In this study, Chen et al. discover that the histone variant H3.3 impairs higher-ordered chromatin folding and promotes gene activation. H3.3 actively marks enhancers and determines the transcriptional potential of retinoid acid (RA)-regulated genes via creating an open chromatin signature that enables the binding of RAR/RXR. RA induction results in the incorporation of H3.3 on promoter regions to activate transcription via counteracting H2A.Z-mediated chromatin compaction.
Keywords: chromatin dynamics, higher-ordered chromatin structure, histone variants, H2A.Z, H3.3, gene transcription
Abstract
The histone variants H3.3 and H2A.Z have recently emerged as two of the most important features in transcriptional regulation, the molecular mechanism of which still remains poorly understood. In this study, we investigated the regulation of H3.3 and H2A.Z on chromatin dynamics during transcriptional activation. Our in vitro biophysical and biochemical investigation showed that H2A.Z promoted chromatin compaction and repressed transcriptional activity. Surprisingly, with only four to five amino acid differences from the canonical H3, H3.3 greatly impaired higher-ordered chromatin folding and promoted gene activation, although it has no significant effect on the stability of mononucleosomes. We further demonstrated that H3.3 actively marks enhancers and determines the transcriptional potential of retinoid acid (RA)-regulated genes via creating an open chromatin signature that enables the binding of RAR/RXR. Additionally, the H3.3-dependent recruitment of H2A.Z on promoter regions resulted in compaction of chromatin to poise transcription, while RA induction results in the incorporation of H3.3 on promoter regions to activate transcription via counteracting H2A.Z-mediated chromatin compaction. Our results provide key insights into the mechanism of how histone variants H3.3 and H2A.Z function together to regulate gene transcription via the modulation of chromatin dynamics over the enhancer and promoter regions.
During DNA replication and gene transcription, the accessibility of eukaryotic DNA is thought to be highly regulated by the dynamic nature of chromatin structures. To date, three main mechanisms have been identified to modulate these dynamic structural changes, including ATP-dependent chromatin remodeling complexes, post-translational modifications of histones, and the replacement of histone variants (Varga-Weisz and Becker 2006). The biological roles of chromatin remodeling factors and histone modifications in the regulation of chromatin dynamics and transcription have been extensively investigated and discussed (Li et al. 2010; Li and Reinberg 2011). The incorporation of histone variants may change the local and global structures of chromatin and create architecturally distinct chromatin states that perform diverse functions. However, the mechanism of this fundamental process and the precise functional implications for gene regulation remain poorly understood.
The variants H2A.Z and H3.3, both of which are highly conserved evolutionarily, have been proposed to play crucial and specific roles in the regulation of chromatin dynamics and transcription. The histone variant H2A.Z shares only an ∼60% sequence identity with canonical H2A and is distinguished by an extended acidic patch on the surface of nucleosomes and a unique C-terminal tail (Suto et al. 2000). H2A.Z has been shown to play contradictory roles in nucleosome stability, gene regulation, and heterochromatin formation. Recent studies have revealed that H2A.Z is enriched at the promoter of inducible genes under repressed or basal expression conditions but is subsequently removed upon transcriptional activation (Guillemette et al. 2005; Li et al. 2005; Raisner et al. 2005; Zhang et al. 2005; Barski et al. 2007; Sutcliffe et al. 2009). Similar studies in mammals demonstrated that H2A.Z is required for gene activation and helps to poise gene promoters for rapid transcriptional activation upon induction (Wong et al. 2007; John et al. 2008; Sutcliffe et al. 2009). Interestingly, a few recent studies have demonstrated that H2A.Z exhibits a repressive role in gene transcription (Farris et al. 2005; Gevry et al. 2007; Kotekar et al. 2008; Dalvai et al. 2012). H3.3 has been largely considered as a mark of transcriptionally activated genes and has been deposited into transcribed genes, promoters, and gene regulatory elements (Schwartz and Ahmad 2005; Jin et al. 2009; Goldberg et al. 2010). Although the induction of gene expression results in the enrichment of H3.3 (Schwartz and Ahmad 2005), the dynamic deposition of H3.3 during transcriptional activation and induction has not been well defined thus far. Most recently, H3.3 has also been found to be linked to transcriptional silencing and is incorporated at regions of the genome that are typically thought to be relatively transcriptionally inactive, such as the telomere and pericentric heterochromatin in mouse embryonic stem (mES) cells (van der Heijden et al. 2007; Williams et al. 2009; Goldberg et al. 2010; Wong et al. 2010). However, it is still unclear whether H3.3-containing chromatins possess unique properties, and how H3.3 affects transcription with only a four- to five-amino-acid residue difference from canonical H3 has not yet been elucidated. Furthermore, genome-wide distribution of nucleosomes containing the double variant H2A.Z/H3.3, which are unusually sensitive to salt-dependent disruption in their native state, have been found to be selectively enriched over “nucleosome-free regions” of active promoters, enhancers, and insulator regions in human cells (Jin and Felsenfeld 2007; Goldberg et al. 2010).
It is clear that the two essential variants H2A.Z and H3.3 are functionally correlated during gene regulation in vivo. They may function together to establish a specialized chromatin conformation and play critical roles in gene transcription. However, the underlying molecular mechanisms still remain unclear. In this study, we described the distinct biophysical effects of H2A.Z and H3.3 on nucleosome stability and higher-ordered chromatin dynamics. In addition, we investigated the effect of these two important histone variants on gene transcription in vitro and in vivo. Moreover, the dynamic deposition and/or replacement of H2A.Z and H3.3 and the corresponding structural changes of chromatin at the enhancer and promoter regions of RAR/RXR targeted genes were further analyzed during gene activation by all-trans retinoid acid (tRA) induction in vivo. Taken together, our results provide new insights into the molecular mechanism of how histone variants function cooperatively to establish featured chromatin structures at enhancer and promoter regions for inducible gene transcription.
Results
H2A.Z enhances the stability of mononucleosomes, but H3.3 does not have any effects
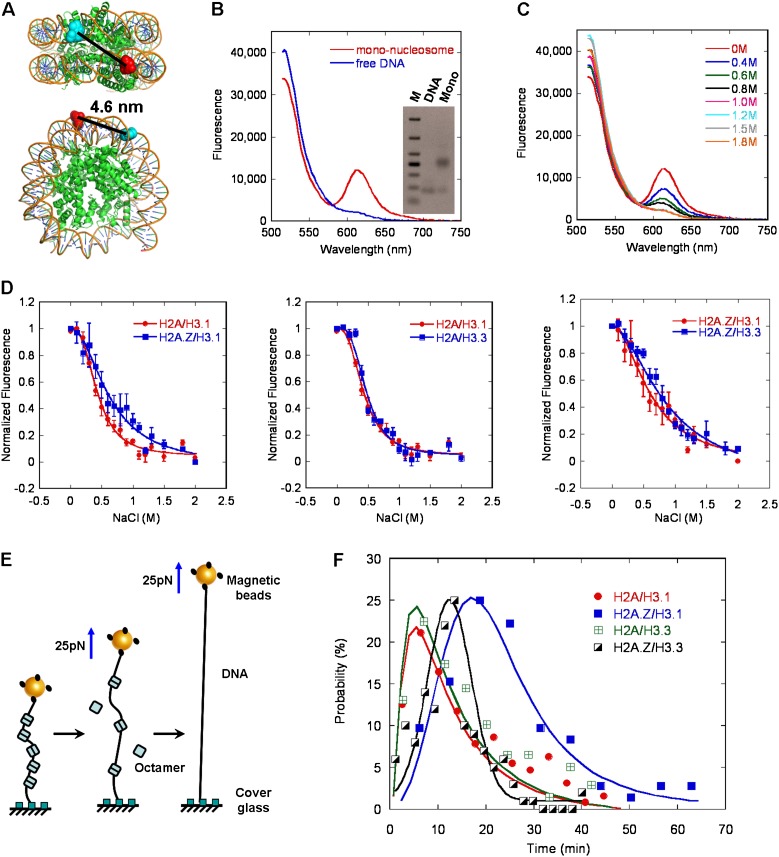
To determine the biophysical effects of H2A.Z and H3.3 on mononucleosomes, the stability of canonical or variant-containing mononucleosomes was characterized by the salt-dependent dissociation analyzed using the fluorescent resonance energy transfer (FRET) assay. In this assay, 601 DNA templates (169 base pairs [bp]) were labeled with the donor Alexa Fluor 488 and the acceptor Alexa Fluor 594 over a 96-bp separation. As expected, the histone-free DNA templates with no significant FRET signals were fully extended as the two dyes separated over 30 nm (Fig. 1B). The well-organized mononucleosomes were reconstituted on the strong nucleosome positioning of the 601-DNA sequence to ensure a high homogeneity and enable efficient FRET, with the dyes approaching each other within 50 Å (Fig. 1A,B). The NaCl-dependent disassembly of mononucleosomes was monitored by the change in quantitative FRET signals as a function of salt concentration (Fig. 1C). The midpoint value for the overall transition process was 0.43 M ± 0.01 M NaCl for canonical H3.1/H2A histones, as shown in Figure 1D. The variant H3.3 did not have a significant effect on the stability of mononucleosomes with the midpoint of 0.47 M ± 0.02 M NaCl. In contrast, the variant H2A.Z greatly enhanced the stability of mononucleosomes by increasing the midpoint to 0.62 M ± 0.02 M NaCl. The stabilization effect of H2A.Z on the nucleosomes was consistent with previously reported results (Park et al. 2004). H3.3/H2A.Z double variant-containing nucleosomes have been shown to be extremely sensitive to salt-dependent disruption in a genome-wide investigation in vivo (Jin and Felsenfeld 2007; Jin et al. 2009). However, under our experimental conditions, the combined incorporation of variant H2A.Z and H3.3 within the same nucleosomes resulted in a salt-dependent stability of the nucleosomes with a midpoint of 0.75 M ± 0.02 M NaCl, which was similar to that observed in H2A.Z-containing nucleosomes (Fig. 1D, right panel). Importantly, some unknown features in the native nucleosomes other than the double variants, including some specific histone modifications or nucleosome-binding factors, may contribute to the unusual instability of the nucleosomes in vivo.
