Nonsense-mediated mRNA decay (NMD) is a quality control mechanism to detect aberrant mRNAs and induce their degradation. Loh et al. now show that mRNA decay enzyme SMG7 directly binds POP2, a catalytic subunit of the CCR4–NOT deadenylase complex, and promotes deadenylation-dependent decapping and decay of NMD targets. The authors further show that mRNA surveillance complex component UPF1 provides multiple binding sites for mRNA decapping factors. This study unveils a missing physical link between NMD and the general mRNA decay machinery.
Keywords: deadenylation, mRNA decay, NMD, UPF1
Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic quality control mechanism that detects aberrant mRNAs containing nonsense codons and induces their rapid degradation. This degradation is mediated by SMG6, an NMD-specific endonuclease, as well as the SMG5 and SMG7 proteins, which recruit general mRNA decay enzymes. However, it remains unknown which specific decay factors are recruited and whether this recruitment is direct. Here, we show that SMG7 binds directly to POP2, a catalytic subunit of the CCR4–NOT deadenylase complex, and elicits deadenylation-dependent decapping and 5′-to-3′ decay of NMD targets. Accordingly, a catalytically inactive POP2 mutant partially suppresses NMD in human cells. The SMG7–POP2 interaction is critical for NMD in cells depleted of SMG6, indicating that SMG7 and SMG6 act redundantly to promote the degradation of NMD targets. We further show that UPF1 provides multiple binding sites for decapping factors. These data unveil a missing direct physical link between NMD and the general mRNA decay machinery and indicate that NMD employs diverse and partially redundant mechanisms to ensure robust degradation of aberrant mRNAs.
The nonsense-mediated mRNA decay (NMD) pathway rids the cell of aberrant mRNAs that have acquired premature translation termination codons (PTCs, or nonsense codons) as a result of mutations or inaccuracies in gene expression that inadvertently generate PTCs. Through this action, NMD prevents the accumulation of truncated proteins, which can be toxic to the cell (Kervestin and Jacobson 2012). In addition to eliminating aberrant mRNAs, NMD post-transcriptionally regulates the abundance of 5%–10% of naturally occurring transcripts that exhibit features recognized by the NMD machinery (Kervestin and Jacobson 2012).
In vertebrates, stop codons trigger efficient NMD when they are located at least 50–55 nucleotides (nt) upstream of an exon–exon boundary, which is marked by the exon junction complex (EJC) (Nagy and Maquat 1998; Le Hir et al. 2000). Alternatively, stop codons can trigger NMD when other features of the mRNA (e.g., long 3′ untranslated regions [UTRs]) may prevent efficient translation termination by interfering with the interaction of the cytoplasmic poly(A)-binding protein (PABPC) with the eukaryotic release factors (eRFs; “faux 3′ UTR”) (Amrani et al. 2004; Behm-Ansmant et al. 2007; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008; Eberle et al. 2009). Aberrant translation termination at a PTC causes the assembly of a “surveillance complex” on the mRNA, which subsequently triggers mRNA degradation (Muhlemann and Lykke-Andersen 2010).
The surveillance complex consists of the evolutionarily conserved proteins UPF1, UPF2, and UPF3. Additional NMD factors include the kinase SMG1, which phosphorylates UPF1, and SMG5, SMG6, and SMG7, which trigger UPF1 dephosphorylation in metazoans (Kervestin and Jacobson 2012; Yamashita 2013).
Current models of NMD suggest that UPF1 and SMG1 are recruited by ribosomes prematurely terminating translation through interactions with eRF1 and eRF3 (Czaplinski et al. 1998; Kashima et al. 2006). UPF1 is phosphorylated by SMG1 when it interacts with UPF2 and/or UPF3, which are generally bound to downstream EJCs (Yamashita 2013). Phosphorylated UPF1 subsequently recruits SMG5, SMG6, and SMG7 to the mRNA (Anders et al. 2003; Chiu et al. 2003; Ohnishi et al. 2003; Okada-Katsuhata et al. 2012). SMG5, SMG6, and SMG7 are three related proteins that bind phosphorylated UPF1 through 14-3-3-like domains and trigger mRNA target degradation, UPF1 dephosphorylation, and the recycling of NMD factors to initiate new rounds of NMD (Ohnishi et al. 2003; Fukuhara et al. 2005; Franks et al. 2010; Okada-Katsuhata et al. 2012; Jonas et al. 2013).
The degradation of NMD targets is known to involve both endonucleolytic and exonucleolytic activities in vertebrates (Muhlemann and Lykke-Andersen 2010). Endonucleolytic degradation is catalyzed by SMG6, which cleaves the mRNA target in the vicinity of the PTC (Gatfield and Izaurralde 2004; Glavan et al. 2006; Huntzinger et al. 2008; Eberle et al. 2009). Exonucleolytic degradation is catalyzed by general mRNA decay factors that promote deadenylation followed by either 3′-to-5′ degradation or decapping and 5′-to-3′ decay (Chen and Shyu 2003; Lejeune et al. 2003; Couttet and Grange 2004; Yamashita et al. 2005). Moreover, deadenylation-independent decapping and 5′-to-3′ degradation also contribute to NMD (Chen and Shyu 2003; Lejeune et al. 2003; Couttet and Grange 2004).
Several lines of evidence indicate that decay enzymes are recruited to NMD targets via interactions with UPF1, SMG5, and/or SMG7. For example, human UPF1 interacts with the decapping enzyme DCP2 and the decapping activators DCP1 and PNRC2 (Lykke-Andersen 2002; Lejeune et al. 2003; Fenger-Grøn et al. 2005; Isken et al. 2008; Cho et al. 2009, 2013; Lai et al. 2012). Because PNRC2 binds directly to DCP1 and UPF1, it has been proposed to bridge the interaction between the surveillance and decapping complexes (Lai et al. 2012; Cho et al. 2013).
Additionally, both SMG5 and SMG7 induce the degradation of bound mRNA in tethering assays (Unterholzner and Izaurralde 2004; Cho et al. 2013). The degradative activity of SMG7 resides in its C-terminal proline-rich region (termed the PC region) and involves the decapping enzyme DCP2 and the 5′-to-3′ exonuclease XRN1 (Unterholzner and Izaurralde 2004). Although SMG5 heterodimerizes with SMG7, the degradative activity of SMG5 in tethering assays is independent of SMG7 and requires UPF1 and PNRC2 instead (Cho et al. 2013). This observation led to the hypothesis that SMG5 acts in NMD independently of SMG7 by interacting with PNRC2 and that the SMG5–PNRC2 interaction dominates over the SMG5–SMG7 interaction (Cho et al. 2013).
Despite the wealth of available information, key questions regarding NMD target degradation remain unanswered. In particular, the identity of the decay factors that are directly recruited to NMD targets remains unknown, along with whether this recruitment is mediated by UPF1, SMG5, and/or SMG7. Moreover, it remains unclear whether the interactions of UPF1 with DCP2 and DCP1 are direct or mediated by PNRC2 and to what extent these interactions contribute to NMD. Similarly, whether the SMG7 PC region interacts directly with decay factors and the relevance of the PC region for NMD have not been addressed. Finally, little is known about the mechanism by which tethered SMG5 triggers mRNA degradation, and it remains unclear whether SMG5 functions in two alternative complexes containing either PNRC2 or SMG7.
To address these questions, we investigated the mechanism of NMD target degradation and the interplay between SMG5, SMG6, and SMG7 in human cells. Here, we show that the SMG7 PC region directly binds POP2, a catalytic subunit of the CCR4–NOT deadenylase complex, and thereby promotes deadenylation and subsequent decapping of NMD targets. The SMG7 PC region is required for NMD in cells depleted of SMG6, indicating that SMG7 and SMG6 act redundantly to promote NMD target degradation. Furthermore, our results do not confirm a role for the SMG5–PNRC2 interaction in NMD but rather indicate that SMG5–SMG7 heterodimerization is critical for NMD. Finally, we show that DCP2 and PNRC2 interact independently with UPF1, perhaps facilitating the decapping of NMD targets. Our data reveal that the surveillance complex has multiple and redundant activities to ensure robust target degradation: It induces endonucleolytic cleavage by SMG6, recruits decapping factors through UPF1, and promotes deadenylation-dependent decay through the direct recruitment of the CCR4–NOT complex by SMG7.
