and is oxidized by the TCA cycle to oxaloacetate, which is converted to aspartate by glutamate–oxaloacetate transaminase. Evidence suggests that aspartate is converted to oxaloacetate by glutamate–oxaloacetate transaminase and then to malate and pyruvate, purportedly producing NADPH (Fig. 1; Son et al. 2013). Glutamine can also be oxidized to malate in mitochondria and transported to the cytoplasm, where it is converted by malic enzyme to pyruvate with the generation of NADPH (Fig. 1). NADPH generation is necessary for production of reduced glutathione, which maintains redox balance (Son et al. 2013). Indeed, inhibiting glutamine metabolism by blocking aspartate transaminase or enzymes downstream sensitizes pancreatic cancer cells to oxidative stress and suppresses tumor growth, demonstrating the functional importance of this pathway (Fig. 1; Son et al. 2013). Thus, these cancer cells rely more on glutamine than glucose and the oxidative arm of the PPP for mitigating toxic reactive oxygen species (ROS). It will be important to determine whether this is a unique attribute of pancreatic cancers or whether other Ras-driven cancers also rely on this pathway. Moreover, as Ras and other cancers, such as those driven by Myc, rely on glutamine and glucose metabolism more so than normal cells, the next question is: Where do they come from?
Scavenging protein and lipid
Ras-driven cancers can obtain the nutrition they need by increasing their delivery and uptake. For example, Ras and its effector pathways promote angiogenesis to increase the tumor blood supply and uptake of soluble glucose and glutamine by up-regulating the expression of transporters. More recently, it has become apparent that Ras-driven cancers acquire additional means to satisfy their nutritional needs through activation of fluid-phase endocytic nutrient uptake by macropinocytosis and self-cannibalization by autophagy (Fig. 1).
Ras stimulates macropinocytosis, in which altered membrane dynamics at the cell surface promote the engulfment of the aqueous extracellular environment, which is captured in vesicles called macropinosomes (Bar-Sagi and Feramisco 1986; Walsh and Bar-Sagi 2001; Porat-Shliom et al. 2008). Macropinosomes then traffic to the lysosomal compartment, where they are degraded (Commisso et al. 2013). One critical cargo for macropinosomes is serum albumin, which is abundant in the tumor microenvironment. Pancreatic cancers with activating mutations in Ras up-regulate macropinocytosis, which is used to uptake albumin to provide an amino acid supply (Commisso et al. 2013). This “albumin eating” by macropinocytosis provides amino acids, particularly glutamine, for multiple metabolic pathways, including TCA cycle anaplerosis and macromolecular synthesis (Fig. 1). Catabolism of serum albumin is sufficient to support the survival of pancreatic cancer cell lines in vitro, and human pancreatic tumors are most depleted of glutamine when compared with normal tissue (Commisso et al. 2013). Thus, like rats in a garbage dump, Ras-driven cancer cells can adapt to meet their nutritional requirements by acquiring the ability to scavenge and catabolize protein from their microenvironment to sufficiently sustain growth and survival. This suggests that targeting the process of macropinocytosis or the downstream metabolic pathways it supplies may be a novel approach to treat pancreatic cancer. It will be of interest to determine the extent and versatility of the scavenging “diet” and what the most critical processes are for which the scavenged material is used.
One clue that serum protein was not the only scavenged material came from the observation from tracer studies—that, distinct from nonnormal cells, Ras-transformed cancer cells synthesize very-long-chain fatty acids from an existing C24 fatty acid backbone rather than from de novo synthesis (Kamphorst et al. 2011). Where the C24 backbone came from was answered by the discovery that Ras-transformed cells bypass de novo lipogenesis by scavenging fatty acids from their environment in the form of lysophospholipids, abundant serum lipids with a single fatty acid tail (Fig. 1; Kamphorst et al. 2013). Why make them if you can take them? In doing so, this overcomes potential limitations in cytosolic acetyl-CoA production and conserves cellular resources.