Figure 1.
The effects of H2A.Z and H3.3 on the stability of mononucleosomes using FRET and magnetic tweezer analyses. (A) The top and side view of the mononucleosome, with the position of the donor (Alexa 488) and acceptor (Alexa 594) indicated. (B) The fluorescence emission spectra of the free-labeled DNA and the well-reconstituted mononucleosomes upon donor excitation at 495 nm the native gel analysis of the same samples is shown in the inset. (C) FRET analysis of NaCl-dependent dissociation of mononucleosomes. (D) The normalized equilibrium dissociation curves for the different variant-containing mononucleosomes, obtained by monitoring the fluorescence difference between the donor and acceptor emissions upon donor excitation at 492 nm. Error bars represent SD (n = 3). (E) Diagram for the force-dependent displacement of octamers from a single nucleosomal array using magnetic tweezers. (F) Probability analysis of the dwell time for the displacement of five nucleosomes containing canonical or variant histones using magnetic tweezers at 25 pN. The solid lines are log-normal fits to the data point with a peak at 5.5 min for the canonical nucleosome, 5.7 min for the nucleosome containing H3.3, 25.2 min for the nucleosome containing H2A.Z, and 12.6 min for the nucleosome containing H2A.Z and H3.3.
To further investigate the effect of histone variants on the stability of the nucleosome, we examined the force-dependent dissociation of canonical or variant-containing mononucleosomes using magnetic tweezers (Fig. 1E). Single nucleosomal arrays were equally reconstituted with canonical or variant-containing histone octamers and tethered onto a magnetic bead and the surface of the cover glass separately. Magnetic tweezers were employed to measure the nucleosome retention within a single nucleosomal array while exposed to a force held at 25 pN. The step-by-step displacement of nucleosomes from the single nucleosomal array could be observed, as shown in Supplemental Figure S1B. Each displacement step was observed with an average extension of 35 nm, and the number of steps represented the number of nucleosomes dissociated from the array. The data for the nucleosomal arrays occupied by nine nucleosomes were selected and used for further statistical analysis. We statistically analyzed the decay time required for the disassembly of five nucleosomes from the canonical and variant-containing nucleosomal arrays (Fig. 1F). Our results showed that H3.3-containing nucleosomes dissociated at a rate of ∼5.7 min per five nucleosomes, which was similar to that observed in canonical nucleosomes, which had a rate of ∼5.5 min per five nucleosomes. In contrast, the dissociation of H2A.Z-containing nucleosomes required ∼25.2 min per five nucleosomes, which was a more than fourfold increase compared with the canonical nucleosomes. The dissociation of double variant H2A.Z/H3.3-containing nucleosomes required ∼12.6 min per five nucleosomes, with a more than twofold increase compared with the canonical nucleosomes (Fig. 1F), Taken together, these results indicated that the incorporation of H2A.Z stabilized the nucleosomes, while H3.3 did not have any distinct effects on the stability of the nucleosomes. The double variant H2A.Z/H3.3-containing nucleosomes were more stable than the canonical nucleosomes, which was consistent with our FRET analysis.
H3.3 greatly impaired the folding of chromatin fibers and counteracted H2A.Z-mediated compaction
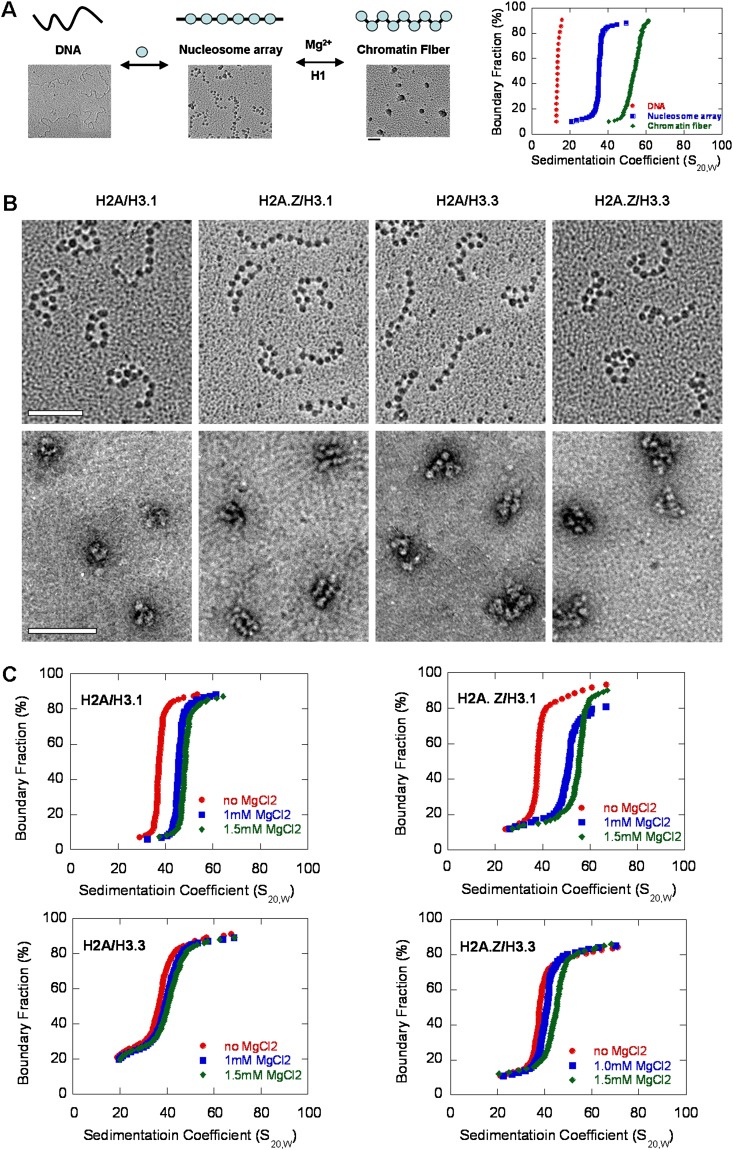
To characterize the effect of H2A.Z and H3.3 on the structure of higher-ordered chromatin fibers, we generated regularly spaced nucleosomal arrays using a DNA template containing 12 tandem repeats of 177-bp “601 nucleosome positioning” sequence. The 12mer–177-bp DNA was assembled into nucleosomal arrays by salt dialysis using recombinant histone octamers. Electron microscopy (EM) imaging was employed to confirm that the canonical and variant-containing nucleosomal arrays had an identical nucleosome density with >90% saturation and adopted an extended beads-on-a-string conformation in the absence of magnesium (Fig. 2A,B). The compact structures formed by canonical or variant-containing nucleosomal arrays in the presence of 1 mM MgCl2 were visualized by negatively stained EM images (Fig. 2B). Compared with the canonical arrays, the H2A.Z-containing arrays folded into a more compact structure in the same condition, while the H3.3-containing arrays adopted a looser and more open structure. Interestingly, we found that >90% of the H2A.Z-containing chromatin molecules adopted a unique “straight ladder-like” structure in the presence of 1 mM MgCl2. Analytical ultracentrifugation (AUC) in sedimentation velocity experiments was performed to quantify the different extent in the compaction of higher-ordered chromatin structures formed by canonical or variant-containing nucleosomal arrays. The sedimentation coefficient profile of histone-free 12mer–177-bp DNA templates in HE buffer was performed as a control (Fig. 2A). In the absence of Mg2+, the well-reconstituted canonical and variant-containing arrays sedimented at 36S and exhibited an extended beads-on-a-string structure in EM images (Fig. 2B). At 1.5 mM MgCl2, the Save for the canonical nucleosomal arrays was 51.5S. H2A.Z-containing nucleosomal arrays compacted more readily with an increasing concentration of MgCl2 and formed a more condensed structure with an Save of up to 57.2S at 1.5 mM MgCl2 (Fig. 2C), which was consistent with our negatively stained EM results. Interestingly, a similar condensation effect of H2A.Z on chromatin was observed by Tremethick's group (Fan et al. 2002). Surprisingly, the H3.3 variant, which was found to have no effects on the stability of mononucleosomes, significantly hindered intrafiber folding of the nucleosomal arrays. The H3.3-containing nucleosomal arrays were much more difficult to compact by Mg2+, with the Save slightly changing to 42.3S at 1.5 mM MgCl2. Moreover, the incorporation of H3.3 also counteracted the enhanced compaction effect of H2A.Z on nucleosomal arrays; the double variant H2A.Z/H3.3-containing nucleosomal arrays adopted a more open conformation compared with the canonical nucleosomal arrays in the presence of MgCl2. We also investigated the compaction of canonical and variant-containing nucleosomal arrays induced by the addition of the linker histone H1 (Supplemental Fig. S2A). The linker histone H1e exhibited a binding affinity similar to the canonical and variant-containing chromatins in our experimental conditions (Supplemental Fig. S2B). At increasing ratios of H1e/nucleosome, the H2A.Z variant still promoted the folding of chromatin arrays, and, intriguingly, the incorporation of H3.3 impaired the intramolecular folding of nucleosomal arrays and counteracted with H2A.Z in the presence of H1e.