Results
SMG5 requires interaction with SMG7 to function in NMD
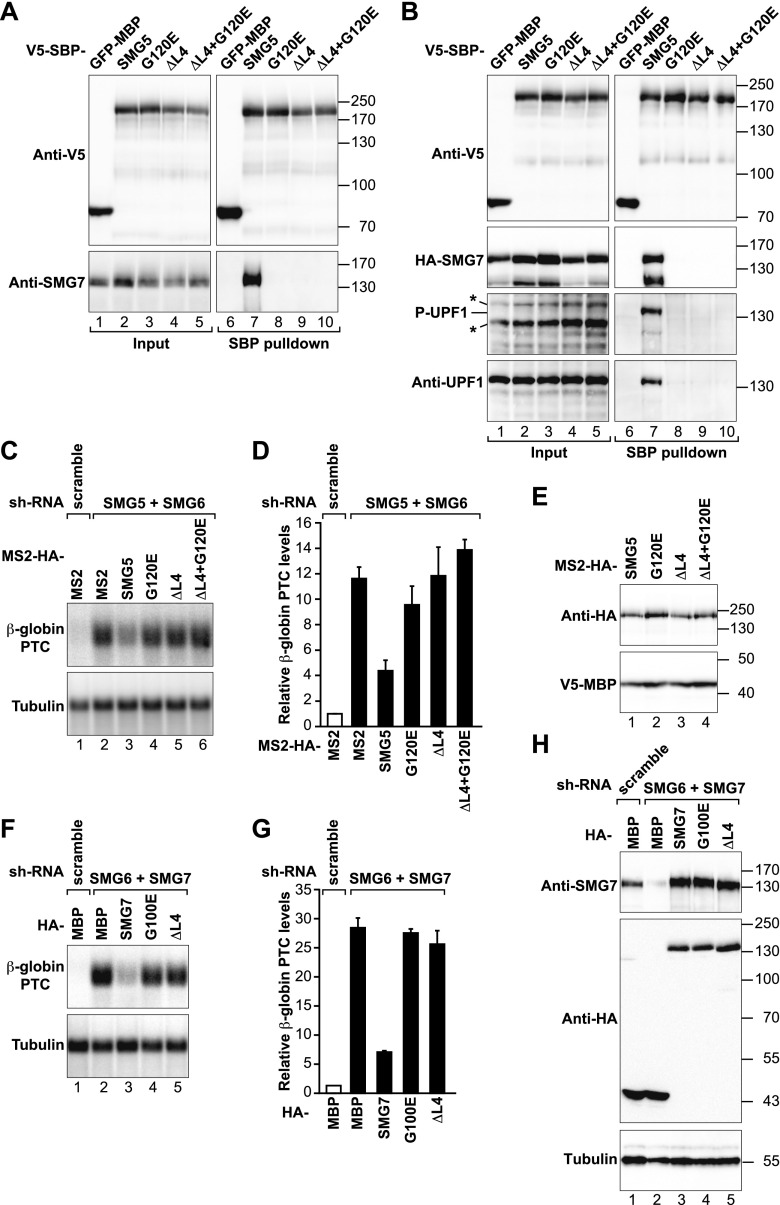
In our previous work, we demonstrated that a SMG5 mutant that does not heterodimerize with SMG7 fails to rescue NMD in human cells depleted of endogenous SMG5, indicating that SMG5 requires interaction with SMG7 to act in NMD (Jonas et al. 2013). In contrast, Cho et al. (2013) reported that SMG5 can function with PNRC2 independently of SMG7. To investigate whether monomeric SMG5 plays a role in NMD, we generated additional SMG5 mutants to disrupt the interaction with SMG7. Based on the structure of the SMG5–SMG7 heterodimer (Jonas et al. 2013), we deleted a SMG5 loop between helices α4 and α5 (loop L4). A symmetrical loop in SMG7 is disordered in the structure of monomeric SMG7 and folds onto the SMG5 surface upon binding, contributing several residues to the heterodimer interface (Supplemental Fig. S1; Fukuhara et al. 2005; Jonas et al. 2013). Therefore, deletion of loop L4 (ΔL4) is predicted to disrupt the interaction between SMG5 and SMG7 without affecting the fold of the SMG5 N-terminal domain. We also combined this deletion with the previously described G120E mutation (Jonas et al. 2013). The SMG5 residue G120 is at the center of the interface with SMG7 and allows for a close packing of the two proteins in the heterodimer (Supplemental Fig. S1; Jonas et al. 2013).
V5-SBP (streptavidin-binding peptide)-tagged wild-type or mutant SMG5 proteins were expressed in human HEK293T cells and examined for their ability to interact with SMG7. Consistent with previous structural studies, the mutations and deletions in SMG5 abolished binding to either endogenous or HA-tagged SMG7 (Fig. 1A,B). The mutations also abolished the interaction with endogenous and phosphorylated UPF1 (Fig. 1B), as previously reported (Jonas et al. 2013). However, the interaction with overexpressed UPF1 was not affected (Supplemental Fig. S2A,B), consistent with the observation that monomeric SMG5 interacts with UPF1 when the two proteins are overexpressed (∼100-fold) in human cells (Ohnishi et al. 2003; Jonas et al. 2013).
Figure 1.
SMG5–SMG7 heterodimerization is required for NMD. (A) Interaction of SMG5-V5-SBP-MBP (wild type or mutants) with endogenous SMG7 in HEK293T cells. A V5-SBP-tagged GFP-MBP fusion served as a negative control. Inputs (1%) and bound fractions (5%) were analyzed by Western blotting. Samples were treated with RNase A before the pull-down. (B) Interaction of SMG5-V5-SBP-MBP (wild type or mutants) with endogenous and phosphorylated UPF1 (P-UPF1) and HA-SMG7 in cell lysates treated with RNase A. Inputs (1%) and bound fractions (2% for the SBP and HA-tagged proteins and 30% for UPF1) were analyzed by Western blotting. Asterisks indicate phosphorylated proteins (distinct from UPF1) recognized by the phospho-(Ser/Thr) ATM/ATR substrate antibody in input samples. The identity of phosphorylated UPF1 in the pull-down was confirmed by reprobing the membrane using anti-UPF1 antibodies. (C–E) HeLa cell lines expressing the β-globin NMD reporter (containing a PTC) were transfected with plasmids expressing the indicated shRNAs. Plasmids expressing shRNA-resistant versions of MS2-HA-SMG5 (wild type or mutants) were included in the transfection mixtures as indicated. MS2-HA served as a negative control. (C) The levels of the PTC-containing β-globin reporter were analyzed by Northern blotting, normalized to those of endogenous β-tubulin mRNA, and set to 1 in control cells (i.e., cells expressing MS2-HA and treated with a scrambled shRNA). D shows mean values ± standard deviations obtained in three independent experiments. (E) Expression of MS2-HA-SMG5 (wild type or mutants) in the complementation assay. V5-SBP-MBP served as a transfection control. (F,G) A complementation assay as described in C and D was performed in cells codepleted of SMG7 and SMG6 and transfected with plasmids expressing shRNA-resistant HA-SMG7 (wild type or mutants). (H) Expression of HA-SMG7 (wild type or mutants) in the complementation assay shown in F and G.
Because the mutations strongly impair SMG5–SMG7 heterodimerization, we next examined the effect of these mutations in NMD using a complementation assay in human cells. Briefly, an shRNA targeting the ORF of the smg5 mRNA was used to deplete endogenous SMG5 in cell lines constitutively expressing a well-characterized NMD reporter based on the β-globin gene (Thermann et al. 1998). Because SMG6 can partially compensate for the absence of SMG5 (Jonas et al. 2013; Metze et al. 2013), we depleted SMG6 in combination with SMG5. SMG5 and SMG6 codepletion resulted in a 12-fold increase in the level of β-globin PTC mRNA (Fig. 1C,D). This codepletion did not affect the expression of the wild-type β-globin reporter (Supplemental Fig. S2C). The SMG5 and SMG6 protein levels were reduced to ∼25% of their control levels by the shRNA (Supplemental Fig. S2D,E), whereas the SMG7 levels remained unchanged (Supplemental Fig. S2F).
In codepleted cells, we then examined the ability of shRNA-resistant versions of SMG5 to rescue NMD. If SMG5 acts independently of SMG7, mutations that disrupt its interaction with SMG7 should not compromise its ability to rescue NMD. However, as shown in Figure 1, C and D, the aforementioned SMG5 mutants were either strongly impaired or inactive in rescuing NMD, whereas wild-type SMG5 did rescue NMD (although not completely because SMG6 is codepleted) (Fig. 1C,D). The levels of the wild-type reporter remained unchanged (Supplemental Fig. S2C). All SMG5 proteins were expressed at similar levels (Fig. 1E). These levels were approximately twofold higher than the levels of endogenous SMG5 in control cells (Supplemental Fig. S2G).
