Short telomere syndromes lead to a number of human degenerative disorders. The telomere syndromes Coats Plus and dyskeratosis congenita (DC) have been linked to mutations in CTC1, a member of the CST telomeric replication complex. Here, Lingner and colleagues identify CTC1 disease mutations that disrupt CST complex assembly, interaction with DNA polymerase α-primase, and telomere association. This study uncovers several molecular activities disrupted by disease-associated CTC1 mutations and defines a novel disease-causing structural defect in telomeric DNA.
Keywords: telomere syndrome, Coats Plus, dyskeratosis congenita, CST complex, DNA polymerase α-primase
Abstract
Mutations in CTC1 lead to the telomere syndromes Coats Plus and dyskeratosis congenita (DC), but the molecular mechanisms involved remain unknown. CTC1 forms with STN1 and TEN1 a trimeric complex termed CST, which binds ssDNA, promotes telomere DNA synthesis, and inhibits telomerase-mediated telomere elongation. Here we identify CTC1 disease mutations that disrupt CST complex formation, the physical interaction with DNA polymerase α-primase (polα-primase), telomeric ssDNA binding in vitro, accumulation in the nucleus, and/or telomere association in vivo. While having diverse molecular defects, CTC1 mutations commonly lead to the accumulation of internal single-stranded gaps of telomeric DNA, suggesting telomere DNA replication defects as a primary cause of the disease. Strikingly, mutations in CTC1 may also unleash telomerase repression and telomere length control. Hence, the telomere defect initiated by CTC1 mutations is distinct from the telomerase insufficiencies seen in classical forms of telomere syndromes, which cause short telomeres due to reduced maintenance of distal telomeric ends by telomerase. Our analysis provides molecular evidence that CST collaborates with DNA polα-primase to promote faithful telomere DNA replication.
Telomeres are specialized heterochromatin structures at the ends of linear chromosomes and are essential for genome stability by preventing chromosome fusion and degradation (de Lange 2009; Jain and Cooper 2010). In mammals, telomeric DNA consists of long segments of tandem (TTAGGG) repeat duplexes with a short single-stranded G-rich 3′ overhang. Telomeric DNA is coated with shelterin proteins, which comprise TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 (de Lange 2009). The length of telomeres determines the protective state and the cellular life span. Progressive telomere shortening occurs in somatic cells due to incomplete replication of chromosome ends by conventional DNA polymerases and nucleolytic processing. Telomerase, a ribonucleoprotein complex with reverse transcriptase activity, lengthens telomeres by adding telomere repeats to their 3′ termini, which is essential for telomere length maintenance in germ cells and stem cells (Nandakumar and Cech 2013). The telomere G-rich sequence, with its repetitive nature, can fold into ssDNA secondary structures such as G-quadruplex structures, which are believed to obstruct efficient telomere DNA replication (Bochman et al. 2012; Vannier et al. 2012).
CST is a conserved trimeric complex composed of CTC1, STN1, and TEN1. Mammalian CST has been shown to associate with telomeres and is implicated in multiple steps of telomere replication (Miyake et al. 2009; Surovtseva et al. 2009; Chen et al. 2012; Gu et al. 2012; Huang et al. 2012; Stewart et al. 2012; Wu et al. 2012). Disrupting CST in human and mouse cells affects telomere duplex replication and telomere 3′ overhang generation. Biochemically, CST was demonstrated to associate with DNA polymerase α-primase (polα-primase) and to be able to stimulate the enzymatic activity of DNA polα-primase (Casteel et al. 2009; Nakaoka et al. 2012). However, studies with cells depleted for CST and with mice deleted for CTC1 suggest that CST may be dispensable for general DNA replication (Gu et al. 2012; Stewart et al. 2012). Mechanistically, it remains to be elucidated how CST cooperates with DNA polα-primase for telomere replication. Apart from its functions in semiconservative DNA replication of telomeres, CST plays a role in constraining telomerase activity for telomere length homeostasis in cancer cells (Chen et al. 2012). CST associates with and sequesters telomeric 3′ overhangs after their elongation by telomerase, thus limiting telomere extension by telomerase.
Recently, mutations in CTC1 were found to cause Coats Plus, dyskeratosis congenita (DC), and related bone marrow failure syndromes (Anderson et al. 2012; Keller et al. 2012; Polvi et al. 2012; Walne et al. 2013). Affected individuals carry biallelic CTC1 mutations, which include point mutations and more severe mutations that presumably lead to a complete loss of function (Supplemental Table S1). Coats Plus is a rare autosomal recessive neurological disorder where patients develop cranial calcifications and cysts (Anderson et al. 2012; Polvi et al. 2012). Significantly, Coats Plus patients also develop features seen with the short telomere syndrome DC, instigated by telomerase defects, such as premature hair graying, anemia, and osteoporosis (Keller et al. 2012; Walne et al. 2013). Lymphocyte telomere length in CTC1 mutant individuals was reported to be short in two studies (Anderson et al. 2012; Keller et al. 2012), but this was not seen in two other reports (Polvi et al. 2012; Walne et al. 2013). Thus, the putative telomere dysfunctions in these patients may not be directly associated with overall telomere length loss seen in typical telomere syndromes caused by telomerase deficiencies (Armanios and Blackburn 2012) but instead might relate to other structural defects of telomeres. Here, we characterize the molecular defects of disease-causing CTC1 and uncover that these mutations give rise to a common telomeric replication defect.