Figure 2.
The effects of H2A.Z and H3.3 on the folding of the chromatin arrays. (A) A schematic diagram with related EM images to show the reconstitution and folding of well-defined nucleosomal arrays on a 12-repeat 177-bp “widom 601 nucleosome positioning” DNA sequence accompanied by their related sedimentation coefficient distribution plots analyzed by the sedimentation velocity in an XL-I analytical ultracentrifuge. (B) EM images of the canonical and H2A.Z-, H3.3-, and double variant H2A.Z/H3.3-containing nucleosomal arrays (by the metal-shadowing method) and their related compact states in 1.0 mM MgCl2 (by negatively stained methods). Bar, 100 nm. (C) Sedimentation coefficient distribution plots for the canonical and H2A.Z-, H3.3-, and double variant H2A.Z/H3.3-containing nucleosomal arrays at 0, 1.0, and 1.5 mM MgCl2.
Effects of unique H3.3 residues on higher-ordered chromatin structures
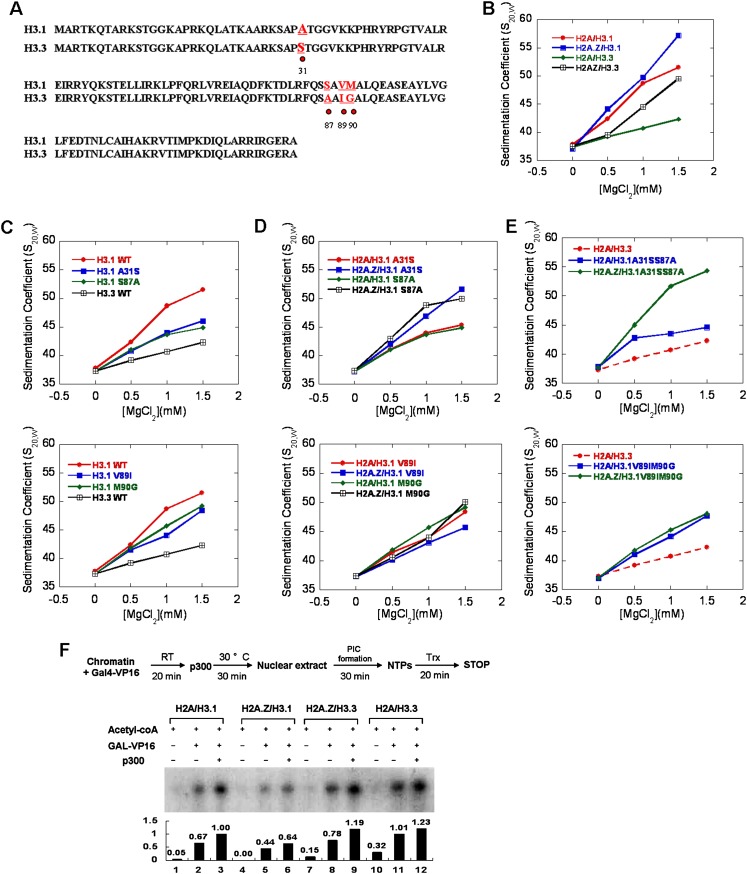
The significant inhibitory effect of H3.3 toward the chromatin folding process warrants further examination. H3.3 is one of the most conserved variants in all eukaryotes and has only a four-amino-acid residue difference from canonical H3, including residue 31 (Ser vs. Ala) in the N-terminal tail and residues 87 (Ala vs. Ser), 89 (Ile vs. Val), and 90 (Gly vs. Met) near the beginning of the α2 helix of histone H3 (Fig. 3A; Szenker et al. 2011). Our aforementioned results have shown that the H3.3 variant did not have any effect on the stability of mononucleosomes but greatly impaired the intrafiber folding of the chromatin fiber and counteracted the compact effect of H2A.Z (Fig. 3B). To determine which residue of these four unique amino acids contributed to the structural regulation of H3.3 on chromatin fibers, we generated a series of single-point mutations over these four residues in Xenopus histone H3, corresponding to the amino acids found on H3.3. We assembled the nucleosomal arrays containing canonical H3 or the A31S, S87A, V89I, and M90G single-point-mutated H3. The Mg2+-dependent compaction of these chromatin fibers was then analyzed using AUC and is shown in Figure 3C and Supplemental Figure S3. Our results showed that all four residues affected the intramolecular folding of the chromatin fibers; the nucleosomal arrays with the single-point-mutated H3 were unable to compact as well as the canonical arrays. Surprisingly, compared with the H2A-containing nucleosomal arrays with H3 mutations, we found that the incorporation of H2A.Z could still facilitate the folding of nucleosomal arrays containing H3A31S or H3S87A but could not enhance the folding of the nucleosomal arrays containing H3V89I or H3M90G (Fig. 3D), which suggested that the Ile89 and Gly90 residues on H3.3 were responsible for antagonizing the enhanced compaction effects of H2A.Z on chromatin fibers. To further investigate this effect, the double mutants (H3A31SS87A and H3V89IM90G) of canonical H3 were generated and reconstituted into the chromatin arrays, respectively. AUC analysis on the Mg2+-dependent compaction of the chromatin fibers containing the double mutations showed that the double mutant H3A31SS87A impaired the chromatin folding but did not affect the compaction effect of H2A.Z, while the other double mutant, H3V89IM90G, significantly counteracted the compaction effect of H2A.Z on chromatin fibers (Fig. 3E). The results agreed well with the investigation on single-point mutations.
Figure 3.
Specific functions of the four unique residues in H3.3 on chromatin folding properties and in vitro transcriptional assays. (A) Sequence alignment of canonical H3.1 (Xenopus) and histone variant H3.3. The amino acid residues that differed between H3.1 and H3.3 are colored red and labeled with dots. (B) The S20,w values of the canonical and variant-containing nucleosomal arrays are shown as a function of MgCl2. (C) The S20,w values of the arrays with point mutations of H3.1 to H3.3 at residue 31 and 87 (top) and at residue 89 and 90 (bottom) compared with wild-type H3.1- and H3.3-containing nucleosomal arrays are shown as a function of MgCl2 concentrations. (D) The S20,w values of the H2A.Z-containing arrays with point mutations of H3.1 to H3.3 at residue 31 and 87 (top) and at residue 89 and 90 (bottom) compared with the wild-type H3.1 and H3.3 are shown as a function of MgCl2 concentrations. (E) The S20,w values of the double-mutant-containing arrays H3.1A31SS87A (top) and H3.1V89IM90G (bottom) in the presence or absence of H2A.Z compared with the H3.3 are shown as a function of MgCl2 concentrations. (F) The effects of H2A.Z, H3.3, and the double variant H2A.Z/H3.3 on the transcriptional activity of chromatin templates. A schematic diagram of the in vitro transcription protocol is shown at the top. The relative transcription levels were quantitated by photoimager and normalized to that for canonical chromatins with all acetyl-CoA, Gal4-VP16, and p300 added. The transcription assays were carried out independently three times.
H3.3 highly antagonizes the inhibition of H2A.Z on chromatin transcriptional activity and promotes gene expression
The dynamic changes of chromatin structure play a critical role in transcriptional regulation. To investigate the correlation between the structural regulation of H2A.Z and H3.3 on chromatin with transcriptional activity, an in vitro transcription assay on the reconstituted chromatin templates was performed (Fig. 3F). The DNA template pG5MLP—containing five tandem Gal4-binding sites, an adenovirus major late core promoter, and a G-less cassette—was assembled into chromatin with purified recombinant canonical histones or histone variants using the ACF/NAP-1 system as described in the Materials and Methods (Supplemental Fig. S4A). Compared with the chromatin assembled using canonical histones, the chromatin containing the H2A.Z variant was found refractory to transcription, which was consistent with our aforementioned findings that the incorporation of H2A.Z stabilized mononucleosomes and facilitated the folding of chromatin fibers to form a highly compact structure. Interestingly, the incorporation of H3.3 highly counteracted the repressive effects of H2A.Z on transcription from the chromatin template, as shown in Figure 3F. Taken together, these results suggest that the presence of H3.3 greatly impaired the folding of the chromatin fibers, antagonizing H2A.Z-mediated chromatin compaction and maintaining chromatin in a relatively loose structure for transcription activation. In addition, we found that the H2A.Z and H3.3 variants exhibited similar effects on the transcriptional activity on chromatins in the presence of the linker histone H1 (Supplemental Fig. S4B). We also investigated the effects of H2A.Z and H3.3 on nucleosome remodeling mediated by ACF, an ISWI-containing and ATP-dependent chromatin remodeling factor (Supplemental Fig. S4C). Interestingly, the variant H2A.Z was found to slightly inhibit the nucleosome sliding by ACF, while the H3.3 did not have any apparent effects on nucleosome sliding by ACF compared with the canonical histones. These effects are consistent with our investigations of the stability of mononucleoosmes by FRET and magnetic tweezers.