To further validate our conclusions, we generated the reciprocal mutations in SMG7. We observed that a SMG7 protein lacking loop L4 did not interact with SMG5 and failed to complement NMD in cells codepleted of SMG6 and SMG7 (Fig. 1F,G; Supplemental Fig. S3A). As a control, a previously described G100E substitution (corresponding to the SMG5 G120E mutant) abolished SMG5 binding and abrogated SMG7's ability to rescue NMD in complementation assays, as reported before (Fig. 1F,G; Supplemental Fig. S3A; Jonas et al. 2013). Importantly, deletion of loop L4 or the G100E substitution did not prevent SMG7 from interacting with UPF1 (Supplemental Fig. S3B), indicating that these mutations do not disrupt the protein fold. The SMG6 and SMG7 protein levels were reduced to ∼25% and 10% of their control levels in depleted cells (Supplemental Fig. S3C,D), whereas the SMG5 levels remained unchanged (Supplemental Fig. S3E). SMG7 wild-type and mutants were expressed at levels comparable with endogenous SMG7 (Fig. 1H). We conclude that SMG5 and SMG7 function as a complex in NMD.
SMG5 does not stably associate with PNRC2
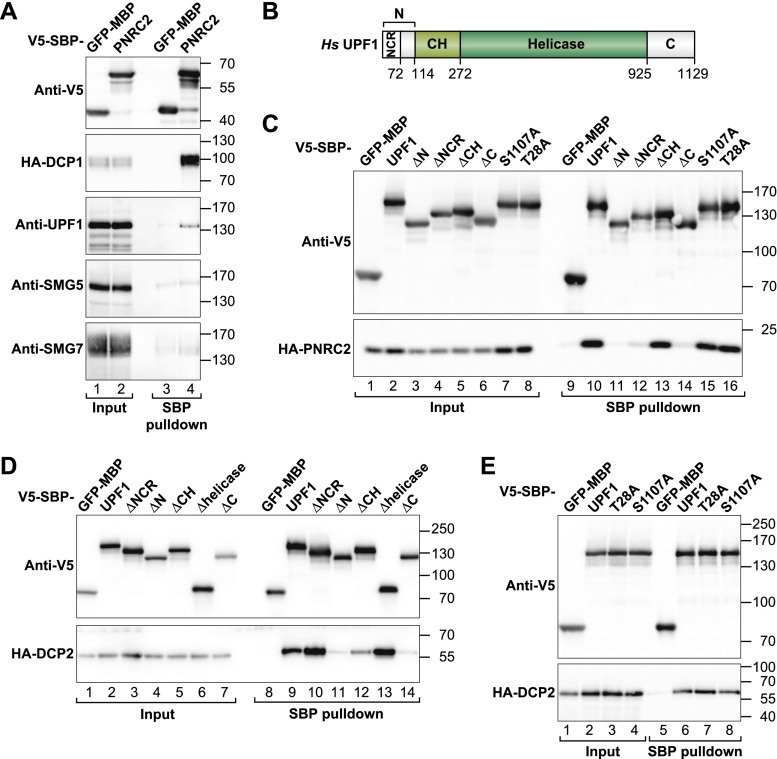
The observation that SMG5 requires interaction with SMG7 to function in NMD contrasts with the suggestion that SMG5 functions with PNRC2 and independently of SMG7 (Cho et al. 2013). One possible explanation for these results is that the mutations in SMG5 that disrupt its binding to SMG7 also disrupt PNRC2 binding. Because the SMG5 residues involved in the interaction with PNRC2 have not been defined, we tested the interaction of wild-type or mutant SMG5 with PNRC2. In contrast to a previous study (Cho et al. 2013), we could not detect an interaction between V5-SBP-SMG5 and HA-PNRC2 in SBP pull-down assays (data not shown).
To clarify this discrepancy, we next tested the interaction of PNRC2 with endogenous SMG5, SMG7, and UPF1. V5-SBP-tagged PNRC2 did not interact with endogenous SMG5 or SMG7 at detectable levels (Fig. 2A, lane 4). Under the same conditions, PNRC2 exhibited a weak (above background) association with endogenous UPF1 and efficiently pulled down HA-DCP1, which was included as a positive control (Figure 2A, lane 4; Lai et al. 2012). Note that these pull-downs were performed in the presence of okadaic acid, which inhibits UPF1 dephosphorylation and has been reported to enhance its binding to PNRC2 and DCP1 (Cho et al. 2009; Lai et al. 2012). From these results, we conclude that SMG5 may interact with PNRC2 only transiently or indirectly, possibly through UPF1.
Figure 2.
UPF1 interacts with decapping factors. (A) V5-SBP-MBP-tagged PNRC2 does not interact with endogenous SMG5 or SMG7 but binds HA-DCP1 and weakly interacts with endogenous UPF1. V5-SBP-MBP served as a negative control. Inputs (1%) and bound fractions (8% for V5-tagged proteins, 15% for HA-tagged proteins, and 35% for UPF1, SMG5, and SMG7) were analyzed by Western blotting. (B) The domain architecture of UPF1. (NCR) N-terminal conserved region; (N) N-terminal tail; (C) C-terminal tail. The globular cysteine–histidine (CH) and helicase domains are indicated. (C–E) Interaction of V5-SBP-UPF1 (wild type or mutants) with HA-PNRC2 (C) and HA-DCP2 (D,E). V5-SBP-GFP-MBP served as a negative control.
UPF1 interacts with DCP2 in a PNRC2-independent manner
UPF1 interacts with PNRC2 through its N-terminal and C-terminal unstructured tails (Cho et al. 2009). UPF1 also interacts with DCP2 and DCP1 (Lykke-Andersen 2002; Fenger-Grøn et al. 2005; Isken et al. 2008), but the domains involved in these interactions have not been defined, and it remains unclear whether these interactions are mediated by PNRC2. Therefore, we tested which domains of UPF1 are required for DCP2 binding.
The UPF1 protein consists of an N-terminal cysteine–histidine (CH) regulatory domain, which provides a binding site for UPF2, and a central helicase domain (Fig. 2B; Cheng et al. 2007; Clerici et al. 2009). These structured domains of UPF1 are flanked by N-terminal and C-terminal tails (∼100 residues) containing several serine/threonine–glutamine (S/T-Q) motifs that are phosphorylated by SMG1 and that serve as binding sites for SMG6 and the SMG5–SMG7 complex (Ohnishi et al. 2003; Okada-Katsuhata et al. 2011). SMG6 and SMG7 have been shown to bind phosphorylated T28 and S1096 (corresponding to S1107 in UPF1 isoform 1), respectively (Okada-Katsuhata et al. 2012).
We observed that both the N-terminal and C-terminal tails of UPF1 are required for PNRC2 binding (Fig. 2C, ΔN and ΔC), in agreement with Cho et al. (2009). Interestingly, deletion of the N-terminal conserved region (ΔNCR) (Ohnishi et al. 2003) abrogated PNRC2 binding (Fig. 2C, lane 12). In contrast, the CH and helicase domains were dispensable (Fig. 2C; data not shown). Additionally, alanine substitutions of residues T28 and S1107 did not prevent PNRC2 binding (Fig. 2C, lanes 15,16), suggesting that UPF1 interacts with PNRC2 independently of SMG6 and SMG7.
Next, we tested the aforementioned UPF1 deletion mutants for their ability to interact with DCP2. Similar to the results obtained with PNRC2, deletion of the UPF1 N-terminal or C-terminal tails abolished DCP2 binding (Fig. 2D, lanes 11,14). Importantly, deletion of the NCR region was ineffectual despite the fact that this deletion abrogated PNRC2 binding (Fig. 2, C vs. D ). This result indicates that DCP2 interacts with UPF1 independently of PNRC2. Additionally, deletion of the helicase domain as well as the T28A and S1107A mutations was ineffectual (Figs. 2D,E). We conclude that UPF1 provides multiple and independent binding sites for decapping factors.