Normally, glucose produces pyruvate, which is converted to mitochondrial acetyl-CoA, and then citrate, which is exported to the cytoplasm, where it is converted to acetyl-CoA for fatty acid synthesis by ATP-citrate lyase. Since Ras greatly stimulates glycolysis but also diversionary side pathways and conversion of pyruvate to lactate, this can limit acetyl-CoA and fatty acid synthesis essential for production of the membranes of new cancer cells. Lipid scavenging rather than synthesis also conserves cellular resources and overcomes a key oxygen-dependent step of desaturation by stearyol-CoA desaturase 1 (SCD1), enabling lipid synthesis in hypoxia (Kamphorst et al. 2013). Ras transformation thereby produces resistance to SCD1 inhibitors, indicating that blockade of lipid scavenging or elongation might be new therapeutic opportunities to target Ras-driven cancers.
Self-cannibalization by autophagy
Ras-driven cancer cells activate and rely on cellular self-cannibalization by autophagy, which can be considered a form of scavenging from within (White 2012). In normal cells, autophagy is activated by starvation and stress, where it captures damaged or superfluous intracellular proteins and entire organelles in vesicles that then fuse with lysosomes, where the cargo is degraded (Fig. 1; Mizushima and Komatsu 2011). The autophagy breakdown products generated in lysosomes are released into the cytoplasm for use in metabolic and biosynthetic pathways (Fig. 1; Rabinowitz and White 2010). Intracellular recycling by autophagy is up-regulated and critical for survival of cells and mice during starvation (Kuma et al. 2004; Mizushima et al. 2004). In starved cells, bulk degradation of intracellular components by autophagy and their recycling maintains homeostasis during interruptions in nutrient availability. One can think of the proteome and organelles as an exhaustible intracellular store of building blocks for biosynthesis (protein, sugars, and lipid) and of metabolic intermediates to sustain energy homeostasis accessed by autophagy in times of need.
In normal cells and tissues, the selective pruning of damaged organelles by a low level of basal autophagy prevents their toxic accumulation and constitutes a quality control mechanism (Mizushima and Komatsu 2011). For example, the selective degradation of defective mitochondria preserves the functioning pool while mitigating production of damaging ROS. Autophagy is similarly important for culling unfolded protein and preventing accumulation of toxic, p62-containing protein aggregates (Komatsu et al. 2007, 2010; Mathew et al. 2009). Thus, by recycling their garbage, cells can conserve resources, balance metabolism, and suppress damage.
Ras transformation induces autophagy, which is required for proliferation, survival in starvation, and tumor growth (Guo et al. 2011; Lock et al. 2011; Yang et al. 2011). Knockout or knockdown of essential autophagy genes causes tumor cells to accumulate respiration-defective mitochondria and display growth and metabolic defects and sensitivity to starvation caused by an energy and metabolic crisis. The requirement for autophagy appears to stem from the need for both a functioning pool of mitochondria and sources of metabolic substrates to fuel metabolic pathways. Knockout of an essential autophagy gene in a mouse model of spontaneous KrasG12D-driven non-small-cell lung cancer (NSCLC) causes accumulation of defective mitochondria, suppresses tumor growth, and diverts the progression of carcinomas to more benign oncocytomas (Guo et al. 2013). Oncocytomas are a benign tumor type characterized by the accumulation of defective mitochondria (Gasparre et al. 2011). This suggests that Ras-driven cancers require autophagy for mitochondrial function and for progression to more aggressive disease. Deletion of an essential autophagy gene in mouse models of spontaneous BrafV600E-driven lung tumorigenesis similarly impaired tumor growth and produced oncocytomas, extending these requirements for autophagy beyond Ras (Strohecker et al. 2013).
The key substrates supplied by autophagy to metabolic pathways remain to be investigated. Autophagy deficiency in both KrasG12D- and BrafV600E-driven tumor cells enhances dependence on glutamine, suggesting that protein degradation by autophagy supplies amino acids and their derivatives to metabolic pathways, of which glutamine is particularly critical (Guo et al. 2013; Strohecker et al. 2013). The role of other amino acids such as serine and glycine in the metabolism of Ras-driven cancers remains to be investigated.