Dynamic incorporation of H2A.Z and H3.3 and chromatin structural dynamics on enhancer and promoter regions during gene activation
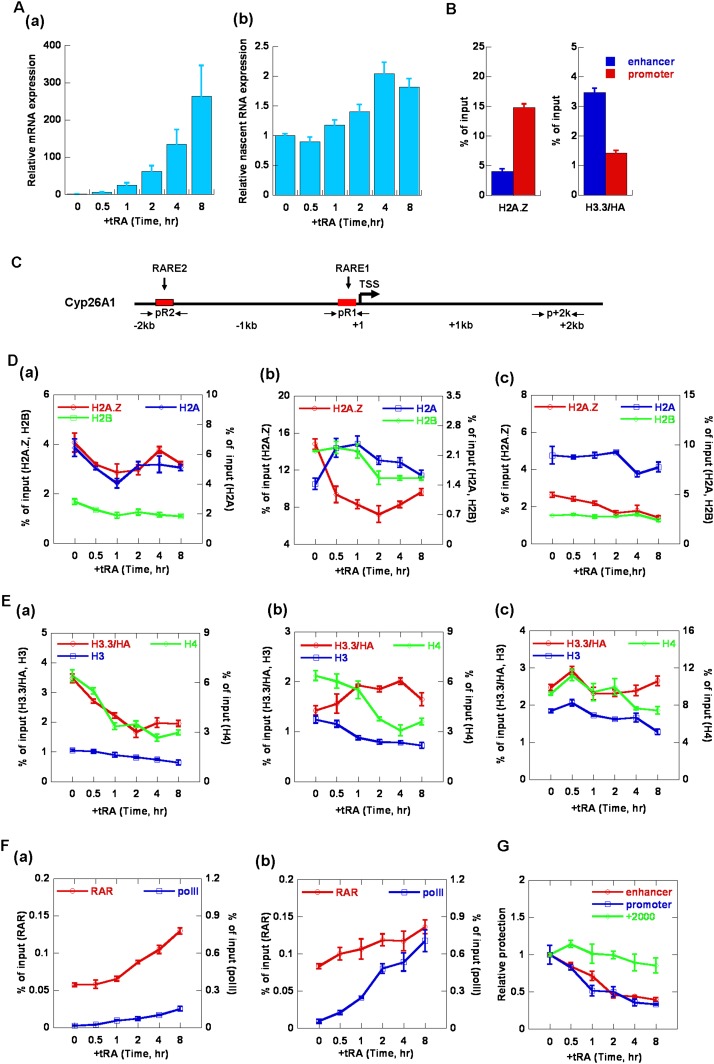
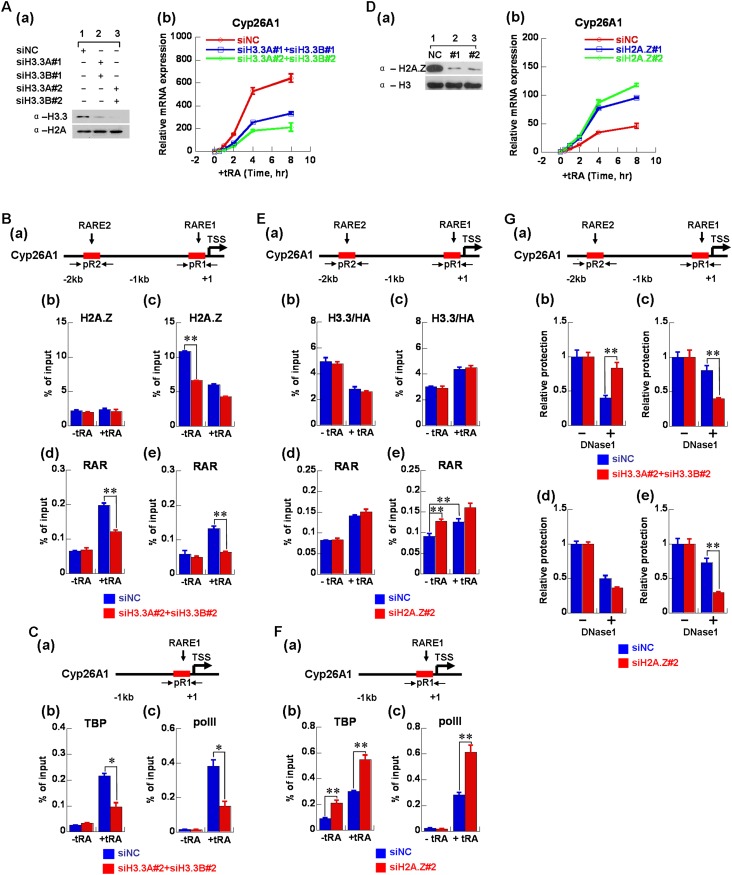
Our in vitro biochemical and biophysical analysis revealed that the incorporation of H2A.Z and H3.3 could directly regulate transcriptional activity via modulation of nucleosome/chromatin dynamics. Previous genome-wide chromatin immunoprecipitation (ChIP) and microarray investigations have also demonstrated that H2A.Z and H3.3 played critical roles in gene regulation. To examine whether our in vitro observations reflect in vivo phenomena, we further investigated the regulation of H2A.Z and H3.3 on chromatin dynamics and transcription in vivo. We examined the dynamic incorporations of H2A.Z and H3.3 on different regions of the transcription unit and the corresponding changes in chromatin structures during the activation of RAR/RXR-regulated genes by tRA induction. As shown in Figure 4A, panel a, the expression of cytochrome P450 26A1 (Cyp26A1) was increased with tRA treatment in a time-dependent manner. Further analysis of the nascent RNA of Cyp26A1 showed that the transcription was fully activated at 4 h after tRA induction (Fig. 4A, panel b). Similar activation was also observed for HoxA1 by tRA induction (Supplemental Fig. S5A).
Figure 4.
Dynamic regulation of H2A.Z and H3.3 on chromatin structures during Cyp26A1 gene activation by tRA in mES cells R1. (A) The relative levels of Cyp26A1 mRNA (panel a) and nascent RNA (panel b) at different time points during tRA induction as measured using real-time RT–PCR. The levels were normalized as n-fold changes relative to the values prior to tRA induction. (B) ChIP analysis of the deposition of H3.3 and H2A.Z on the enhancer and promoter regions of Cyp26A1 in mES cells (R1). The positions of the primer pairs used in ChIP are indicated in the schematic diagram in C. (C) A schematic diagram of the positions of the primers used in ChIP assay on the Cyp26A1 gene. The primer pair of pR2 amplified the enhancer region of Cyp26A1 where RARE2 is located. pR1 amplified the promoter region where RARE1 is located near the transcription start site (TSS). p+2k amplified the gene body region around Cyp26A1+2000. (D–F) ChIP analysis of the level of H2A.Z, H2A, and H2B (D); H3.3, H3, and H4 (E); and RARα and Pol II (F) on the enhancer (panel a), promoter (panel b), and gene body regions (panel c) of Cyp26A1 during tRA induction. The primer pairs used in real-time PCR are shown in the schematic diagram in C. (G) EpiQ analysis of the accessibility of chromatin on the enhancer, promoter, and gene body regions of the Cyp26A1 gene during tRA induction. The cells were treated with DNase I, and the protection was quantified using real-time PCR. The results were normalized to the reference Rho gene. All of the data shown are expressed as the mean ± SD (standard deviation) of three independent biological replicates.
Time-course ChIP was employed to detect the dynamic recruitments of H2A.Z and H3.3 on the transcribed genes during transcriptional activation. The spatiotemporal dynamics of H2A.Z and H3.3 on three regions, including the enhancer, promoter, and gene body, of the gene Cyp26A1 during tRA addition were analyzed (Fig. 4C–E). Interestingly, we found that H3.3 was predominantly accumulated on the enhancer region of Cyp26A1 in the resting state but was rapidly depleted, together with H4, after tRA induction (Fig. 4E, panel a). However, no significant enrichment of H2A.Z was observed at the enhancer regions prior to or after tRA addition, which indicated that H2A.Z did not function on the enhancer region during Cyp26A1 gene activation by tRA (Fig. 4D, panel a). In addition, our time-course ChIP analysis revealed that the level for all other histones examined, including H2A, H2B, and H4, dropped quickly within 1 h after tRA induction, which was concomitant with the rapid opening of the chromatin structure at this region, as shown by the DNase I digestion (EpiQ) assay (Fig. 4G). Interestingly, RAR was also recruited to the enhancer region at almost the same time when the chromatin was opened, which suggested that the nucleosomes marked by H3.3 on the enhancer region were displaced immediately after tRA addition in the recruitment of RAR/RXR.
Most strikingly, H2A.Z was highly enriched on the promoter region prior to tRA induction, with a low level of H3.3 detected (Fig. 4B,D,E [panel b]). During transcriptional activation by tRA, the level of H2A.Z at the promoter rapidly dropped down, accompanied by rapidly increased canonical H2A, within 1 h after tRA addition, while the H2B levels did not display any apparent change (Fig. 4D, panel b). Furthermore, we also observed a rapid incorporation of H3.3, while the H4 and canonical H3 levels decreased during this time course (1 h induction by tRA) (Fig. 4E, panel b), suggesting an apparent exchange of H3.3 with H3 into the promoter region, which is concomitant with a slight nucleosome eviction in the early stage of gene activation by tRA. The accompanying opening of the chromatin structure around this region was also demonstrated using time-course EpiQ assays (Fig. 4G). As shown in Figure 4A, transcription began after 1 h of tRA induction, while the levels of histone H2B and H4 began to quickly decrease, indicating the quick eviction of nucleosomes on the promoter region of the Cyp26A1 gene after 1 h of tRA induction. In addition, our time-course ChIP analysis revealed that RAR and RNA polymerase II (Pol II) were increasingly recruited to the promoter region, which was accompanied by the opening of the chromatin and transcriptional activation during tRA induction (Fig. 4A,F,G). In contrast, our time-course ChIP and EpiQ analysis showed that the levels of H2A.Z, H2A, H3.3, H3, H2B, H4, and chromatin structure on the gene body region did not have significant changes during the time course of tRA induction examined (Fig. 4D–G). Moreover, the dynamics of H2A.Z and H3.3 and chromatin structures were also analyzed for another gene, HoxA1, which was also highly regulated by RA signaling. The time-course ChIP and EpiQ analysis revealed that similar dynamic replacements of the histone variants H2A.Z and H3.3 and chromatin dynamics were observed on the promoter and enhancer regions of HoxA1 during tRA induction (Supplemental Fig. S5).