SMG7 but not SMG5 triggers degradation of bound mRNAs independently of additional NMD factors
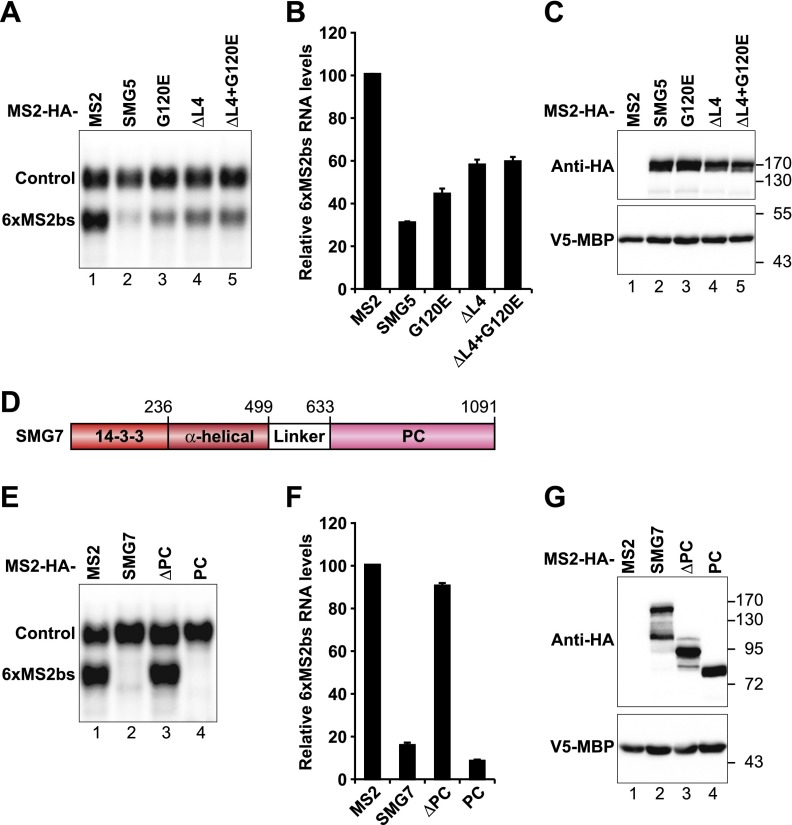
Using an MS2-based tethering system, Cho et al. (2013) reported that SMG5 is active in tethering assays and that this activity requires UPF1 and PNRC2 but is independent of SMG7. In contrast, using a λN-based reporter assay, we observed that tethered SMG5 only promotes mRNA decay when it is coexpressed with SMG7 (Unterholzner and Izaurralde 2004; Jonas et al. 2013). To address this apparent discrepancy, we compared the activity of tethered SMG5 and SMG7 using the MS2 tethering system (Lykke Andersen et al. 2000).
Tethered MS2-SMG5 promoted the degradation of a β-globin reporter containing six copies of the high-affinity binding site for the MS2 viral coat protein in its 3′ UTR (6xMS2bs) (Fig. 3A–C), as reported by Cho et al. (2013). SMG5 activity was partially dependent on SMG7 because the aforementioned mutations that prevent binding to SMG7 reduced, but did not abolish, SMG5 activity in tethering assays, although the mutant proteins were expressed at levels similar to wild-type levels (Fig. 3A–C). The residual activity of the SMG5 mutants is most likely due to their interactions with UPF1 because UPF1 depletion suppresses the activity of tethered MS2-SMG5 (Cho et al. 2013). The reason for the partial different results obtained with the MS2 and λN reporters was not further investigated.
Figure 3.
SMG5 and SMG7 degrade bound mRNAs. (A–C,E–G) Human HEK293T cells were transfected with a mixture of three plasmids: one expressing β-globin-6xMS2bs mRNA, another expressing MS2-HA or the indicated MS2-HA-tagged proteins, and a third expressing a transfection control containing the β-globin gene fused to the GAPDH 3′ UTR but lacking MS2-binding sites (β-globin-GAP; Control). β-Globin-6xMS2bs mRNA levels were normalized to those of the control mRNA. The normalized values of the β-globin-6xMS2bs mRNA levels were set to 100 in the presence of MS2-HA. Mean values ± standard deviations from three independent experiments are shown in B and F. A and E show Northern blots of representative RNA samples. (C,G) Western blot analysis showing equivalent expression of the MS2-HA-tagged proteins used in the corresponding tethering assays. (D) The domain architecture of SMG7 consists of an N-terminal 14-3-3-like domain, a middle α-helical domain, and a C-terminal PC region.
SMG7 consists of an N-terminal 14-3-3-like domain, a middle α-helical domain, and a PC region (Fig 3D). Using the MS2 system, we observed that the SMG7 PC region is both necessary and sufficient to promote the degradation of the mRNA reporter (Fig. 3E–G). In contrast, the SMG7 N-terminal region (denoted ΔPC mutant) was inactive, consistent with our previous studies (Unterholzner and Izaurralde 2004; Jonas et al. 2013). Thus, although the SMG7 N-terminal region interacts with both SMG5 and UPF1, this region does not lead to mRNA degradation in isolation. In contrast, the PC region, which does not interact with SMG5 or UPF1 (Anders et al. 2003; Jonas et al. 2013), is sufficient to cause degradation. Our results, together with the observation that SMG7 activity in tethering assays is not affected by UPF1 or PNRC2 depletion (Cho et al. 2013), indicate that SMG7 causes mRNA degradation independently of UPF1, SMG5, and PNRC2. We conclude that SMG7 recruits decay enzymes independently of any other NMD factor, whereas SMG5 degradative activity depends on SMG7 and UPF1.
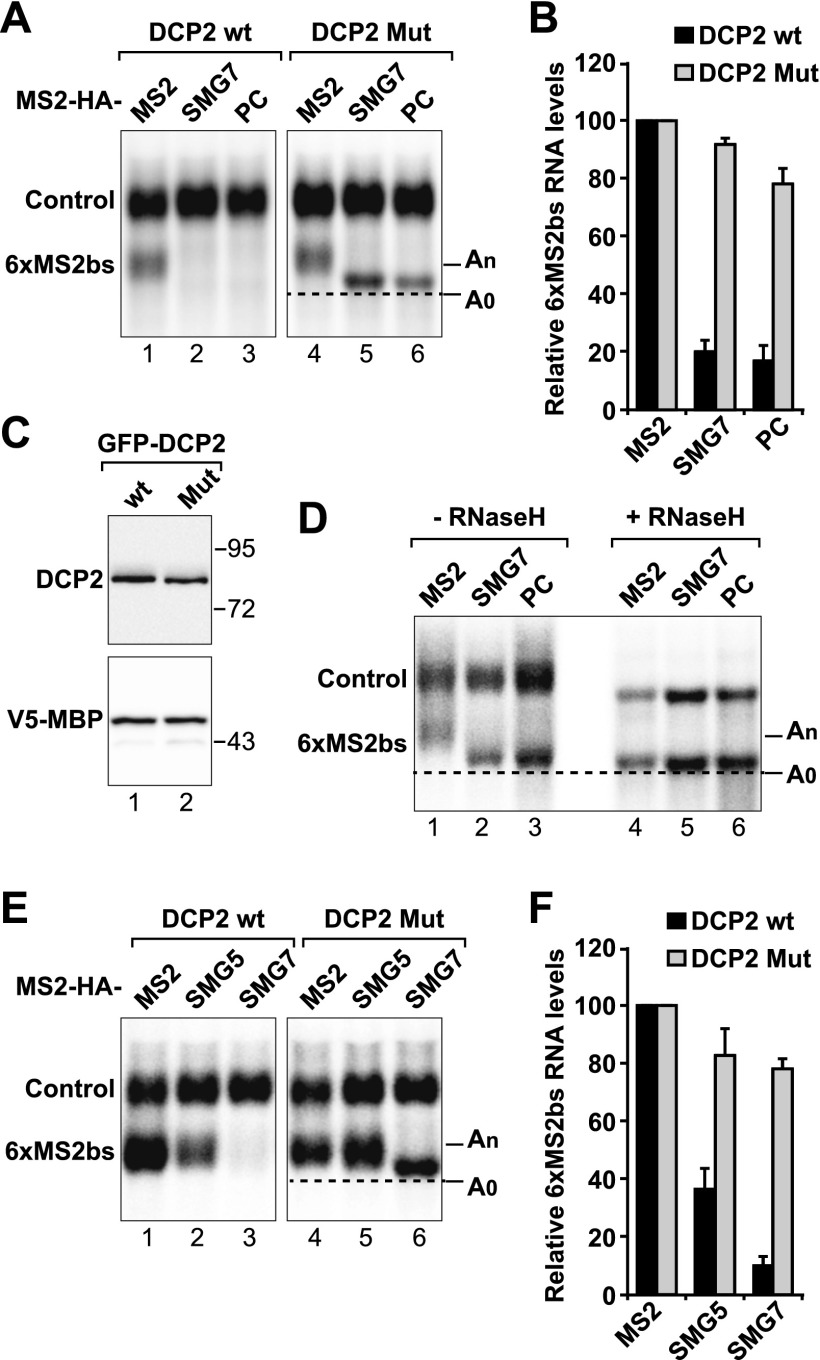
The SMG7 PC region promotes deadenylation-dependent decapping
SMG7-mediated degradation is inhibited in cells depleted of the decapping enzyme DCP2 or the 5′-to-3′ exonuclease XRN1 (Unterholzner and Izaurralde 2004). However, it remains unknown whether decapping is dependent on prior deadenylation and whether SMG5 uses a similar mechanism to degrade bound mRNAs. If deadenylation precedes decapping and 5′-to-3′ mRNA degradation, then deadenylated mRNA decay intermediates are expected to accumulate in cells in which decapping is inhibited. Consistent with this expectation, degradation of the β-globin-6xMS2bs reporter by full-length SMG7 or the PC fragment was inhibited in cells overexpressing a catalytically inactive DCP2 mutant (DCP2 Mut; E148Q) (Fig. 4A–C). The reporter accumulated in a fast-migrating form, corresponding to the deadenylated decay intermediate (A0). Indeed, the mobility of the fast-migrating form did not change after oligo(dT)-directed RNase H cleavage (Fig. 4D). Thus, the PC region of SMG7 elicits mRNA decay by triggering deadenylation and then decapping.