In the setting of Ras activation and p53 deficiency in NSCLC, autophagy is also required to preserve mitochondrial function for fatty acid oxidation (FAO) and lipid homeostasis (Guo et al. 2013). Loss of p53 promotes glycolysis and lipid storage while suppressing FAO (Cheung and Vousden 2010; Goldstein and Rotter 2012). Autophagy-preserved mitochondrial function is critical for retaining FAO, without which tumor cells fail to metabolize and thus accumulate lipids and are exquisitely sensitive to further inhibition of FAO (Guo et al. 2013). Thus, inhibiting autophagy in Ras-driven cancers can cut off the vital glutamine fuel supply, block critical fatty acid fuel consumption, and compromise tumor growth. Clinical trials to test this concept by inhibiting lysosomal function and autophagy cargo degradation with hydroxychloroquine are under way in various cancers (Amaravadi et al. 2011; White 2012).
Therapeutically exploiting altered metabolism
The concepts of inhibiting enzymes in the pathways of glycolysis and glutaminolysis in cancer have been, and continue to be, investigated (Vander Heiden et al. 2009; Dang 2012). Drilling down into the details using elegant cancer models has revealed new potential metabolic enzymes to target in Ras-driven cancers. These include enzymes involved in ribose production for nucleic acid biosynthesis and those for glutamine-derived NADPH production for antioxidant defense (Fig. 1). Recent evidence also shows that Ras profoundly alters nutrient acquisition by promoting protein and lipid scavenging and autophagy. By doing so, this provides additional sources of nutrients within cells and from the tumor microenvironment necessary for growth, metabolic robustness, and stress tolerance. Thus, inhibiting scavenging and autophagy also represents new approaches to compromise the growth and survival of Ras-driven cancers.
These new findings raise additional questions. Will blocking nucleotide biosynthesis be selective and active enough? Will compromising NADPH production require combination with promotion of oxidative damage to be effective? What is the complete extent of the tumor cell “diet”? For what essential processes are the scavenged and autophagized material used? How do scavenged serum proteins get to lysosomes? How are lipids recycled? What is the complete mechanism of macropinocytosis? How do scavenged lipids get into cells? What are the essential autophagy cargos? What are the key intracellular substrates supplied by autophagy? Will inhibition of scavenging or autophagy be effective therapeutically for cancer? Do other cancers rely on scavenging or autophagy? What is the potential metabolic plasticity of Ras-transformed cells? Specifically, when one blocks glucose metabolism, is it possible that macropinocytosis becomes increasingly important? There is also currently no quantitative assessment of the contributions of various metabolic pathways to Ras-mediated alterations in metabolism. For example, under a defined condition, how much do glucose, glutamine, and macropinocytosis contribute to the overall bioenergetic needs of Ras-transformed cells? How might this change under different conditions found in the tumor microenvironment?
Knowing that scavenging and autophagy are important functions for Ras-driven cancers, it will be important to determine the metabolic functions that they support. Scavenged serum proteins provide glutamine for anaplerosis and perhaps also ROS detoxification. Scavenging serum lipids may reduce the demand for NADPH required for lipid synthesis, making more available for antioxidant defense. Autophagy may also contribute to ROS detoxification by supplying glutamine but also through the elimination of defective mitochondria, thereby reducing the demand for NADPH and ROS detoxification. Mitigating ROS can promote survival, but also ROS produced from respiration can trigger oncogene-induced senescence, a barrier to tumorigenesis (Kaplon et al. 2013). Combining inhibitors of autophagy and glutamine metabolism with agents that increase oxidative stress may be useful.