The incorporation of H3.3 is important for activation of Cyp26A1 by tRA
Our results revealed that H3.3 was actively deposited into the enhancer region prior to gene induction, and H3.3 was rapidly depleted from the enhancer region but deposited into the promoter region during gene activation by tRA induction, which indicated that the H3.3 variant may play a critical role in the activation of inducible genes. Thus, we generated H3.3 knockdown mES cells to investigate the functions of H3.3 on the activation of Cyp26A1 by tRA. As shown in Figure 5A, H3.3 knockdown greatly impaired the expression of Cyp26A1 during tRA induction, which indicated that H3.3 was important for tRA-mediated activation of Cyp26A1. We also assessed the dynamic changes of H2A.Z and RAR in the enhancer and promoter regions, respectively, during tRA induction after H3.3 knockdown (Fig. 5B). Knockdown of H3.3 did not have any obvious effects on the binding of H2A.Z on the enhancer region (Fig. 5B, panel b) because the level of H2A.Z was very low at the enhancer region of Cyp26A1 in wild-type mES cells. However, knockdown of H3.3 could reduce the binding of RAR to the enhancer region during tRA induction (Fig. 5B, panel d). In addition, our EpiQ assays demonstrated that knockdown of H3.3 resulted in compaction of the chromatin structure at the enhancer region of the Cyp26A1 gene (Fig. 5G, panel b), which indicated that deposition of H3.3 at enhancer regions impaired the folding of chromatin and enabled the binding of the transcription factor (RAR/RXR). For the promoter region of the Cyp26A1 gene, knockdown of H3.3 greatly impaired the deposition of H2A.Z before tRA induction (Fig. 5B, panel c), resulted in the opening of the chromatin structure (Fig. 5G, panel c), and weakened the stimulated binding of RAR, TBP, and Pol II on the promoter region during tRA induction (Fig. 5B [panel e], C). These findings strongly suggested that marking the enhancer region by H3.3 prior to tRA induction was essential for the subsequent binding of RAR, TBP, and Pol II on the promoter region during tRA induction. Moreover, the deposition of H2A.Z at the promoter region was also dependent on the incorporation of H3.3 at the enhancer region of Cyp26A1. Interestingly, further investigation showed that the knockdown of H3.3 inhibited the recruitment of the histone acetyltransferase complex Tip60 and chromatin remodelers (SRCAP and BRG1), which may be involved in H2A.Z deposition at the promoter regions of Cyp26A1 and HoxA1 (Supplemental Fig. S6). Interestingly, we did not observe the apparent recruitment of chromatin remodeler SNF2H at the promoter regions in our experimental conditions (Supplemental Fig. S6). The results indicated that the H3.3 on the enhancer regions may help to recruit the H2A.Z-specific histone acetyltransferase complex Tip60 and chromatin remodelers to deposit H2A.Z onto the promoter regions.
Figure 5.
The incorporation of H3.3 is important for Cyp26A1 activation by tRA in mES cells R1. (A, panel a) The level of H3.3 protein was clearly reduced by siRNA-mediated interference. (Panel b) The effect of H3.3 knockdown on the relative levels of Cyp26A1 mRNA at different time points during tRA induction in mES cells (R1). (B, panel a) A schematic diagram of the primer pairs in the enhancer (RARE2) and promoter (RARE1) regions on the Cyp26A1 gene. The effect of H3.3 knockdown on the enrichment of H2A.Z (panels b,c) and RARα (panels d,e) on the enhancer and promoter regions of the Cyp26A1 gene during tRA induction in mES cells (R1) using ChIP assays. (C, panel a) A schematic diagram of the primer pair in the promoter (RARE1) region on the Cyp26A1 gene. The effect of H3.3 knockdown on the recruitment of TBP (panel b) and Pol II (panel c) on the promoter region of the Cyp26A1 gene during tRA induction in mES cells (R1) using ChIP assays. (D, panel a) The level of H2A.Z protein was clearly reduced by siRNA-mediated interference. (Panel b) The effect of H2A.Z knockdown on the Cyp26A1 mRNA at different time points during tRA induction in mES cells (R1). (E, panel a) A schematic diagram of the primer pairs in the enhancer (RARE2) and promoter (RARE1) regions on the Cyp26A1 gene. The effect of H2A.Z knockdown on the enrichment of H3.3 (panels b,c) and RARα (panels d,e) on the enhancer and promoter regions of the Cyp26A1 gene during tRA induction in mES cells (R1) using ChIP assays. (F, panel a) A schematic diagram of the primer pairs in the promoter (RARE1) region on the Cyp26A1 gene. The effect of H2A.Z knockdown on the recruitment of TBP (panel b) and Pol II (panel c) on the promoter region of the Cyp26A1 gene during tRA induction in mES cells (R1) using ChIP assays. (G, panel a) A schematic diagram of the primer pairs in the enhancer (RARE2) and promoter (RARE1) regions on the Cyp26A1 gene. EpiQ analysis of the accessibility of chromatin on the enhancer (panels b,d) and promoter (panels c,e) regions of the Cyp26A1 gene before tRA induction in H3.3 (panels b,c) and H2A.Z (panels d,e) knockdown mES cells (R1). The results were normalized to the reference Rho gene. Statistical analysis in the experiment was performed using two-tailed Student's t-test; (*) P < 0.05; (**) P < 0.01. All of the data shown are expressed as the mean ± SD of three independent biological replicates.
H2A.Z knockdown mES cells were also generated to investigate the functions of H2A.Z on the activation of the Cyp26A1 gene by tRA. As shown in Figure 5D, H2A.Z knockdown promoted the expression of Cyp26A1 during tRA induction, which indicated that H2A.Z exhibited a repressive role in tRA-induced transcription of the Cyp26A1 gene. We also assessed the effects of H2A.Z knockdown on the dynamic changes of H3.3, RAR, TBP, and Pol II in the enhancer and promoter regions during tRA induction (Fig. 5E,F). H2A.Z knockdown did not have any obvious effect on either the recruitment of H3.3 on the enhancer region before tRA induction or the dynamic changes of H3.3 on the enhancer and promoter regions during tRA induction (Fig. 5E, panels b,c). These results indicated that the deposition of H3.3 was independent of the incorporation of H2A.Z. In addition, because the level of H2A.Z was very low at the enhancer region of the Cyp26A1 gene in wild-type mES cells, H2A.Z knockdown did not affect the binding of RAR to the enhancer region during tRA induction (Fig. 5E, panel d). However, at the promoter region, EpiQ assays showed that the knockdown of H2A.Z resulted in the opening of the chromatin structure (Fig. 5G, panel e) and enhanced the binding of RAR and TBP to the promoter prior to tRA induction (Fig. 5E [panel e], F [panel b]). In addition, the eviction of H2A.Z from the promoter region could enhance the stimulated binding of TBP and Pol II to the promoter during tRA induction (Fig. 5F). The functions of H2A.Z and H3.3 on the activation of another gene, HoxA1, by tRA were also examined, as shown in Supplemental Figure S7, which revealed a phenomenon similar to that observed on the Cyp26A1 gene.
Genome-wide investigation of the correlation of incorporation of H2A.Z and H3.3 with chromatin structures
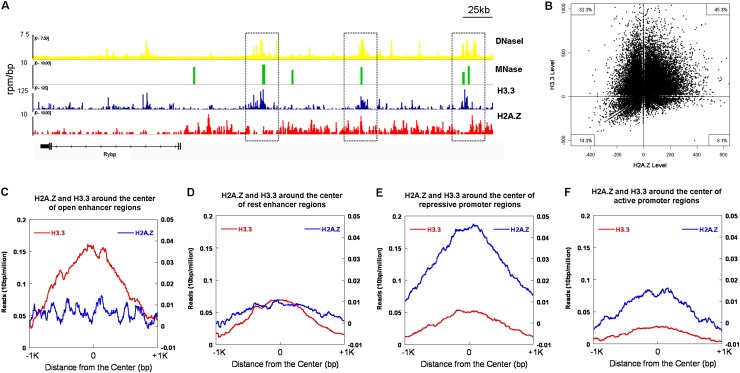
Using RAR/RXR targeted genes as a model, we revealed that the activation of gene transcription was accompanied by the depletion of the histone variant H2A.Z and concomitant deposition of H3.3 to open chromatin structures at the promoter region to initiate transcription. To map the chromatin structures across the whole genome, MNase sequencing (MNase-seq) was developed to analyze the chromatin structures on the basis of the accessibility of the genome to MNase digestion. As shown in Figure 6, A and B, most of the open chromatin regions (MNase-sensitive sites) are enriched with histone variant H3.3 (32.3% for H3.3 only or 45.3% for double variants H3.3 and H2A.Z), which was consistent with our aforementioned findings that H3.3 can open chromatin via preventing the folding of chromatin fibers and/or counteracting H2A.Z-mediated chromatin compaction. We further analyzed the correlation of H3.3 and H2A.Z enrichment with chromatin structure at the predicted enhancer and promoter regions. The open degree of chromatin structure was positively correlated with the enrichment of H3.3, but negatively correlated with the level of H2A.Z, over the intergenic regulatory regions (most likely representing the enhancer regions) (Supplemental Fig. S8A). However, the highly opened chromatin on the regions within the transcription start site (TSS) ± 500 bp (promoter regions) was negatively correlated with very low levels of both H3.3 and H2A.Z (Supplemental Fig. S8B), which is very similar to the correlation pattern of FAIRE-positive nucleosome-free regions with the H3.3 and H2A.Z levels (Pchelintsev et al. 2013), indicating that the highly opened chromatin structure at promoter regions may result from nucleosome depletion. In addition, correlation of the distribution of H2A.Z and H3.3 on the predicted enhancer and promoter regions with chromatin structures and their expression levels, respectively, was also analyzed across the entire genome. As shown in Figure 6, C and D, genome-wide analysis showed that a low level of H2A.Z was found in the predicted enhancer regions. The most open enhancer regions were highly enriched with H3.3 compared with the rest enhancer regions, which indicated that the open degree of chromatin structure was positively correlated with the enrichment of H3.3 at the enhancer regions. We further analyzed the correlation between enrichments of H2A.Z and H3.3 at the promoter regions and gene expression genome-wide. To this end, we subgrouped the promoter regions into active promoters and repressive promoters based on their expression levels, where the active promoters referred to the predicted promoters for the top one-quarter of expression genes, and the repressive promoters referred to the predicted promoters for the bottom one-quarter of expression genes. As shown in Figure 6, E and F, a higher level of H2A.Z was found at the repressive promoter regions compared with the active promoters, while H3.3 showed low levels on both active promoters and repressive promoters. These results were mutually consistent with our findings from in vivo experiments on specific genes and in vitro biochemical and biophysical assays.