Figure 4.
SMG5 and SMG7 use distinct mechanisms to degrade bound mRNA in tethering assays. (A–F) Tethering assays using the β-globin-6xMS2bs reporter were performed as described in Figure 3, with the exception that plasmids expressing wild-type DCP2 or a catalytically inactive mutant (Mut; E148Q) were included in the transfection mixtures as indicated. A and E show Northern blots of representative RNA samples. β-Globin-6xMS2bs mRNA levels were normalized to those of the control β-globin-GAP mRNA. These normalized values were set to 100 in the presence of MS2-HA. Mean values ± standard deviations from three independent experiments are shown in B and F. Black and gray bars show the values obtained in cells expressing DCP2 wild type (wt) and mutants (Mut), respectively. (C) Western blot analysis showing equivalent expression of the DCP2 proteins. (D) Samples corresponding to lanes 4–6 of A were treated with RNase H in the presence of oligo(dT).
Notably, although overexpression of the DCP2 inactive mutant also inhibited SMG5-mediated decay in tethering assays, the reporter accumulated in the polyadenylated form (Fig. 4E,F). These results indicate that tethered SMG5 triggers decay by a mechanism other than SMG7, as SMG5-mediated decay involves decapping in the absence of deadenylation.
In summary, our results confirm that the SMG5 activity in tethering assays can be independent of SMG7, although in complementation assays, SMG5 strictly depends on SMG7. The simplest hypothesis that explains these observations is that in complementation assays, SMG5 requires interaction with SMG7 to be recruited to NMD targets, a step that is bypassed in tethering assays (see the Discussion).
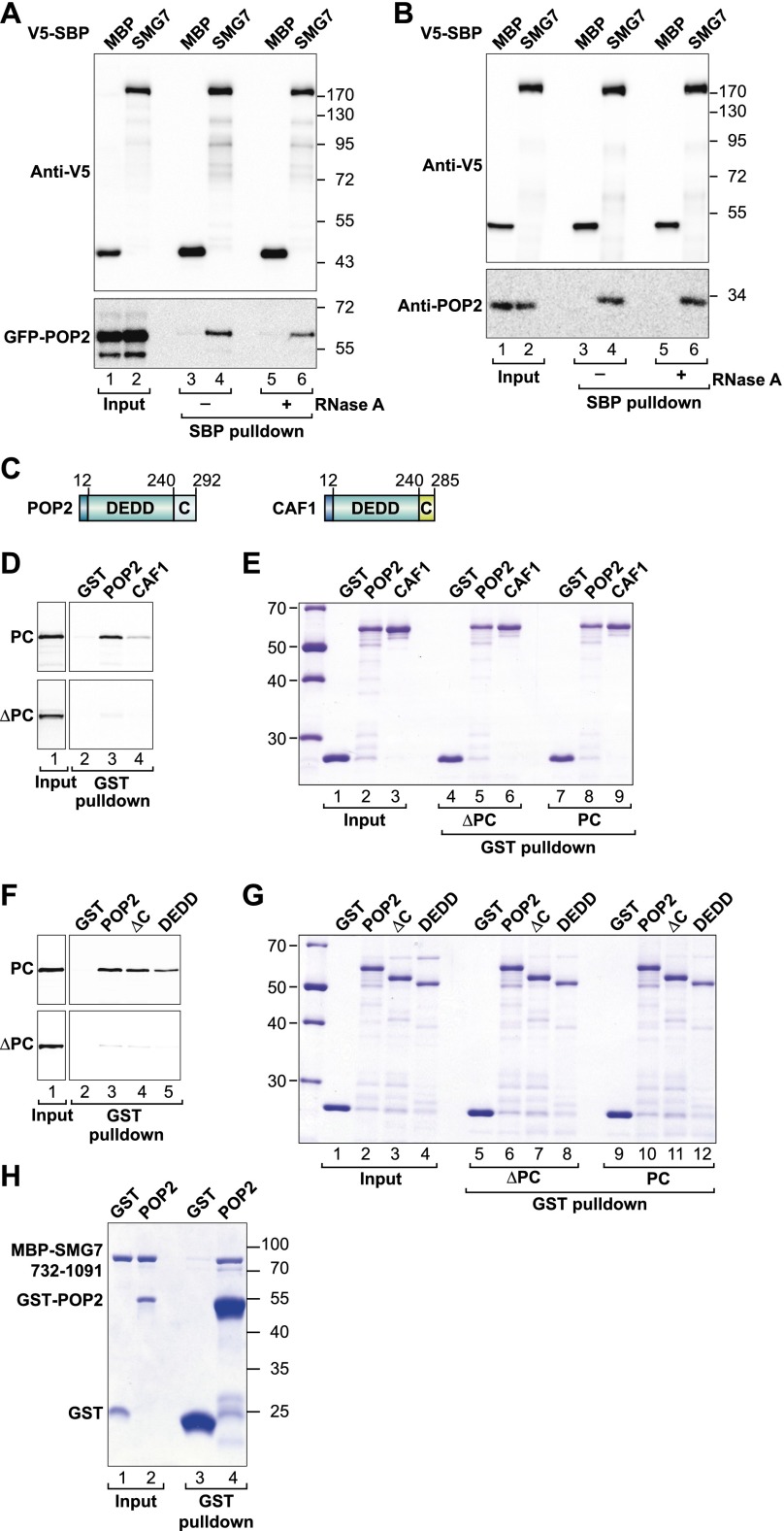
The SMG7 PC region interacts with POP2
Having established that the PC region of SMG7 promotes deadenylation-dependent decapping, we next sought to identify potential interaction partners. To achieve this, we performed tandem affinity purification (TAP) (Rigaut et al. 1999) in human HEK293T cells using a TAP tag consisting of protein A, a PreScission protease cleavage site, and the SBP. Remarkably, the TAP-tagged SMG7 PC region copurified with the entire CCR4–NOT deadenylase complex, which consists of 10 subunits (Supplemental Table S1). No other decay factors were identified as potential binding partners (Supplemental Table S1).
To validate the interaction between SMG7 and the CCR4–NOT complex and identify the subunits of this complex that may interact directly with the SMG7 PC region, we expressed the individual subunits together with SBP-tagged SMG7 in HEK293T cells and performed pull-down assays. The core of the human CCR4–NOT complex consists of six subunits (CNOT1, CNOT2, CNOT3, CNOT9, CNOT10, and CNOT11) and two catalytically active subunits (CCR4 and CAF1). In human cells, there are two alternative paralogs for each catalytic subunit: CCR4a or CCR4b (also known as CNOT6 or CNOT6-L) and CAF1 or POP2 (also known as CNOT7 or CNOT8) (Lau et al. 2009).
Remarkably, only POP2 (CNOT8) interacted with SMG7 in pull-down assays (Fig. 5A; Supplemental Fig. S4). This result is in contrast to the TAP tag selection, in which the endogenous CCR4–NOT complex associates with SMG7. The simplest explanation for these results is that in SBP pull-down assays, the isolated subunits are overexpressed and may not be efficiently incorporated into endogenous complexes. Accordingly, SMG7 did not interact with the human-specific subunit TAB182 (Supplemental Fig. S4; Lau et al. 2009). These results indicate that the SBP pull-down assays using transiently expressed proteins detected binary interactions, although the SBP-tagged SMG7 was expressed at levels comparable with endogenous SMG7 (Supplemental Fig. S5A). Furthermore, SBP-tagged SMG7 did not interact with the subunits of the PAN2–PAN3 deadenylase complex (Supplemental Fig. S4) or with PNRC2 (Fig. 2A; Supplemental Table S1), underscoring the specificity of the interaction with POP2. Importantly, the interaction of SMG7 with POP2 was insensitive to RNase A treatment, suggesting that this interaction is not mediated by RNA (Fig. 5A).