Results
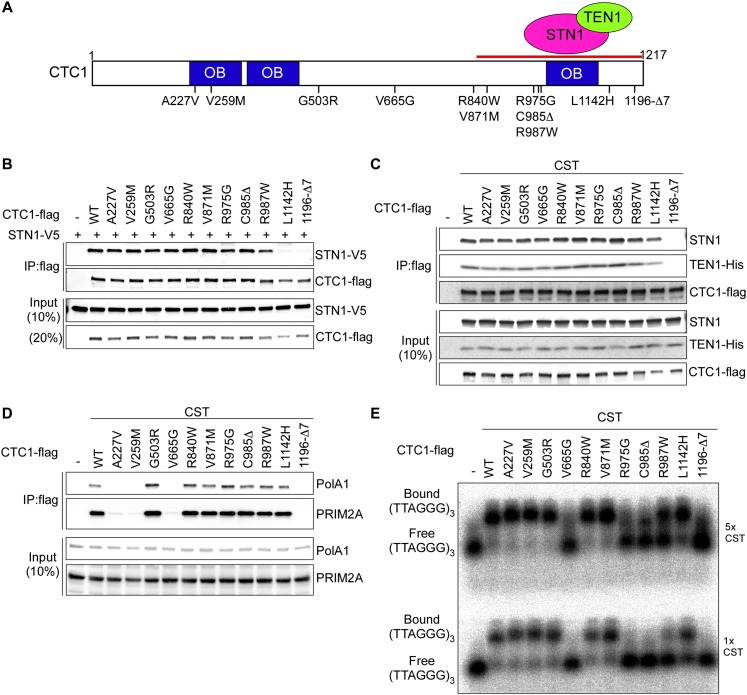
To elucidate the molecular defects caused by CTC1 mutations at telomeres, we generated 11 mutations in human CTC1 cDNA that were reported in Coats Plus/DC patients to produce CTC1 polypeptides carrying point mutations or small deletions (Fig. 1A; Anderson et al. 2012; Keller et al. 2012; Polvi et al. 2012; Walne et al. 2013). CTC1 forms the CST complex with STN1 and TEN1 through a direct physical interaction with STN1. To assess the interaction between CTC1 and STN1, V5-tagged STN1 (STN5-V5) and wild-type or mutant Flag-tagged CTC1 (CTC1-Flag) were coexpressed upon transient transfection in HEK293T cells. Association of STN1-V5 with CTC1-Flag was detected upon immunoprecipitation with anti-Flag antibodies (Fig. 1B). The C-terminal disease mutations CTC1-L1142H and CTC1-1196-Δ7 (deletion of amino acid residues 1196–1202) disrupted the ability of CTC1 to bind to STN1. This is consistent with our previous finding that the C-terminal region of CTC1 mediates STN1 interaction (Chen et al. 2012; Chen and Lingner 2013). Coexpression of TEN1 with CTC1-Flag and STN1 partially rescued CST complex formation of CTC1-L1142H but not CTC1-1196-Δ7 (Fig. 1C; Supplemental Fig. S1A). Therefore, CTC1 assembly into the CST complex involves critical residues of the CTC1 C terminus that promote interactions with STN1. The enhancement of the CTC1–STN1 interaction by TEN1 concurred with a stabilization of the STN1 and TEN1 polypeptides upon concomitant expression of the two factors (Supplemental Fig. S1A).
Figure 1.
CTC1 disease mutations cause various defects in molecular interactions. (A) Summary of characterized mutations in CTC1. The C-terminal STN1–TEN1-interacting domain and putative oligonucleotide/oligosaccharide binding (OB) folds in CTC1 are indicated. (B) Identification of mutations affecting STN1 interaction. STN1-V5 and wild-type or mutant CTC1-Flag were transiently coexpressed in 293T cells. Co-IP was done with anti-Flag antibodies followed by Western blot analysis for CTC1-Flag and STN1-V5 using anti-Flag and anti-V5 antibodies, respectively. (C,D). Identification of mutations affecting CST complex formation and DNA polα-primase interaction. STN1, TEN1-His, and wild-type or mutant CTC1-Flag were transiently coexpressed for co-IP as described in B, followed by Western blot analyses using anti-STN1, anti-his tag, and anti-Flag antibodies in C and using anti-PolA1 and anti-Prim2A antibodies in D. (E) Identification of mutations affecting telomeric G-strand ssDNA binding. Wild-type and CTC1 mutant CST complexes were purified from 293T cells transiently expressing CTC1-Flag, STN1, and TEN1-His as described in the Materials and Methods. Binding of 5× or 1× CST to 0.1 nM (TTAGGG)3 was analyzed by EMSA on agarose gels. The purified CST was analyzed by the Western blot in B.
CTC1 and STN1 are thought to contribute to semiconservative DNA replication of telomeric DNA (Gu et al. 2012; Huang et al. 2012; Stewart et al. 2012). They were shown to associate with DNA polα-primase and increase the affinity of DNA polα-primase for template DNA (Casteel et al. 2009). We therefore assessed by coimmunoprecipitation (co-IP) the physical interaction of CTC1-Flag with the catalytic subunit PolA1 of DNA polα and the regulatory subunit PRIM2A of DNA primase in HEK293T cells in which STN1 and TEN1 were coexpressed (Fig. 1D). Three CTC1 point mutations (A227V, V259M, and V665G) abolished association with endogenous DNA polα-primase. A227 and V259 are part of the most N-terminal of three putative OB (oligonucleotide/oligosaccharide-binding) folds in CTC1, whereas V665 resides in an unrecognized domain of CTC1. In addition, the CST complex-defective CTC1-1196-Δ7 was unable to interact with DNA polα-primase. For CTC1-L1142H, the interaction with DNA polα-primase occurred only upon CST complex formation, which was dependent on ectopic coexpression of STN1 and TEN1 (cf. Fig. 1D and Supplemental Fig. S1B). Together, these data suggest that the physical contacts of both CTC1 and STN1/TEN1 with DNA polα-primase contribute to a stable association of CST with DNA polα-primase. CST was also known to physically interact with the shelterin components POT1–TPP1 (Wan et al. 2009; Chen et al. 2012; Wu et al. 2012), but none of the CTC1 mutations tested here abolished the interaction with TPP1 or POT1 as measured in co-IP experiments (Supplemental Fig. S2).
CST binds with preference to the G-rich telomeric ssDNA (Chen et al. 2012). Therefore, we expressed and partially purified wild-type and mutant CST complexes upon transient transfection in HEK293T cells (Fig. 1C) for electromobility shift assays (EMSAs) with the telomeric G-strand oligonucleotide (TTAGGG)3 (Fig. 1E). EMSA with two different concentrations of CST revealed defects in telomeric ssDNA binding of CTC1-V665G, CTC1-R975G, CTC1-C985Δ, CTC1-R987W, and CTC1-1196-Δ7. DNA binding by CTC1-L1142H was also reduced. Therefore, in vitro binding of CST with telomeric G-strand ssDNA relies on critical residues comprising the most C-terminal 230 amino acids of CTC1. Defects in ssDNA binding of CTC1-L1142 and CTC1-1197-Δ7, which also show diminished ability of STN1 interaction, are consistent with our previous study that CTC1 on its own does not bind ssDNA (Chen et al. 2012). These data support the notion that forming a trimeric complex is crucial for high-affinity binding of telomeric G-rich ssDNA by CST.