Figure 6.
Genome-wide distributions of H2A.Z and H3.3 and their correlation with chromatin structures. (A,B) Correlation of the enrichment of H3.3 and/or H2A.Z with open chromatin regions (MNase-sensitive sites). (A) Genome browser tracks show the correlation of open chromatin regions (DNaseI- and MNase-sensitive sites) with the histone variant H3.3 and H2A.Z occupancy (Goldberg et al. 2010; Xiao et al. 2012). (B) Quantitative analysis of the genome-wide correlation of H3.3 (Y-axis) or H2A.Z (X-axis) with MNase-sensitive regions (total number 34,142). (C,D) The distribution of H2A.Z and H3.3 at the open and rest enhancer regions across the entire genome. H3.3 was highly enriched in the open enhancer regions with very low levels of H2A.Z (C), while there were relatively low levels of H3.3 and H2A.Z localized at the rest enhancer regions (D). (E,F) The distribution of H2A.Z and H3.3 at the repressive and active promoter regions across the entire genome. H2A.Z was highly enriched in the repressive promoter regions with very low levels of H3.3 (E), while both relatively low levels of H2A.Z and H3.3 were observed at the active promoter regions (F).
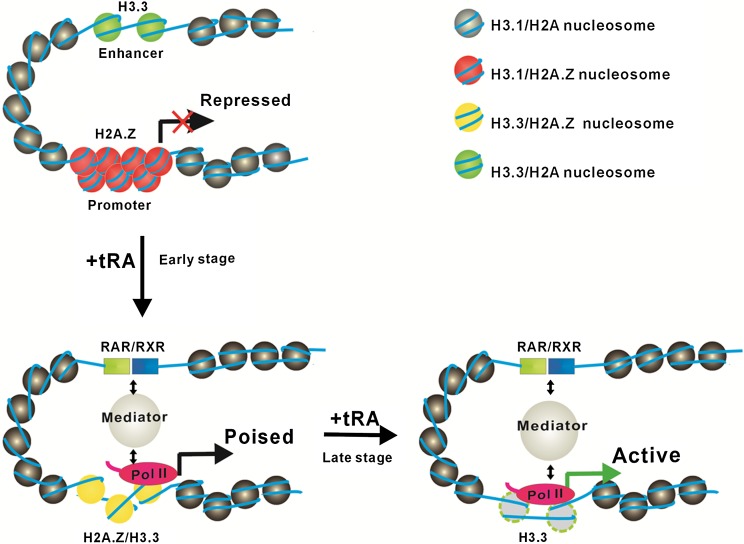
In summary, the results obtained from our in vitro biochemical and biophysical assays and the studies in vivo on both specific genes and the genome-wide level suggest that the incorporation of H2A.Z on the promoter would result in compaction of chromatin to repress transcription, while the incorporation of H3.3 impaired chromatin folding to allow the enhancer and promoter regions to adopt a relatively open conformation for the binding of transcription factors and cofactors to activate transcription (Fig. 7).
Figure 7.
A model for the dynamic regulation of H2A.Z and H3.3 on gene activation. When a gene is ready to activate, H3.3 is highly enriched on the enhancer region to maintain a relatively open chromatin structure that is accessible for the binding of transcription factors (RAR/RXR); meanwhile, at the promoter region, the H3.3-dependent enrichment of H2A.Z compacts chromatin to poise gene transcription. When the gene is activated by the addition of tRA, the H3.3-containing nucleosomes at the enhancer region are immediately evicted for RAR/RXR binding; at the same time, H2A.Z is selectively replaced by canonical H2A accompanied by the deposition of H3.3 to open the chromatin structure for bindings of transcription factors and transcriptional machinery at the promoter region (the early stage of gene activation by tRA). Subsequently, the nucleosomes at the promoter region are also evicted for the full gene activation (the late stage of gene activation by tRA).
Discussion
Here we investigated the structural and transcriptional regulation of two variants, H2A.Z and H3.3, at the chromatin level in vitro and in vivo. Our results revealed that H2A.Z could repress transcription via stabilizing mononucleosomes and facilitating chromatins to generate more compact structures. However, the incorporation of H3.3 could activate gene transcription via hindering the compaction of chromatin fibers regardless of the presence of H2A.Z. Moreover, we further demonstrated that the dynamic depositions and/or replacement of H2A.Z and H3.3 resulted in the corresponding structural changes of chromatin at the enhancer and promoter regions of RAR/RXR targeted genes during gene activation by tRA. Our results shed light on the molecular mechanisms of how histone variants function together to prime gene transcription via modulating nucleosome/chromatin dynamics over enhancer and promoter regions.
Effect of the histone variants H2A.Z and H3.3 on nucleosome/chromatin dynamics and transcriptional regulation
It has been shown that chromatin structure exhibits a highly dynamic equilibrium between an open conformation and a compacted 30-nm fiber, which may play a key role in transcriptional regulation (Li and Reinberg 2011). In this study, we demonstrated that H2A.Z itself can stabilize mononucleosomes in experiments using FRET and single-molecule magnetic tweezers, which was consistent with recent findings that H2A.Z-containing nucleosomes harbored a more stable structure when compared with canonical nucleosomes (Park et al. 2004; Thambirajah et al. 2006). In addition, the incorporation of H2A.Z facilitated the folding of nucleosomal arrays in the presence of MgCl2 or H1. These highly compacted structures, resulting from the H2A.Z-containing chromatins, were refractory to transcription by RNA Pol II. We further analyzed the effect of H3.3 on nucleosome/chromatin dynamics and transcriptional regulation. Our FRET and magnetic tweezer experiments demonstrated that the incorporation of H3.3 only showed little effect on the stability of mononucleosomes, which was consistent with previous findings that incorporation of H3.3 did not affect nucleosome stability in the absence of H2A.Z (Flaus et al. 2004; Thakar et al. 2009). Jin and Felsenfeld (2007) have shown that coexistence of H2A.Z and H3.3 resulted in destabilization of “native” nucleosomes. However, our in vitro biochemical and biophysical results revealed that the incorporation of double variant H2A.Z/H3.3 in the same nucleosome resulted in a more stable property compared with those observed with canonical histones. These apparently contradicting data may result from the context of pre-existing higher-order chromatin, post-translational modifications of histones and DNA, and/or the additional actions of ATP-dependent chromatin remodeling factors and histone chaperones on the different histone variants. Intriguingly, our AUC and EM analyses demonstrated that the incorporation of H3.3 greatly impaired the folding of the chromatin arrays, which indicated the H3.3-containing chromatin tended to adopt a relatively open state. Moreover, the incorporation of H3.3 could counteract the promoting effects of H2A.Z on the folding of nucleosomal arrays. The impairment of H3.3 on higher-ordered chromatin structures may partially contribute to the combined effect of H2A.Z and H3.3 on nucleosome dynamics observed in vivo (Jin and Felsenfeld 2007). In addition, using assembled chromatin as a template, we found that H3.3 could antagonize the inhibitory effects of H2A.Z on chromatin transcription by RNA Pol II, which partially resulted from the counteractivity of compaction by H2A.Z. This suggests that H3.3 may play a dominant role in regulation of the dynamics of higher-ordered chromatin and transcriptional activity in the chromatin context. Importantly, using prokaryotic T7 RNA polymerase and reconstituted mononucleosomes, Zlatanova and colleagues (Thakar et al. 2010) observed a similar inhibitory effect of H2A.Z on transcription by T7 RNA polymerase, while only H3.3 incorporation onto nucleosomes exhibited no effect on transcription in vitro. Interestingly, they also found that the hybrid nucleosome particle containing both H2A.Z and H3.3 inhibited transcription, similar to H2A.Z. Their results were inconsistent with the properties of H2A.Z and H3.3 that we observed on the chromatin level, suggesting that at the mononucleosome level, transcription activity is controlled mainly by the properties of mononucleosomes, while at the chromatin level, the higher-ordered structure of chromatins might play a dominant role in transcriptional regulation.