Figure 5.
SMG7 interacts directly with POP2. (A,B) Interaction of SMG7-V5-SBP-MBP with GFP-POP2 (A) or endogenous POP2/CAF1 (B) in the absence or presence of RNase A. A V5-SBP-MBP served as a negative control. (C) The domain architectures of POP2 and CAF1. POP2 and CAF1 contain a central catalytic domain of the DEDD family exhibiting 84% identity. The C-terminal extensions are more divergent in sequence and length. (D–G) GST-tagged CAF1 or POP2 (wild type or mutants) or GST was used to pull down [35S]-methionine-labeled in vitro translated SMG7 PC and ΔPC fragments. (D,F) Inputs (1%) and bound fractions (16%) were analyzed by 10% SDS-PAGE followed by fluorography. The corresponding Coomassie-stained gels are shown in E and G. (H) GST, GST-POP2, and MBP-SMG7 were expressed in E. coli and purified. The purified proteins were mixed in a 1:1 ratio (inputs), pulled down using glutathione agarose beads, and analyzed by SDS-PAGE followed by Coomassie staining.
To further validate the SMG7–POP2 interaction, we pulled down SMG7 and identified endogenous CAF1/POP2 in the bound fractions using an antibody that does not discriminate between POP2 and CAF1 (Fig. 5B). Collectively, these results indicate that SMG7 associates with the CCR4–NOT complex through interactions with POP2.
SMG7 discriminates between the catalytic domains of POP2 and CAF1
POP2 and CAF1 are one-domain proteins that adopt an RNase D-like fold (Daugeron et al. 2001) and exhibit 74% overall identity with each other (Fig. 5C). The main divergence between these two proteins is in their short C-terminal extensions. Given the high identity between these paralogs, it is surprising that SMG7 interacts with POP2 but not with CAF1. To investigate the basis for this selectivity, we performed GST (glutathione S transferase) pull-down assays with recombinant POP2 and CAF1 proteins expressed in Escherichia coli as GST fusions. The SMG7 PC fragment was translated in vitro in wheat germ extracts to minimize potential indirect interactions mediated by other proteins. We observed that the PC fragment preferentially interacted with GST-POP2 and exhibited a much weaker interaction with GST-CAF1 (Fig. 5D,E). The interaction was specific because the PC fragment was not pulled down with GST alone (Fig. 5D,E). Furthermore, a SMG7 mutant lacking the PC region (ΔPC) did not interact with any of the proteins tested at detectable levels (Fig. 5D,E).
We next investigated whether binding to the SMG7 PC region required the POP2 C-terminal extension, which is the sequence exhibiting the greatest divergence between POP2 and CAF1. Unexpectedly, the PC region interacted with the POP2 catalytic domain and did not require N-terminal or C-terminal extensions (Fig. 5F,G). We conclude that SMG7 has an exquisite preference for the catalytic domain of POP2 despite the fact that this domain is 84% identical to the equivalent domain in CAF1.
The SMG7 PC region interacts directly with POP2
To investigate whether the POP2–SMG7 interaction is direct, we expressed and purified the SMG7 PC fragment in E. coli fused to maltose-binding protein (MBP). GST-POP2 pulled down MBP-SMG7-PC, demonstrating that the interaction is indeed direct (Fig. 5H, lane 4).
The catalytic domains of CAF1/POP2 interact with CNOT1, CCR4, and the anti-proliferative protein TOB simultaneously, with nonoverlapping binding surfaces. Structural studies identified critical surface residues that disrupt CAF1/POP2 binding to CNOT1, CCR4, and TOB when mutated (Supplemental Table S2). Therefore, we tested these mutants for binding to SMG7. We observed that POP2 mutants that do not bind CNOT1, CCR4a,b, or TOB still interacted with SMG7 (Supplemental Fig. S5B,C). Together, these results indicate that the binding surface of SMG7 on POP2 does not overlap with the previously characterized binding surfaces for CNOT1, CCR4a,b, or TOB. Accordingly, SMG7 copurifies with the entire CCR4–NOT complex and thus does not compete with CNOT1 and CCR4 for binding to POP2.
A POP2 catalytically inactive mutant suppresses SMG7-mediated mRNA decay and NMD
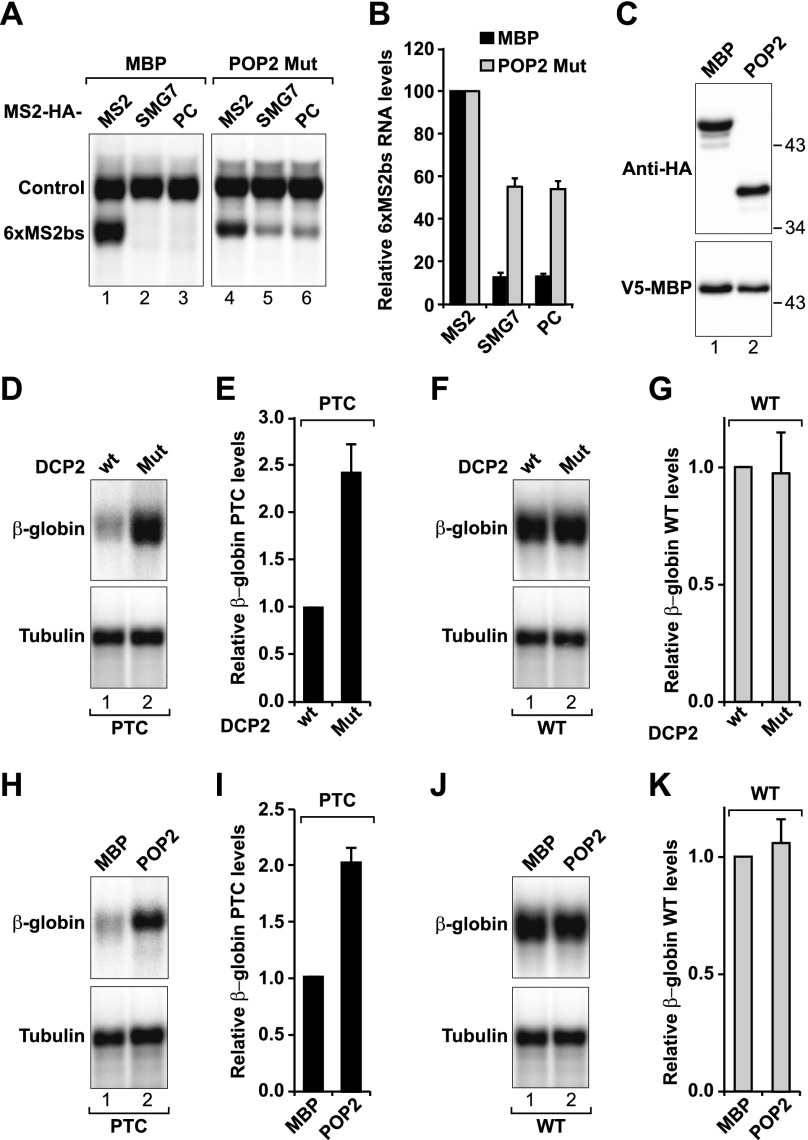
To further establish the role of POP2 in SMG7-mediated decay, we used a tethering assay and overexpressed a catalytically inactive POP2 mutant (POP2 Mut; D40A,E42A). The POP2 mutant inhibited the degradation of the reporter by SMG7 or the PC region (Fig. 6A–C). In contrast to the results obtained with the DCP2 mutant (Fig. 4A), the POP2 mutant led to the accumulation of the mRNA reporter in the polyadenylated form, as expected (Fig. 6A).
Figure 6.
A POP2 catalytically inactive mutant inhibits SMG7-mediated decay and NMD in a dominant-negative manner. (A–C) Tethering assays using the β-globin-6xMS2bs reporter were performed as described in Figure 3, with the exception that plasmids expressing HA-MBP or a HA-POP2 catalytically inactive mutant (Mut; D40A,E42A) were included in the transfection mixtures as indicated. A presents a Northern blot of representative RNA samples. The β-globin-6xMS2bs mRNA levels were normalized to those of the control β-globin-GAP mRNA. These normalized values were set to 100 in the presence of MS2-HA. Mean values ± standard deviations of three independent experiments are shown in B. Black and gray bars represent the values obtained in cells expressing MBP and the POP2 mutant, respectively. (C) Western blot analysis demonstrating equivalent expression of the transfected proteins. (D–K) HeLa cells constitutively expressing a wild-type (WT) or PTC-containing β-globin reporter were transfected with plasmids expressing DCP2 (wild type or mutant) (D–G) or MBP and POP2 mutant (H–K). D, F, H, and J show Northern blots of representative RNA samples. The levels of wild-type or PTC-containing β-globin reporters were normalized to those of β-tubulin mRNA and set to 1 in cells expressing DCP2 wild type (E,G) or MBP (I,K). Mean values ± standard deviations of three independent experiments are shown in E, G, I, and K.