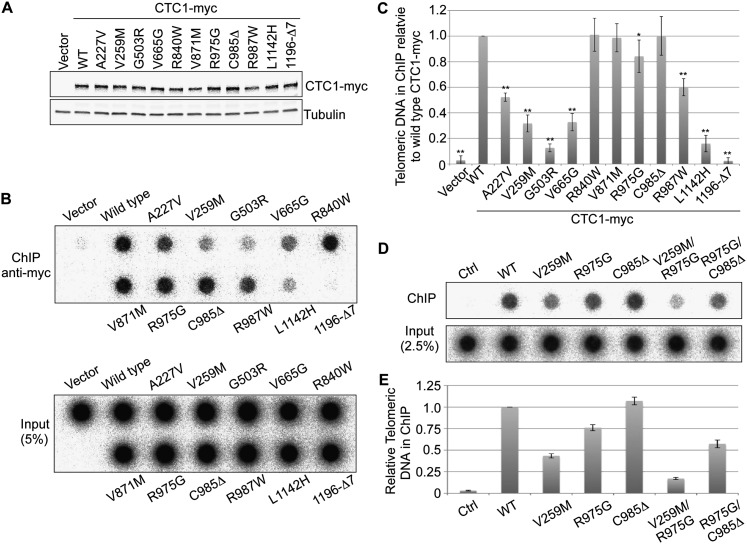
To test whether the disease mutations affect CTC1 telomere association and cellular localization, we stably expressed myc-tagged CTC1 in HT1080 cells (Fig. 2A). Telomere association was measured by chromatin immunoprecipitation (ChIP) using anti-myc antibodies (Fig. 2B–E). We found that the telomere association in vivo required critical residues in the N-terminal and central part of the CTC1 polypeptide (A227, V259, and V665) interfacing the interaction with DNA polα-primase and also an intact C terminus mediating ssDNA binding and CST complex formation. This result suggests that telomere association in vivo depends on the formation of a trimeric CST complex that is both proficient in telomeric ssDNA binding and interacting with DNA polα-primase. Consistently, an additive defect in telomere association was seen when combining CTC1 mutations that affected ssDNA binding with mutations that reduced the interaction with DNA polα-primase (Fig. 2D,E). Intriguingly, the G503R CTC1 disease mutant showed the functional molecular activities examined above except telomere association. This suggests that residue G503 of CTC1 defines an unexplored molecular interaction or post-translational modification required for CST association with telomeres. TRF1 association with telomeres was not affected by expression of mutant CTC1, as determined in ChIP experiments (Supplemental Fig. S3). By immunofluorescence, we also detected colocalization of myc-tagged CTC1 with the telomeric RAP1 protein upon pre-extraction of nuclei with detergent-containing buffer (Supplemental Fig. S4), although this assay did not allow consistent quantification.
Figure 2.
CTC1 disease mutations perturb telomere localization. (A) HT1080 cells stably expressing the wild-type (WT) or mutant myc-tagged CTC1. The expression levels of CTC1-myc and α-tubulin (loading control) were analyzed on Western blots using anti-myc and anti-tubulin antibodies. (B–E) Identification of mutations affecting telomere association. The indicated cells were fixed by formaldehyde and subjected to ChIP using anti-myc antibodies followed by dot blot analysis using a (CCCTAA)n probe. The signals of telomeric DNA in ChIP were quantified and normalized to the corresponding total telomeric signal (Input) and plotted against wild-type CTC1-myc in C and E (mean ± SD; n = 4; Student's t-test with respect to V871M: [*] P ≤ 0.05; [**] P ≤ 0.01).
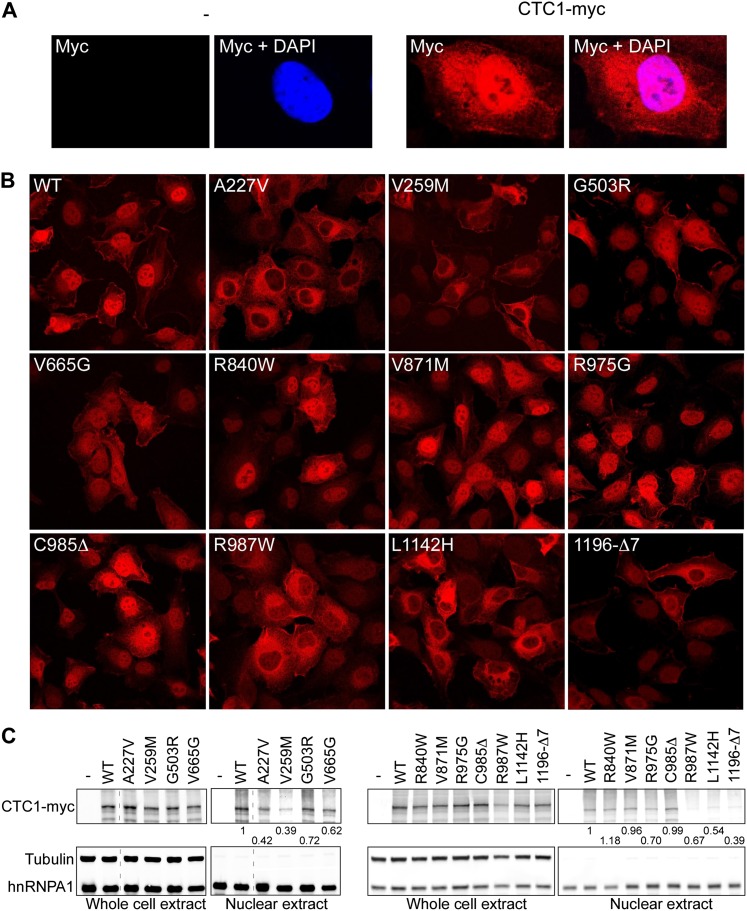
Immunofluorescence staining using anti-myc antibodies without pre-extraction allowed the predominant detection of wild-type CTC1-myc protein in the nucleus (Fig. 3A). In contrast, mutations of residues A227, V259, R987, and L1142 and the C-terminal deletion of CTC1 caused a remarkable reduction of the proteins in the nucleus and accumulation in the cytoplasm (Fig. 3B). Cellular fractionation studies confirmed the reduced presence of the above mutants in the nuclear fraction, which, as expected, was enriched for the nuclear protein hnRNPA1 and depleted from the cytoplasmic tubulin (Fig. 3C). As expected, mutants with reduced nuclear localization were also defective in telomere association as described above, providing an additional explanation for their impairment in telomere binding.