These recent insights into the metabolism of Ras-driven cancers have illuminated new metabolic vulnerabilities and potentially new treatment modalities for cancer. Targeting macropinocytosis or autophagy has the potential to starve Ras-driven cancers, and both processes may be enhanced by inhibition of angiogenesis. Since scavenging serum albumin or lipid is functionally important, can it be used to deliver a toxic payload? This may indeed be the case with nanoparticle albumin-bound (nab)-paclitaxel (Abraxane), which increases the median overall survival in combination with gemcitabine in pancreatic cancer over gemcitabine alone (Von Hoff et al. 2011, 2013). It will be important to determine which tumors scavenge extracellular proteins, whether serum albumin is the sole source of glutamine, and exactly which metabolic pathways require it most. Blocking macropinocytosis in Ras-driven cancers needs to be explored. Targeting the enzymes essential for lipid metabolism in the altered scavenging and autophagy setting may be worthwhile. Autophagy is a survival mechanism for Ras-driven cancers, so inhibiting the genesis of autophagosomes or cargo degradation is under investigation. Identification of the substrates that are provided by autophagy and the exact metabolic mechanisms they support may also be valuable. Autophagy will likely be a resistance mechanism to metabolic pathway inhibition, although this remains to be formally tested. Whether inhibition of scavenging or autophagy in cancers has the selectivity and robustness to affect cancer over normal cells, whether there is tissue and oncogene and tumor suppressor gene specificity, and whether there are mechanisms of resistance remain to be determined.
Acknowledgments
I thank Dr. R. Mathew, Dr. J.Y. Guo, Dr. A.M. Strohecker, Dr. S. Joshi, Dr. X. Xie, and Ms. G. Karsli-Uzunbas for helpful comments and discussions. The White laboratory acknowledges support from the NIH grants R01CA163591 and RO1CA130893, the Department of Defense (W81XWH-09-01-0394), the Val Skinner Foundation, the New Jersey Commission for Cancer Research, and the Rutgers Cancer Institute of New Jersey.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.228122.113.
References
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E 2011. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 17: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco JR 1986. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 233: 1061–1068 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK 2011. Molecular mechanisms and clinical applications of angiogenesis. Nature 473: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, Vousden KH 2010. The role of p53 in glucose metabolism. Curr Opin Cell Biol 22: 186–191 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC 2008a. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC 2008b. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452: 181–186 [DOI] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497: 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012. Links between metabolism and cancer. Genes Dev 26: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB 2007. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier JS, Mueckler MM, Usher P, Lodish HF 1987. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science 235: 1492–1495 [DOI] [PubMed] [Google Scholar]
- Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F 2009. Glutamine deprivation induces abortive S-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS ONE 4: e4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparre G, Romeo G, Rugolo M, Porcelli AM 2011. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta 1807: 633–642 [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rotter V 2012. Regulation of lipid metabolism by p53—fighting two villains with one sword. Trends Endocrinol Metab 23: 567–575 [DOI] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. 2013. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev 27: 1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Fan J, Lu W, White E, Rabinowitz JD 2011. Liquid chromatography–high resolution mass spectrometry analysis of fatty acid metabolism. Anal Chem 83: 9114–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD 2013. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci 110: 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, et al. 2013. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498: 109–112 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223 [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J 2011. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. 2012. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M 2011. Autophagy: Renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ 2012. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat-Shliom N, Kloog Y, Donaldson JG 2008. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell 19: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. 2011. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476: 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racker E, Resnick RJ, Feldman R 1985. Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc Natl Acad Sci 82: 3535–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL 2012. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 33: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. 2013. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496: 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E 2013. Autophagy sustains mitochondrial glutamine metabolism and growth of BRAFV600E-driven lung tumors. Cancer Discov doi: 10.1158/2159-8290.CD-13-0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, Korn RL, Desai N, Trieu V, Iglesias JL et al. 2011. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: A phase I/II trial. J Clin Oncol 29: 4548–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Ervin TJ, Arena FP, Chiorean EG, Infante JR, Moore MJ, Seay TE, Tjulandin S, Ma WW, Saleh MN, et al. 2013. Randomized phase III study of weekly nab-paclitaxel plus gemcitabine versus gemcitabine alone in patients with metastatic adenocarcinoma of the pancreas (MPACT). J Clin Oncol 31: LBA148 [Google Scholar]
- Walsh AB, Bar-Sagi D 2001. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J Biol Chem 276: 15609–15615 [DOI] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev 24: 2784–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB 2011. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci 108: 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Arimura GK, Yunis AA 1978. Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase. Int J Cancer 22: 728–733 [DOI] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149: 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, et al. 2009. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 325: 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y 2007. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]