The H3.3 variant differs from canonical H3 at only four amino acid residues, three of which are hidden inside the nucleosome core particle in region 87–90, and one residue Ser31 is exposed outside of the nucleosome core particle. Interestingly, conversion of the H3.3 variant region 87–90 (AAIG) to the canonical H3 sequence “SAVM” abolished replication-independent incorporation (Szenker et al. 2011). However, a single replacement of “S” with “A” at position 31 of the histone H3 tail resulted in no effect on the deposition pathway, suggesting that H3.3 S31 and its phosphorylation did not play a role in H3.3 incorporation (Hake et al. 2005). Most recently, we and others showed that Ala87 and Gly90 in H3.3 play critical roles in DAXX recognition (Elsasser et al. 2012; Liu et al. 2012). However, it still remains unclear whether H3.3 S31 and the region 87–90 function in the regulation of chromatin dynamics. In this study, all four unique residues in H3.3 were shown to hinder the compaction of the chromatin arrays, while residues 89 and 90 were mainly responsible for the counteractivity of H2A.Z-mediated chromatin compaction. Using in vitro cross-linking experiments in a model of reconstituted nucleosomal arrays, Hayes and colleagues (Zheng et al. 2005) demonstrated that the entire N-terminal tail of H3 participated in internucleosomal interactions in highly compacted chromatin fiber induced by MgCl2. Thus, it was likely that the substitution of reside 31 in H3.3 disrupted the internucleosomal interaction mediated by the N-terminal tail of H3 and impaired the compaction of chromatin fibers. However, according to the crystal structure of the nucleosome core particle, none of these four amino acids appear to directly interact with H2A.Z or H2A, and the substitutions of the three amino acid residues located in beginning of the α2 helix of H3.3 will not result in any major changes in intranucleosomal or internucleosomal interactions. Thus, the participation of H3.3 in regulating chromatin dynamics is difficult to rationalize.
H3.3 and H2A.Z function together to determine the transcriptional potential of RA-regulated genes via regulating chromatin dynamics over the enhancer and promoter regions
The dynamic incorporation of H2A.Z and H3.3 and their corresponding regulation on chromatin structure and transcription have been investigated during the activation of the nuclear receptors RAR/RXR by tRA induction. In summary, H2A.Z exhibits a repressive role in tRA-induced transcription of Cyp26A1 and HoxA1 genes, while the incorporation of H3.3 was essential for the gene activation. Knockdown of H2A.Z in cells promoted the expression of Cyp26A1 and HoxA1, but H3.3 knockdown inhibited gene expression during tRA induction. It has been reported that the incorporation of H3.3 was found at distal promoter elements, upstream enhancers, and other regulatory elements that occurred relatively distant from the genes (Jin and Felsenfeld 2006). Our time-course ChIP analysis revealed that H3.3 was actively deposited to enhancers prior to gene induction and was depleted rapidly after gene activation. We also found that structural alteration occurred on the enhancer regions during gene activation. Our EpiQ and genome-wide MNase-seq analysis demonstrated that the chromatin in the enhancer regions enriched with H3.3 displayed a relatively open conformation prior to gene activation, which may be partially due to the impaired effect of H3.3 on chromatin folding, similar to what we observed in vitro. Furthermore, the nucleosomes together with the histone variant H3.3 were rapidly displaced from the enhancer regions shortly after tRA induction. These results indicated that the incorporation of H3.3 may decorate the chromatin architecture at the enhancer regions and allow for the recognition and binding of transcriptional activators, such as nuclear receptors and transcription factors, which subsequently recruit the ATP-dependent chromatin remodeling complex SWI/SNF and/or p300 to remodel the nucleosome architecture, as previously reported (Li et al. 2010). We also observed that knockdown of H3.3 resulted in chromatin compaction and impaired the subsequent binding of RAR, TBP, and Pol II on the enhancer and promoter regions during tRA induction. Henikoff et al. (2009) have shown that H3.3-enriched chromatins were relatively soluble in low-salt conditions, which suggested that chromatin containing H3.3 adopted a more open structure for MNase digestion and low-salt extraction. In addition, the deposition of H3.3 in the enhancer region was independent of the H2A.Z variant. Knockdown of H2A.Z did not affect the enrichment and dynamic changes in H3.3 in the enhancer regions during gene activation. Despite observations of an association between transcription and H3.3 incorporation in the enhancer regions of the genes studied, the mechanism that preferentially deposits H3.3 into nucleosomes at transcribed genes is still not well understood; for example, it is still unclear which histone chaperone is responsible for recruiting H3.3 to the enhancer regions prior to gene induction.
The dynamics of histone variants on promoter regions are very different from what occurs on the enhancer regions. Our time-course ChIP results clearly showed a two-step chromatin remodeling event at the promoter regions of both Cyp26A1 and HoxA1 genes (Fig. 4; Supplemental Fig. S5). The histone replacement step occurred first within 1 h of induction by tRA, followed by the subsequent nucleosome eviction step. Instead of H3.3, H2A.Z is actively recruited to promoters prior to RNA Pol II and is rapidly replaced during transcriptional activation by tRA induction. Accordingly, our EpiQ analysis indicated that the chromatin in the promoter region of the Cyp26A1 gene was also relatively more compacted compared with the enhancer region (data not shown), but the compaction state could be greatly impaired by H2A.Z knockdown, which was consistent with our in vitro finding that H2A.Z could facilitate chromatin compaction and stabilize nucleosome structures. In addition, the histone variant H2A.Z was rapidly replaced by H2A during early time points (within 1 h) of tRA induction, which was accompanied by a significant increase of H3.3. However, it is still unclear whether the H3.3 deposition in this region was a consequence of or a prerequisite for transcriptional activation. We also showed that knockdown of H3.3 greatly impaired the incorporation of H2A.Z in the promoter region of Cyp26A1 before tRA induction, which suggested that the recruitment of H2A.Z to the promoter region prior to gene activation was dependent on the existence of H3.3 on the enhancer region. However, knockdown of H2A.Z did not affect the incorporation of H3.3 into the promoter region after tRA induction, which suggested that the recruitment of H3.3 to the promoter region after gene activation was independent of the pre-existence of H2A.Z on the same region. Interestingly, we also found that H3.3 at the enhancer can facilitate the recruitment of the histone acetyltransferase complex and chromatin remodelers (such as Tip60 and SRCAP) to deposit H2A.Z at the promoter regions. Two recent studies assessing inducible gene expression have also found that incorporation of H3.3 promoted initial gene activation (Placek et al. 2009; Tamura et al. 2009). The incorporation of H3.3 may be important to remove repressive histone modifications such as H3K9 methylation and/or replacement with activating marks, as suggested by others (Ahmad and Henikoff 2002). However, although nucleosomal H3.3 is enriched in activating modifications such as H3K4me3, our time-course ChIP results revealed that H3K4me3 appears to be established only after histone replacements and coupled to active transcription (data not shown). Accordingly, our results suggested that although the incorporation of H3.3 onto enhancer regions prior to induction and onto promoter regions after induction may result from distinct mechanisms, the incorporation of H3.3 on these regions facilitates the chromatins to form loose structures for the binding of transcription factors (RAR/RXR), chromatin remodeling complexes, and histone-modifying enzymes and the subsequent recruitment of transcriptional machinery upon gene induction. Taken together, our results revealed that the extensive H3.3 deposition accompanying the exchange of H2A.Z on the enhancer and promoter regions of active genes will create featured chromatin signatures to allow inducible genes to be able to rapidly respond to environmental stimulation.
Materials and methods
Protein, DNA, antibodies, and cell culture
For protein, DNA, antibodies, and cell culture, see the Supplemental Material.
RNAi-mediated gene silencing in mES cells (R1)
Validated siRNA duplexes were used to knock down H3.3A, H3.3B, and H2A.Z. The oligonucleotide sequences were as follows: siH3.3A#1, 5′-TGAGTTGTCCTACATACAA-3′; siH3.3A#2, 5′-GCCAAACGTGTAACAATTA-3′; siH3.3B#1, 5′-GCGTATTAAACCTTGCATA-3′; siH3.3B#2, 5′-GCATGTTTCTGTATGTTAA-3′; siH2A.Z#1, 5′-GGTAAGGCTGGAAAGGACT-3′; and siH2A.Z#2, 5′-TGGAGATGAAGAATTGGAT-3′.
siRNA duplexes were transfected into mES cells using Lipofectamine 2000. Cells were harvested after 72 h of transfection.
Nucleosome and chromatin reconstitution
The respective histone octamers and chromatins were reconstituted as previously described (Dyer et al. 2004; Li et al. 2010). The experimental details are described in the Supplemental Material.
EM
The metal shadowing with tungsten and negative staining experiment for EM study were performed as described in the Supplemental Material. The samples were examined using a FEI Tecnai G2 Spirit 120-kV transmission electron microscope.
Sedimentation velocity AUC
The chromatin samples were prepared in measurement buffer (10 mM HEPES at pH 8.0, 0.1 mM EDTA) using various concentrations of MgCl2. Sedimentation experiments were performed on a Beckman Coulter ProteomeLab XL-I using a four-hole An-60Ti rotor. Samples with an initial absorbance at 260 nm of ∼0.5–0.8 were equilibrated for 2 h at 20°C under vacuum in a centrifuge prior to sedimentation. The absorbance at 260 nm was measured in a continuous scan mode during sedimentation at 32,000g in 12-mm double-sector cells. The data were analyzed using enhanced van Holde-Weischet analysis and the Ultrascan II 9.9 revision 1504. The S20,w values (sedimentation coefficient corrected for water at 20°C) were calculated with a partial specific volume of 0.622 mL/g for chromatin, and the buffer density and viscosity were adjusted. The average sedimentation Save coefficients were determined at the boundary midpoint.