Figure 3.
CTC1 disease mutations affect intracellular localization. (A,B) HT1080 cells stably expressing wild-type (WT) or mutant myc-tagged CTC1 were subjected to immunofluorescence staining using anti-myc (red) antibody. Staining of cellular DNA with 4′, 6′-diamidino-2-phenylindole (DAPI) (blue) allowed visualization of the nuclei. (C) HT1080 cells stably expressing vector, wild-type (WT), or mutant myc-tagged CTC1 were subjected to cellular fractionation. Anti-myc, anti-tubulin, and anti-hnRNPA1 antibodies were used in Western blots. Relative nuclear distributions of CTC1-myc were determined by quantification of the signals and normalization to total CTC1-myc levels. Tubulin and hnRNPA1 were used as loading controls and served as cytoplasmic and nuclear markers, respectively.
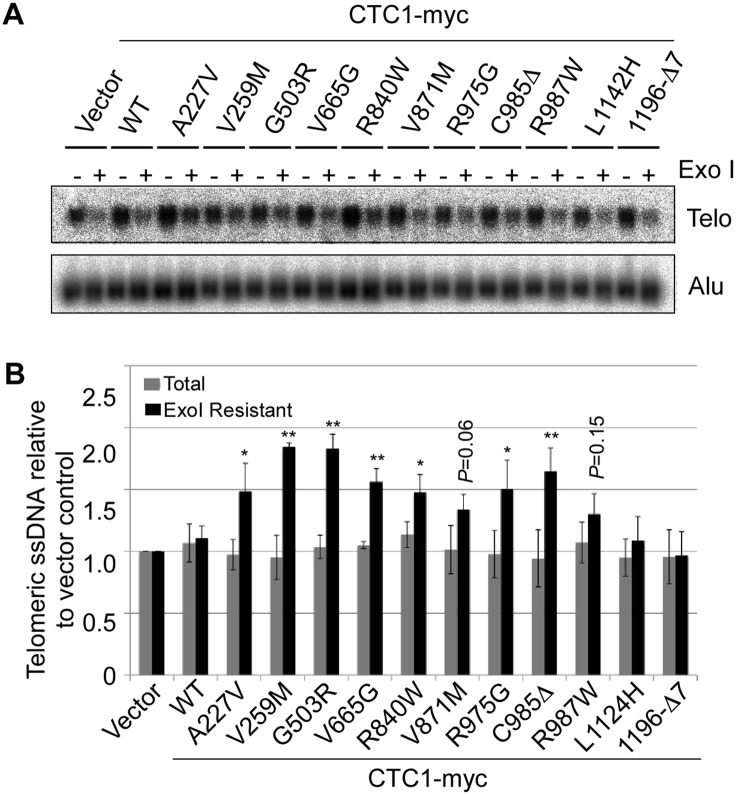
CST has been implicated in the replication of the bulk of telomeric DNA and in C-strand fill-in synthesis upon telomere elongation by telomerase (Huang et al. 2012; Wang et al. 2012; Wu et al. 2012). To measure possible defects of CTC1 disease mutations in the maintenance of telomeric DNA, we expressed wild-type and mutant CTC1 from a retroviral long terminal repeat (LTR) promoter upon retroviral transduction in telomerase-positive HT1080 cells. The presence of telomeric G-rich ssDNA was detected by in-gel hybridization of restriction enzyme-digested nondenatured DNA that was fractionated on agarose gels (Fig. 4A). The same assay was previously employed to detect single-stranded G-rich 3′ overhangs and internal single-stranded G-rich DNA that accumulates in CTC1 and STN1-deficient cells (Miyake et al. 2009; Surovtseva et al. 2009; Huang et al. 2012; Wu et al. 2012). In vector control or wild-type CTC1-expressing cells, most of the telomeric ssDNA signal was abolished upon prior treatment with saturating amounts of Escherichia coli exonuclease I (Exo I) (Fig. 4; Supplemental Fig. S5). Since Exo I digests 3′ overhangs, this indicates that most single-stranded G-rich telomeric sequences were present as terminal 3′ overhangs, as expected (McElligott and Wellinger 1997). However, the fraction of Exo I-resistant ssDNA signals of G-rich telomere sequences increased in CTC1 disease mutant-expressing cells (Fig. 4A,B; Supplemental Fig. S5), uncovering that CTC1 disease mutations lead to accumulation of internal stretches of telomeric DNA, presumably during lagging strand synthesis, which highlights the importance of the functional interaction between CST and DNA polα-primase described above. This result therefore suggests that disease-associated CTC1 mutations cause defects in semiconservative DNA replication of telomeric DNA. Cell cycle distribution analysis by flow cytometry did not reveal significant perturbations in cell cycle progression for the expressions of CTC1 mutants compared with the wild-type CTC1 (Supplemental Fig. S6). ChIP experiments with antibodies against the DNA damage marker γ-H2AX did not reveal a telomeric DNA damage response in CTC1 mutant-expressing cells, whereas checkpoint activation occurred as expected upon depletion of the shelterin component TPP1 (Supplemental Fig. S3). This therefore indicates that the internal G-rich DNA that accumulates in CTC1 mutant-expressing cells does not elicit strong checkpoint activation. Notably, STN1 depletion also did not elicit a telomeric DNA damage response in a previous study (Miyake et al. 2009). Expression of the two C-terminal mutant CTC1 polypeptides (L1124H and 1196-Δ7) did not show defects in telomeric DNA replication in Figure 4. These C-terminal mutations disrupt CST complex formation and cause multiple functional defects (Table 1). Thus, we suspect that they represent null mutations that, in contrast to the other tested CTC1 mutations, could not interfere with the function of endogenous CTC1.