To investigate the contribution of the 5′-to-3′ mRNA decay pathway and POP2 to NMD target degradation, we tested whether the catalytically inactive DCP2 and POP2 mutants suppressed NMD. We observed that the levels of the PTC-containing reporter increased 2.5-fold and twofold in cells overexpressing DCP2 and POP2 mutants, respectively (Fig. 6D–K). In the presence of the DCP2 mutant, the reporter migrated as a broad band (Fig. 6D, lane 2), suggesting that a fraction of the mRNA is deadenylated. In contrast, in cells expressing the POP2 mutant, it migrated as a sharp band (Fig. 6H, lane 2), suggesting that it is fully polyadenylated in this case. The levels of the wild-type reporter did not change (Fig. 6F,G,J,K). The twofold increase observed in PTC mRNA levels when deadenylation or decapping is blocked is in the same range as the effects observed in cells depleted of SMG5 or SMG7 (see Fig. 7; Jonas et al. 2013).
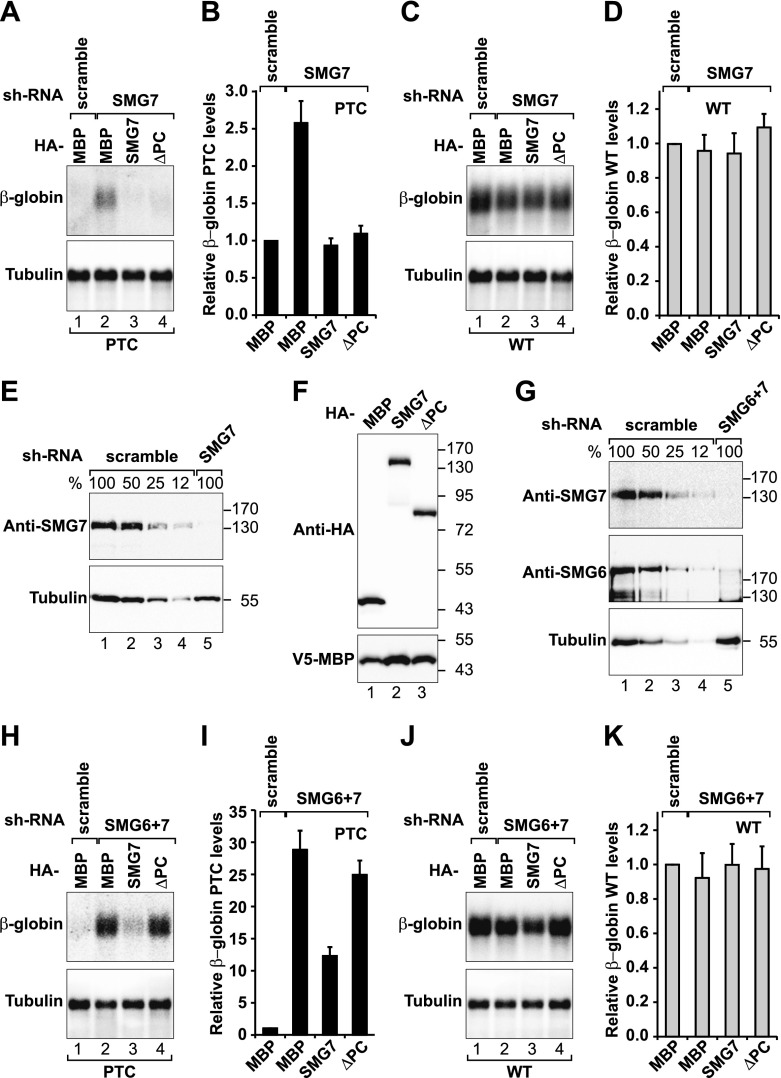
Figure 7.
The SMG7 PC region is required for NMD in the absence of SMG6. (A–F) HeLa cell lines expressing the β-globin reporter (wild type [WT] or with PTC) were transfected with plasmids expressing the indicated shRNAs and shRNA-resistant forms of SMG7 full-length or ΔPC. (A,C) The levels of the PTC-containing or wild-type β-globin reporters were analyzed by Northern blot, normalized to those of endogenous β-tubulin mRNA, and set to 1 in control cells (i.e., cells treated with a scrambled shRNA and expressing HA-MBP). B and D show the mean values ± standard deviations obtained in three independent experiments. (E) Western blot showing the efficiency of SMG7 depletion. (F) Expression levels of transiently expressed HA-tagged SMG7 proteins. (G–K) A complementation assay with plasmids expressing an shRNA-resistant version of HA-SMG7 (wild type or ΔPC) was performed in cells codepleted of SMG6 and SMG7. (G) Western blot showing the efficiency of SMG6 and SMG7 codepletion. Northern blot analyses of representative RNA samples are shown in H and J. The relative levels of PTC-containing or wild-type β-globin reporters in three independent experiments are shown in I and K.
The SMG7 PC region functions redundantly with SMG6
To investigate the functional relevance of the SMG7 PC region for NMD target degradation, we performed complementation assays, as described above. First, cells were depleted of endogenous SMG7 and reconstituted with shRNA-resistant versions of wild-type SMG7 or the SMG7-ΔPC mutant. SMG7 depletion caused a 2.5-fold increase in the PTC-containing β-globin reporter levels (Fig. 7A,B). Reintroduction of SMG7 restored NMD (Fig. 7A,B). Unexpectedly, we found that the SMG7-ΔPC mutant was also fully competent in restoring NMD (Fig. 7A,B). The expression levels of the corresponding wild-type β-globin mRNA were unaltered (Fig. 7C,D). Endogenous SMG7 protein levels were reduced below 12% by the shRNA expression (Fig. 7E). The shRNA-resistant versions of SMG7 wild-type and ΔPC were expressed at similar levels (Fig. 7F, lanes 2,3); these levels were comparable with the expression of endogenous SMG7 in control cells (Supplemental Fig. S5D). We conclude that the PC region of SMG7 is not required for the degradation of the β-globin PTC mRNA.
Because SMG6 can compensate, at least in part, for the lack of SMG7, we next repeated the complementation assay in cells codepleted of SMG6 and SMG7 (Fig. 7G). We observed that the codepletion of both proteins resulted in a synergistic inhibition of NMD, leading to a 30-fold increase in the β-globin PTC reporter levels, as previously reported (Luke et al. 2007; Jonas et al. 2013; Metze et al. 2013). This effect was prevented by the expression of a shRNA-resistant version of SMG7, which partially restored NMD (Fig. 7H,I). In contrast, the ability of the ΔPC mutant to rescue NMD was strongly impaired, indicating that the PC region of SMG7 is required for NMD in the absence of the SMG6 endonuclease. The expression levels of the wild-type β-globin mRNA remain unchanged (Fig. 7J,K).
Discussion
NMD targets are degraded by multiple mechanisms in vertebrates. These mechanisms include endonucleolytic cleavage by SMG6, deadenylation-dependent decapping, accelerated decapping of partially deadenylated mRNAs, and exosome-mediated degradation (Muhlemann and Lykke-Andersen 2010). Despite this wealth of information, the direct physical interactions that link the surveillance complex to the general mRNA decay machinery have not been described. In this study, we demonstrate that SMG7 directly recruits the CCR4–NOT deadenylase complex to NMD targets and promotes deadenylation-dependent decapping. We further show that this recruitment is mediated by direct interactions between the PC region of SMG7 and POP2, a catalytic subunit of the CCR4–NOT complex, the major cytoplasmic deadenylase complex in eukaryotes. The ability of SMG7 to discriminate between POP2 and CAF1, which are mutually exclusive subunits of the CCR4–NOT complex, provides an unprecedented example for how substrate specificity for alternative CCR4–NOT complexes can be achieved.
SMG5 requires heterodimerization with SMG7 to function in NMD
SMG5 and SMG7 heterodimerize through interactions mediated by their 14-3-3-like domains. Several lines of evidence indicate that they function as a complex in NMD (Anders et al. 2003; Ohnishi et al. 2003; Okada-Katsuhata et al. 2012; Jonas et al. 2013). First, they form highly stable heterodimers in vivo and in vitro (Jonas et al. 2013). Second, SMG5 or SMG7 mutants that do not interact with one another do not rescue NMD in cells depleted of SMG5 or SMG7, respectively (Fig. 1; Jonas et al. 2013). Furthermore, codepletion of SMG5 and SMG7 does not exacerbate the effects of the individual depletions, suggesting an epistatic interaction. In contrast, codepletion of SMG6 with either SMG5 or SMG7 results in a synergistic inhibition of NMD (Luke et al. 2007; Jonas et al. 2013; Metze et al. 2013), suggesting that the SMG5–SMG7 complex acts redundantly with SMG6 to promote the degradation of NMD targets.