Steady-state FRET analysis
Double-fluorescence-labeled 169-bp 601 DNA fragments were prepared by PCR using primers labeled with the dye pair of Alexa 488 and Alexa 594 with a Förster distance of ∼54 Å from a plasmid containing the 601 positioning sequence. The sequences for the primers (Invitrogen) used were as follows: FWD, 5′-ACAGTACTGGCCGCCCTGGAGAATCCCGGTGCCGAGGCCGCT(Alexa488)CAATTG-3′; and REV, 5′-TACATGCACAGGATGTATATATCTGACACGTGCCTGGAGACT(Alexa594)AGGGAG-3′.
The labeled DNA templates were reconstituted into mononucleosomes using the salt dialysis method as previously described. Steady-state fluorescence experiments were performed at 20°C on a fluorescence spectrometer (EmSpire, PerkinElmer). For the salt-dependent dissociation study, samples with different concentrations of NaCl were excited at 492 nm, and the emission was recorded from 510 nm to 750 nm. The difference in the fluorescence intensity between the donor and acceptor emissions was plotted against the concentration of NaCl, which generated the salt-dependent dissociation curves for the canonical and variant-containing mononucleosomes.
Single-molecule magnetic tweezer analysis
Single molecular manipulation of a nucleosomal array was performed on magnetic tweezers (Pico Twist Company) as described previously (Gosse and Croquette 2002; Meglio et al. 2009). Additional details are provided in the Supplemental Material.
In vitro transcription assay
Transcription assays using a pG5MLP template were performed as previously described (Loyola et al. 2001). Briefly, 50 ng of naked DNA or an equimolar amount of chromatin assembled with recombinant canonical or variant-containing histone octamers was incubated with 20 ng of Gal4-VP16 for 15 min at room temperature. Next, 50 ng of p300 and 1.5 μM acetyl-CoA was added as indicated and incubated for 30 min at 30°C. One-hundred micrograms of HeLa cell nuclear extract was added into each reaction and incubated for another 20 min at 30°C. Transcription was carried out by adding ATP, CTP, GTP, and 32P-labeled UTP for 20 min at 30°C. The transcription reaction was stopped, and the RNA transcripts were extracted with phenol-chloroform, precipitated with ethanol, and analyzed by 5% urea-PAGE.
Isolation of nascent RNA and mRNA and real-time PCR analysis
Nascent RNA was extracted as previously described (Khodor et al. 2011). The total RNA was extracted using TRIzol reagent (Invitrogen), and the first strand of cDNA was reverse-transcribed using 2 μg of RNA. cDNA products were used for quantitative real-time PCR using the SYBR Premix Ex Taq (Takara). Additional experimental details are described in the Supplemental Material.
Chromatin structure analysis in vivo
The chromatin structure was analyzed using the EPiQ chromatin analysis kit (Bio-Rad). Briefly, chromatin was digested using DNase I in situ for 1 h. The digested and undigested chromatin DNA was purified and quantified using the Nanodrop 2000 (Thermo Scientific). The samples were analyzed using real-time PCR with the following primer sequences: Cyp26A1 enhancer (sense, 5′-CCGGGCTCATGTTGTAAAC-3′; antisense, 5′- GCTGCCACTGTCATATCTTGTA-3′), Cyp26A1 promoter (sense, 5′-AGGAAAGAGGTGTTCCTAGTCC-3′; antisense, 5′-ATAAGGCCGCCCAGGTTA-3′), Cyp26A1+2000 (sense, 5′-CACGAGGAACCGTATTAAAGG-3′; antisense, 5′- CATGACCACCAAAGAGGAGC-3′), HoxA1 promoter (sense, 5′-GCCACTGAAACGGTGATCC-3′; antisense, 5′-AGAGTCGCCACTGCCAAG-3′), and HoxA1 enhancer (sense, 5′-TTTTGGAGGCTATTCAGATGC-3′; antisense, 5′-TCCCGAAGAGGAATGGAAC-3′).
ChIP
ChIPs and quantitative real-time PCR were performed as previously described (Margueron et al. 2008). The experimental details are provided in the Supplemental Material.
MNase-seq analysis
The genome of mES cells (Milli Trace Nanog GFP Reporter mES cells, SCR089) was digested with MNase at different time points. Subsequently, the genomic DNA from the MNase digestion was extracted with phenol-chloroform, precipitated with ethanol, and analyzed on a 1.0% agarose gel. Two digestion conditions, a mild digestion in which a small fraction of the genome was digested into a mononucleosome and an extensive digestion in which most of the genome (>80%) was digested into a mononucleosome, were chosen accordingly. The DNA fragments corresponding to mononucleosomal sizes of the two selected digestion conditions were isolated and purified from the gel, and the resulting DNAs were subjected to sequencing using the HiSeq 2000 sequencing system. FASTQ sequences at 100 bp in length were aligned to the Mus musculus reference genome (mm9) using BWA with default parameters (Li and Durbin 2010). Only the uniquely mapped reads with a quality >10 were considered for genome-wide data analysis. The genome-wide data analyses were performed as described in the Supplemental Material. The MNase-seq data are deposited in Gene Expression Omnibus under accession number GSE 50706.
Acknowledgments
We are grateful to Dr. Eric Campos, Dr. Bing Zhu, and Dr. Raphael Margueron for critical reading and discussion of our manuscript. The work was supported by grants from the Ministry of Science and Technology (2011CB966303 to G.L., and 2009CB825503 to P.C.), Chinese Academy of Sciences (CAS) Strategic Priority Research Program (XDA01010304 to G.L., and XDA01010305 to Q.W.) and the Key Research Program (KSZD-EW-Z-009 to P.C.), the National Natural Science Foundation of China (91019007, 31071147, and 91219202 to G.L.; 31000566 to P.C.; and 61275192 to M.L.), and the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (to P.C.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.222174.113.
References
- Ahmad K, Henikoff S 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Dalvai M, Bellucci L, Fleury L, Lavigne AC, Moutahir F, Bystricky K 2012. H2A.Z-dependent crosstalk between enhancer and promoter regulates Cyclin D1 expression. Oncogene 32: 4243–4251 [DOI] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K 2004. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375: 23–44 [DOI] [PubMed] [Google Scholar]
- Elsasser SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ 2012. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 491: 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A 2005. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J Biol Chem 280: 25298–25303 [DOI] [PubMed] [Google Scholar]
- Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T 2004. Sin mutations alter inherent nucleosome mobility. EMBO J 23: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev 21: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosse C, Croquette V 2002. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys J 82: 3314–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, Allis CD 2005. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci 102: 6344–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K 2009. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res 19: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G 2006. Distribution of histone H3.3 in hematopoietic cell lineages. Proc Natl Acad Sci 103: 574–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G 2007. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al. 2008. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, Rosbash M 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotekar AS, Weissman JD, Gegonne A, Cohen H, Singer DS 2008. Histone modifications, but not nucleosomal positioning, correlate with major histocompatibility complex class I promoter activity in different tissues in vivo. Mol Cell Biol 28: 7323–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Reinberg D 2011. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci 102: 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Hu G, Stokes D, Wang YH, Reinberg D 2010. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell 38: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CP, Xiong C, Wang M, Yu Z, Yang N, Chen P, Zhang Z, Li G, Xu RM 2012. Structure of the variant histone H3.3–H4 heterodimer in complex with its chaperone DAXX. Nat Struct Mol Biol 19: 1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, LeRoy G, Wang YH, Reinberg D 2001. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev 15: 2837–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meglio A, Praly E, Ding F, Allemand JF, Bensimon D, Croquette V 2009. Single DNA/protein studies with magnetic traps. Curr Opin Struct Biol 19: 615–622 [DOI] [PubMed] [Google Scholar]
- Park YJ, Dyer PN, Tremethick DJ, Luger K 2004. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem 279: 24274–24282 [DOI] [PubMed] [Google Scholar]
- Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, Adams PD 2013. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3: 1012–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol 83: 1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, Milburn PJ, Parish CR, Tremethick DJ, Rao S 2009. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol 29: 1972–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol 7: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Szenker E, Ray-Gallet D, Almouzni G 2011. The double face of the histone variant H3.3. Cell Res 21: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Smith M, Kanno T, Dasenbrock H, Nishiyama A, Ozato K 2009. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem 284: 12217–12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, Fukui K, Tomschik M, Ausio J, Zlatanova J 2009. H2A.Z and H3.3 histone variants affect nucleosome structure: Biochemical and biophysical studies. Biochemistry 48: 10852–10857 [DOI] [PubMed] [Google Scholar]
- Thakar A, Gupta P, McAllister WT, Zlatanova J 2010. Histone variant H2A.Z inhibits transcription in reconstituted nucleosomes. Biochemistry 49: 4018–4026 [DOI] [PubMed] [Google Scholar]
- Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J 2006. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J Biol Chem 281: 20036–20044 [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P 2007. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 39: 251–258 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB 2006. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 16: 151–156 [DOI] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell 33: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MM, Cox LK, Chrivia JC 2007. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J Biol Chem 282: 26132–26139 [DOI] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res 20: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, et al. 2012. Comparative epigenomic annotation of regulatory DNA. Cell 149: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Lu X, Hansen JC, Hayes JJ 2005. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J Biol Chem 280: 33552–33557 [DOI] [PubMed] [Google Scholar]