Figure 4.
CTC1 disease mutations affect telomere replication. (A,B) Identification of mutations affecting telomeric DNA replication. Genomic DNA extracted from the indicated HT1080 cells was treated with or without Exo I for telomeric G-rich ssDNA detection by native in-gel hybridization using a (CCCTAA)n probe. Alu DNA (loading control) was detected after denaturation and hybridization to an Alu probe. The signals of total and internal (Exo I-insensitive) telomeric G-rich ssDNA were quantified and normalized to the corresponding Alu DNA signal and plotted against mock expression in B (mean ± SD; n = 3; Student's t-test with respect to wild type: [*] P ≤ 0.05; [**] P ≤ 0.01).
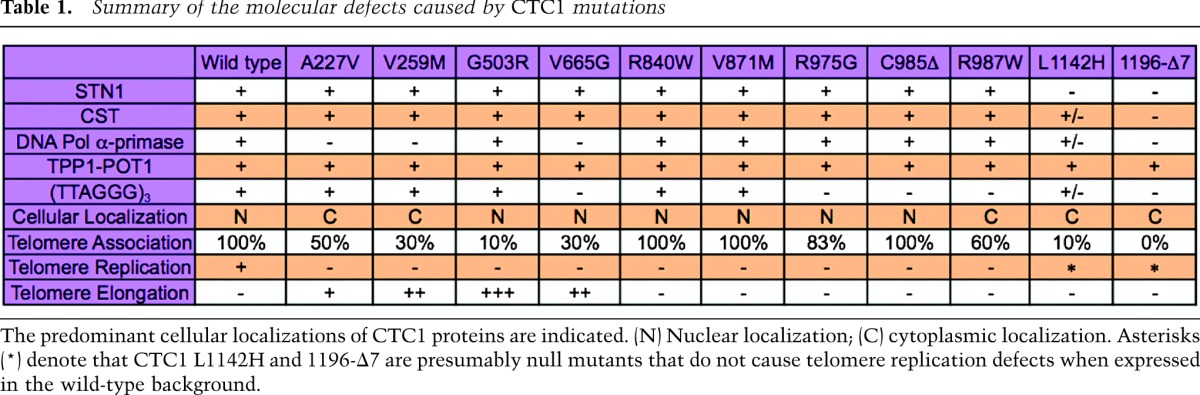
Table 1.
Summary of the molecular defects caused by CTC1 mutations
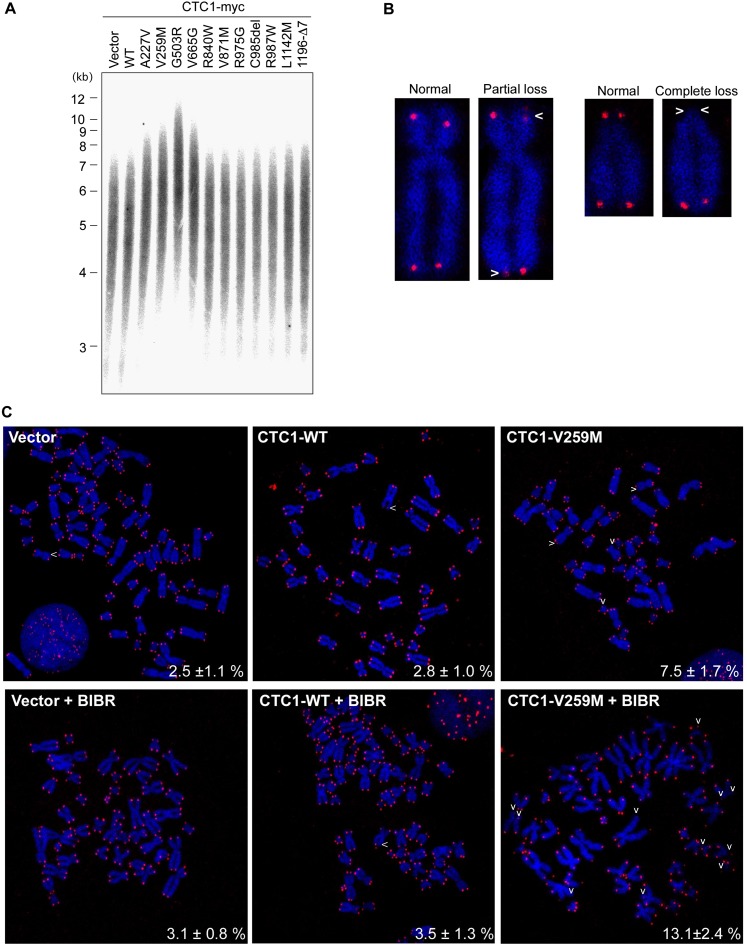
Since CST constrains telomerase activity to limit telomere lengthening, we also carried out telomere restriction fragment analysis to determine whether expression of CTC1 disease mutants affects telomere length (Fig. 5A). We stably transduced HT1080 cells with the retroviral CTC1-myc constructs and collected the cells 29 d post-transduction. During the period in culture, all HT1080 lines showed a similar growth rate. Cells expressing wild-type or several of the mutant CTC1 polypeptides had the same telomere length as vector control cells. However, expression of CTC1-A227V, CTC1-V259M, CTC1-G503R, and CTC1-V665G resulted in telomere elongation, which is a phenotype observed when telomerase regulation is disrupted upon CST dysfunction (Chen et al. 2012). Telomerase enzymatic activity was not affected in vitro, as determined in a telomeric repeat amplification protocol (TRAP) assay (Supplemental Fig. S7). While causing replication defects of telomeric DNA, expression of CTC-R840W, CTC-V871M, CTC-R975G, CTC-C985Δ, and CTC-R987W did not lead to notable telomere length changes. However, we suspected that telomerase activity at chromosome ends was elevated in the HT1080 cells expressing CTC1 mutants to compensate for the telomere loss events that may ensue at a subset of chromosome ends with telomere replication defects. To test this notion more directly, we analyzed telomere maintenance at single chromosome ends in metaphase chromosomes by fluorescent in situ hybridization (FISH) using telomere-specific probes (Fig. 5B,C). Empty vector, wild-type CTC1, or CTC1-V259M retroviral constructs were transduced into HT1080 cells, selected for 3 d, and then treated or not treated for three additional days with the telomerase inhibitor BIBR1532 (Damm et al. 2001). Partial or complete telomere loss events occurred at chromosomes in vector and CTC1 wild-type-expressing cells at a frequency of 2.5% and 2.8%. Telomere loss events increased to 7.5% (P < 0.005) in CTC1-V259M-expressing cells. Furthermore, the telomere loss frequency nearly doubled to 13.1% (P < 0.005) in CTC1-V259M-expressing cells upon treatment with BIBR1532 for 3 d. This therefore indicates that telomerase efficiently counteracts the stochastic telomere loss events that occur in cells expressing CTC1-V259M. CTC1 loss is also known to lead to fragile telomeres, which are characterized by the presence of multiple telomeric signals instead of one signal per chromosome end (Stewart et al. 2012). However, this phenotype was reported for a HeLa cell clone that carries unusually long telomeres (>20 kb), and the telomeres of the HT1080 cells analyzed here were probably too short to detect such events.
Figure 5.
CTC1 mutations affect telomerase control and telomere maintenance. (A) Identification of mutations affecting telomere length homeostasis. Indicated CTC1 proteins or mock-expressing HT1080 cells were collected 29 d after puromycin selection for telomere length analysis by terminal restriction fragment assay. (B,C) Telomere FISH analysis of HT1080 cells stably expressing vector, wild-type CTC1, or CTC1-V259M in the presence or absence of BIBR1532 (15 μm, 72 h). Metaphase chromosome spreads were hybridized with a Cy3-(CCCTAA)4 PNA probe (red) and stained with DAPI (blue). Representative normal and defective mitotic telomere structures upon expression of CTC1-V259M are shown in B. Representative metaphase chromosome spreads and quantification of chromosome aberrations (mean of the percentage of defective chromosomes per metaphase; n = 20–30) are shown in C. (White arrows) Telomere loss sites.
Discussion
Our analysis of disease mutant CTC1 polypeptides identifies molecular defects that underlie CST dysfunction in Coats Plus/DC. We identify CTC1 mutations that affect CST complex formation, interaction with DNA polα-primase, binding of telomeric ssDNA, and/or cellular localization in vivo (Table 1). Our systematic analysis also suggests that all of the characterized molecular interactions that were identified as defective in mutant polypeptides are crucial for CST function in telomere replication. On the other hand, most of the point mutant CTC1 alleles studied here may not represent true null alleles but instead are hypomorphic alleles. First, they all elicit a dominant-negative effect on telomere replication in CTC1 wild-type cells when able to assemble into the CST complex and when strongly expressed from viral vectors (this study). Second, the point mutant CTC1 alleles are often combined in patients with presumed complete loss-of-function alleles that contain premature stop codons and whose expression should be suppressed by the nonsense-mediated mRNA decay pathway (Supplemental Table 1). Notably, however, none of the described patients carried two complete loss-of-function alleles, indicating that such a combination would be lethal. On the other hand, the CTC1 mutants carrying C-terminal mutations (L1142H and 1196-Δ7) exhibited primary defects in STN1 interaction and CST complex formation. They were also defective in telomeric ssDNA binding, DNA polα-primase interaction, and nuclear/telomere localization. Intriguingly, expression of the C-terminal CTC1 mutants did not perturb telomere replication or length maintenance, all of which suggests that they represent null mutants. Consistently, the C-terminal mutants, like other loss-of-function alleles, are causing disease in combination with presumed hypomorphic alleles (Supplemental Table 1).
The pleiotropy of the molecular defects of CTC1 point mutants may be unexpected. However, with the exception of the complex formation-defective CTC1 (L1142H and 1196-Δ7), expression of all CTC1 mutants caused accumulation of internal stretches of telomeric G-rich DNA. This therefore indicates that disruption of a great variety of physical interactions leads to a common telomere defect manifested by incomplete lagging strand synthesis during telomeric DNA replication. Our analysis suggests that biogenesis of functional CTC1 involves, as an early step, the assembly into a complex with STN1 and TEN1. This may occur in the cytoplasm, as complex-defective CTC1 variants are cytoplasmic. Nuclear import or retention of CST appears to rely on complex formation and involve critical amino acids in the CTC1 N terminus (A227 and V259). In the nucleus, telomere association may be reinforced by DNA polα-primase, as DNA polα-primase interaction mutants show reduced telomere association of CTC1. As yeast CST is thought to recruit DNA polα-primase, it will be interesting to test whether telomere association-defective CTC1 also impairs the recruitment of DNA polα-primase. The ability to bind single-stranded telomeric DNA may also enhance association with telomeric chromatin, but this activity does not seem necessary (C985Δ) or sufficient (V259M). Finally, since none of the mutants showed a defect in the interaction with POT1–TPP1, it seems that POT1–TPP1 per se is not able to recruit or retain CST at telomeres.
The internal stretches of telomeric ssDNA associated with telomere replication defects did not elicit a detectable DNA damage response. However, incomplete lagging strand synthesis is expected to lead to rapid telomere-shortening events in subsequent rounds of DNA replication if gapped C strands serve as replication templates. In addition, telomeric G-rich ssDNA can fold into G-quadruplex structures that impede DNA replication and may cause telomere-shortening events (Paeschke et al. 2010; Vannier et al. 2012). Consistent with this notion, we observed accumulation of telomere-free ends in cells that expressed mutant CTC1 (V259M). We therefore propose that Coats Plus/DC-causing CTC1 mutations initiate telomere syndrome by affecting semiconservative replication of telomeric DNA.
Our data also indicate that telomerase can efficiently counteract the telomere loss events that occur upon CTC1 dysfunction. Indeed, telomerase inhibition during 3 d nearly doubled the occurrence of telomere-free ends in CTC1-V259M cells. In addition, we observed that expression of certain CTC1 mutations caused telomere lengthening in telomerase-positive cells, consistent with the inhibitory role of CST in telomerase regulation (Chen et al. 2012). The mutations that caused telomere elongation all showed reduced association with telomeric DNA, suggesting that a reduced presence of CST at telomeres prevented telomerase inhibition (Chen et al. 2012). A reduced interaction of mutant CST with DNA polα-primase may also promote telomerase, as seen in Saccharomyces cerevisiae mutants in which a reduced interaction between Cdc13 and DNA polα gives rise to longer telomeres (Qi and Zakian 2000; Grossi et al. 2004). However, telomere elongation was only seen with a subset of CTC1 mutants with defective telomere replication, indicating that defective telomere replication per se does not stimulate telomere elongation in human cells.
In the presence of telomerase, telomere replication defects and telomere loss events caused by CTC1 mutations are at least partially compensated by enhanced telomerase action at truncated telomeres. This suggests that, in humans, CTC1 mutations may differentially affect various cell types, depending on their telomerase status. For instance, somatic cells in adults that lack telomerase activity may be more susceptible to CTC1 mutations than stems cells that express significant amounts of telomerase. This may explain the discrepancy of telomere-shortening phenotypes recently reported in patients with CTC1 mutations. In summary, this study reveals that disease-causing CTC1 mutations cause telomere replication defects, which corresponds to a new type of telomere syndrome that is strikingly different from the well-characterized “classical” telomere syndromes, which show defects in telomerase biogenesis or activity.
While this study was under review, a report was published on mouse CTC1 that contained the disease-associated human CTC1 mutations at the corresponding positions (Gu and Chang 2013). Notably, several mouse CTC1 mutations did not fully recapitulate the defects observed for mutant human CTC1. For instance, the corresponding CTC1-G503R, CTC1-V665G, and CTC1-R840W mutations rendered the mouse CTC1 protein unstable. Therefore, an assessment of localization and molecular interaction defects was not possible. Furthermore, we found that residues A227, V259, and V665 of human CTC1 are critical for the physical interaction with DNA polα-primase, while the corresponding mutations in mouse CTC1 did not affect this interaction. In addition, Gu and Chang (2013) showed that mutations in the central part of mouse CTC1 disrupt interaction with STN1. However, we found that the same mutations in human CTC1 preserve the abilities of CTC1 to bind STN1 and form a CST complex. Instead, the disease mutations at the C terminus of human CTC1, including L1142H and 1196-Δ7, prevent its interaction with STN1. Therefore, our analysis indicates that it is crucial to analyze the human CTC1 disease mutations with human CTC1 to decipher the defects of the disease. Our molecular and functional characterization of disease mutations of human CTC1 is a first step toward understanding the disease mechanisms, which may facilitate future developments of therapeutic interventions.
Materials and methods
Protein expression vectors and stable cell line generation
cDNAs of CTC1, STN1, and TEN1 were subcloned into pEAK8 and retroviral-based pCL vectors for mammalian expression of tagged or untagged proteins as indicated. CTC1 point mutations and CTC1-C985Δ were generated by site-directed mutagenesis (Agilent, Inc.). For CTC1-1196-Δ7 (deletion of amino acid residues 1196–1202), two overlapping cDNA fragments that skipped the deletion region were generated by PCR and cloned into the expression vectors by In-Fusion Cloning (Clontech). Stable cell lines were generated by viral transduction and puromycin selection (1 μg mL−1). All human cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% FCS and penicillin/streptomycin.
Co-IP
Cells were lysed in NETN buffer (40 mM Tris-HCl at pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with protease inhibitor cocktail (Sigma) for immunoprecipitation with anti-Flag M2 affinity gel (Sigma) and subsequent Western blot analysis as previously described (Chen et al. 2012).
Cellular fractionation
HT1080 cells were grown in 10-cm dishes and collected by trypsinization. After washing with PBS, cells were partially permeabilized for 3 min on ice with 60 μL of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT) supplemented with 0.1% Triton X-100. Nuclear fractions were obtained by centrifugation at 1500g for 5 min. Cell fractionation was validated by Western blot analysis using tubulin and hnRNPA1 as cytoplasmic and nuclear markers, respectively.
CST complex purification and EMSA
Protein expression and purification and EMSA were carried out as previously described (Chen et al. 2012) with minor modifications. In brief, 293T cells in a 6-cm dish were transfected with 2 μg of pEAK8-CTC1-Flag, 1 μg of pEAK8-STN1, and 1 μg of pEAK8-TEN1-His plasmids using Lipofectamine 2000 (Invitrogen). Three days after transfection, cells were collected for immunoprecipitation using anti-Flag M2 affinity gel, and the CST complex was eluted with 60 μL of NETN buffer containing 200 μg/mL Flag peptide. For EMSA, 20-μL reactions containing 1 μL of eluate and 4 μL of eluate buffer or 5 μL of eluate, 0.1 nM [32P]-labeled (TTAGGG)3 oligonucleotide, 50 nM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM DTT, and 5% glycerol were loaded on 2.5% agarose gels for electrophoresis. Gels were dried and analyzed on a PhosphorImager (FLA-3000, Fujifilm).
ChIP
Stable HT1080 cell lines expressing wild-type and mutant myc-tagged CTC1 protein were fixed with 1% formaldehyde, sonicated, and subjected to analysis as previously described (Chen et al. 2012).
Immunofluorescence staining
Cells fixed with 2% paraformaldehyde were permeabilized with 0.5% Triton X-100 for immunodetection of CTC1-myc and RAP1 using α-mouse anti-myc (9E10) and α-rabbit anti-RAP1 primary antibodies (NB100-292, Novus), respectively. After washing with phosphate-buffered saline, secondary antibodies against mouse IgG conjugated with Alexa-Fluor 663 and rabbit IgG conjugated with Alexa-Fluor 488 were used to recognize the primary antibodies for visualization. Cell nuclei were visualized by DNA staining with 4′, 6′-diamidino-2-phenylindole (DAPI) dye. Z-stack images of the samples were obtained using an LSM700 confocal microscope (Zeiss) and processed with ImageJ software.
Terminal restriction fragment assay and telomeric G-rich ssDNA detection
Genomic DNA from HT1080 cells was extracted using the Wizard Genomic DNA purification kit (Promega). Genomic DNA (2.5 μg) was subjected to restriction digestion with RsaI and HinfI overnight at 37°C for agarose gel electrophoresis. Gels were dried for 1 h at 50°C, denatured with 0.8 M NaOH and 150 mM NaCl, and hybridized to a [32P]-labeled (CCCTAA)n probe for detection of telomeric DNA. The radioisotope-labeled (CCCTAA)n probe was generated by random prime labeling in the presence of [α32P]-dCTP using telomeric repeat sequences with a size of ∼600 base pairs (bp) as template. The telomeric repeat template was produced by self-amplification in 28 cycles of PCR using (TTAGGG)5 and (CCCTAA)5 oligonucleotides. To detect telomeric G-rich ssDNA, gels were dried after electrophoresis and subjected to hybridization with a [32P]-labeled (CCCTAA)n probe. To remove telomeric 3′ overhang sequences, 1 μg of genomic DNA was treated with 10 U of E. coli Exo I for 3 h before restriction digestion with RsaI and HinfI. To detect Alu DNA, the gels were denatured and hybridized with a [32P]-labeled 5′-GTGATCCGCCCGCCTCGGCCTCCCAAAGTG-3′ probe.
TRAP assay
Cell extracts were prepared by cells lysis in CHAPS buffer (10 mM Tris-HCl at pH 7.5, 1 mM MgCl2, 1 mM EGTA, 0.5% CHAPS, 10% glycerol, 5 mM β-mercaptoethanol, protease inhibitors) and centrifugation at 12,000g for 20 min. Telomerase reactions were performed for 30 min at 30°C and contained in 50-μL volumes of cell extract (0.1 μg or 0.3 μg), 0.1 μg of TS oligonucleotide (5′-AATCCGTCGAGCAGAGTT-3′), 20 mM Tris-HCl (pH 8.3), 6.5 mM MgCl2, 63 mM KCl, 0.005% Tween 20, 1 mM EGTA, 0.1 mg/mL BSA, 0.25-μm dNTPs, and 0.2 μL of [α-32P]dGTP (3000 Ci/mmol). Subsequently, 0.2 U of Taq polymerase and 0.1 μg of ACX oligonucleotide (5′-GCGCGGCTTACCCTTACCCTTACCTAACC-3′) were added to the telomerase reactions for 28 cycles of PCR (cycles of 30 sec at 94°C and 30 sec at 60°C). TRAP reactions were separated on 12.5% polyacrylamide gels and analyzed on a PhosphorImager (FLA-3000, Fujifilm).
Telomere FISH
HT1080 cells were treated with 0.05 μg/mL Demecolcine (Sigma) for 1 h, harvested by trypsinization, swollen in 0.056 M KCl, and resuspended in fixative (methanol:acetic acid = 3:1) for metaphase spread on glass slides. Following denaturation for 3 min at 80°C, hybridization was performed for 3 h at 25°C in 70% formamide, 0.5% blocking reagent (Roche), 200 μM Cy3-OO-(CCCTAA)4 PNA probe (Eurogentec), and 10 mM Tris-HCl (pH 7.4). Slides were washed first with 10 mM Tris-HCl (pH 7.4) and 70% formamide and subsequently with 0.1 M Tris-HCl (pH 7.4), 0.15 M NaCl, and 0.08% Tween-20. DNA was counterstained with DAPI, and slides were mounted in ProLong Gold (Invitrogen) anti-fade reagent.
Acknowledgments
Research in the laboratory was supported by the Swiss National Science Foundation, a European Research Council advanced investigator grant (grant agreement no. 232812), an Initial Training Network (ITN) grant (CodeAge) from the European Commission's Seventh Framework Programme (grant agreement no. 316354), the Swiss Cancer League, and EPFL. L.-Y.C. performed most of the experiments with assistance from J.M., and L.-Y.C. and J.L. designed the study and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.222893.113.
References
- Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O'Sullivan J, et al. 2012. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 44: 338–342 [DOI] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH 2012. The telomere syndromes. Nat Rev Genet 13: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA 2012. DNA secondary structures: Stability and function of G-quadruplex structures. Nat Rev Genet 13: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB 2009. A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Lingner J 2013. CST for the grand finale of telomere replication. Nucleus 4: 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J 2012. The human CST complex is a terminator of telomerase activity. Nature 488: 540–544 [DOI] [PubMed] [Google Scholar]
- Damm K, Hemmann U, Garin-Chesa P, Hauel N, Kauffmann I, Priepke H, Niestroj C, Daiber C, Enenkel B, Guilliard B, et al. 2001. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J 20: 6958–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T 2009. How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D 2004. Pol12, the B subunit of DNA polymerase α, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Chang S 2013. Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats Plus. Aging Cell doi: 10.1111/acel.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S 2012. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J 31: 2309–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Dai X, Chai W 2012. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res 22: 1681–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP 2010. Telomeric strategies: Means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Keller RB, Gagne KE, Usmani GN, Asdourian GK, Williams DA, Hofmann I, Agarwal S 2012. CTC1 mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer 59: 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ 1997. The terminal DNA structure of mammalian chromosomes. EMBO J 16: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F 2009. RPA-like mammalian Ctc1–Stn1–Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F 2012. Xenopus laevis Ctc1–Stn1–Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR 2013. Finding the end: Recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 14: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, McDonald KR, Zakian VA 2010. Telomeres: Structures in need of unwinding. FEBS Lett 584: 3760–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polvi A, Linnankivi T, Kivela T, Herva R, Keating JP, Makitie O, Pareyson D, Vainionpaa L, Lahtinen J, Hovatta I, et al. 2012. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet 90: 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD 2nd, Wright WE, Price CM 2012. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 31: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806 [DOI] [PubMed] [Google Scholar]
- Walne A, Bhagat T, Kirwan M, Gitaux C, Desguerre I, Leonard N, Nogales E, Vulliamy T, Dokal I 2013. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica 98: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D 2009. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284: 26725–26731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM 2012. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep 2: 1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T 2012. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]