In contrast to these results, which were obtained in complementation assays using a genuine NMD target, SMG5 mutants that do not interact with SMG7 can nevertheless trigger mRNA decay in tethering assays, although less efficiently than wild-type SMG5 (Fig. 1). SMG5 activity in tethering assays is suppressed by PNRC2 or UPF1 depletions (Cho et al. 2013). These observations were interpreted as evidence that SMG5 acts in NMD independently of SMG7 through its interaction with PNRC2 and that the SMG5–PNRC2 interaction is dominant over the SMG5–SMG7 interaction (Cho et al. 2013). Our study challenges this view and shows that although MS2-based tethering assays bypass the requirement for SMG5–SMG7 interaction, in complementation assays, SMG5 requires interaction with SMG7 to function in NMD.
Although we cannot rule out the possibility that SMG5 acts independently of SMG7 under some circumstances or for some specific targets, we found that SMG5 does not form a stable complex with PNRC2. Thus, the observation that PNRC2 depletion suppresses SMG5-mediated degradation in tethering assays could simply be explained as a general inhibition of decapping without invoking the formation of a complex with SMG5. Similarly, a catalytically inactive DCP2 mutant prevents SMG5-mediated decay (Fig. 4) without directly interacting with SMG5. In other words, it appears possible that PNRC2 is a decapping factor rather than a bona fide NMD factor. Remarkably, Drosophila melanogaster lacks both SMG7 (Gatfield et al. 2003) and PNRC2, raising the question of whether alternative SMG5 partners are present in this organism.
How does SMG5 trigger decay in tethering assays? The activity of SMG5 depends on UPF1 (Cho et al. 2013), which in turn interacts with decapping factors, suggesting that mRNA degradation is caused by UPF1. The dependence of SMG5 on UPF1 in tethering assays contrasts with the sequential recruitment of NMD factors to bona fide NMD targets in which UPF1 acts upstream of SMG5 and suggests that tethering assays bypass this sequential assembly and may not faithfully recapitulate NMD. Thus, although tethering assays are a powerful tool for studying the activity of proteins in isolation, they are bypassing important recruitment and assembly steps. Therefore, conclusions drawn from these assays require additional insight and validation in complementation assays.
Role of decapping in NMD
Decapping is normally coupled to deadenylation and may occur by default as a consequence of this coupling. However, the observation that UPF1 interacts with decapping factors suggests that these factors could be recruited to NMD targets independently of deadenylation. Indeed, UPF1 interacts with DCP1, DCP2, and PNRC2, and these interactions are enhanced by UPF1 phosphorylation (Fig. 2; Lykke-Andersen 2002; Lejeune et al. 2003; Fenger-Grøn et al. 2005; Isken et al. 2008; Cho et al. 2009; Lai et al. 2012). While PNRC2 was proposed to bridge the interactions between UPF1 and the decapping complex, in this study, we show that DCP2 interacts with UPF1 independently of PNRC2. Notably, UPF1 also interacts with DCP2 and the decapping factors Edc3 and Pat1 in yeast (He and Jacobson 2001; Swisher and Parker 2011) despite the fact that yeast lacks PNRC2.
How do the multiple and potentially redundant interactions of UPF1 with decapping factors contribute to the degradation of NMD targets? UPF1 may simply increase the local concentration of decapping factors on NMD targets, facilitating their degradation, and contribute to the accelerated decapping observed for bona fide NMD targets (Supplemental Fig. S6; Couttet and Grange 2004). Nevertheless, decapping factors could also be indirectly recruited to NMD targets by SMG7 through the CCR4–NOT complex.
An important challenge for future studies is the elucidation of the molecular basis for the interaction of UPF1 with decapping factors and its regulation by phosphorylation. One particularly intriguing observation is that both the N-terminal and C-terminal unstructured tails are required for PNRC2 and DCP2 binding (Fig. 2; Cho et al. 2009; Lai et al. 2012). One possible explanation is that in the context of full-length UPF1, these extensions are in close proximity and cooperate to form a common binding site. For example, the UPF1 C-terminal tail interacts with the helicase domain (Fiorini et al. 2013). However, the observation that deletion of the helicase domain is not detrimental for decapping factor binding (Fig. 2) suggests that a predefined three-dimensional conformation of these extensions is not required for binding. Alternatively, the N-terminal and C-terminal tails may provide low-affinity binding sites for decapping factors, in which case both will be required for full binding affinity because of additive/avidity effects.
In yeast, decapping plays a major role in the degradation of NMD targets, which are rapidly decapped without any requirement for prior deadenylation (Muhlrad and Parker 1994; Bond et al. 2001; He et al. 2003). A minor pathway involving accelerated deadenylation followed by exosome-mediated 3′-to-5′ decay is also observed, particularly when decapping is impaired (Cao and Parker 2003; Mitchell and Tollervey 2003; Takahashi et al. 2003). Thus, it appears that the basic mechanisms of NMD target degradation have been maintained throughout evolution. However, in metazoans, additional mechanisms for recruiting decay factors and degrading mRNA targets have been appended through the acquisition of SMG6, a specific NMD endonuclease, and the SMG5–SMG7 complex, which directly interacts with the CCR4–NOT complex (Supplemental Fig. S6). As a consequence, tighter regulation of mRNA target degradation is possible in these organisms. Indeed, SMG6 and the SMG5–SMG7 complex are only recruited to NMD targets upon UPF1 phosphorylation, enabling the sequential assembly of the surveillance complex and step-wise recruitment of decay factors. In this context, it is important to note that UPF1 also associates with mRNAs that are not destined for degradation by NMD (Hogg and Goff 2010; Hurt et al. 2013; Kurosaki and Maquat 2013; Zünd et al. 2013), and thus a tight regulation of UPF1 phosphorylation and the subsequent recruitment of decay factors are required to prevent unscheduled degradation. In contrast, in yeast, UPF1 is not phosphorylated and is thought to recruit decay factors to the target mRNA (He and Jacobson 2001; Swisher and Parker 2011). Thus, serendipitous UPF1 binding might lead to undesired degradation of normal mRNAs in yeast, which is unlikely to occur in metazoan.
In summary, together with previously published data (Muhlemann and Lykke-Andersen 2010), our results indicate that the surveillance complex disposes of multiple and redundant activities to ensure robust target degradation: It uses the endonucleolytic activity of SMG6, the deadenylase activity of the CCR4–NOT complex through direct interaction with SMG7, and the decapping activity of DCP2 through interactions with UPF1 and/or SMG7.
Materials and methods
DNA constructs
Plasmids for the expression of SMG5 and SMG7 and deadenylation factors have been described previously (Braun et al. 2011; Jonas et al. 2013). SMG7 ΔPC carries a deletion of amino acids 633–1091. For the TAP tag selection, SMG7 cDNA encoding the PC region (residues 633–1091) was inserted into the pCMV-TAPtagN-PrP-SBP vector using XhoI and Acc651 restriction sites. The TAP tag consists of protein A, a PreScission protease cleavage site, and the SBP. For in vitro translation, cDNAs encoding SMG7 fragments (PC and ΔPC) were cloned into XhoI–NotI sites of the pClneo-λNHA vector. For the expression of SMG5 and SMG7 with MS2-HA tags, the corresponding cDNAs were inserted into the XhoI–NotI sites of pCN-MS2 (Lykke-Andersen et al. 2000), which was modified by the insertion of an HA tag C-terminal to the MS2. SMG5 and SMG7 mutants were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene) and the appropriate oligonucleotide sequences. Mutants used in this study are described in Supplemental Table S2. To express PNRC2 with an N-terminal V5-SBP-MBP tag, the cDNA was inserted into the XhoI–NotI restriction sites of the pCIneo-V5-SBP-MBP vector. Plasmids for expression of recombinant proteins in E. coli are described in the Supplemental Material.
TAP tag purification
For the TAP tag purification, HEK293T cells were grown in 145-mm dishes and transfected with 30 μg of plasmid per dish using the calcium phosphate method. Cells were harvested 48 h post-transfection and lysed on ice in lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1% Triton-X-100, 1 mM EDTA, 0.5 mM DTT, 10% glycerol with 2.5 mL per 84 × 106 cells) supplemented with Complete protease inhibitor (Roche). Cell lysates were spun at 18,000g for 15 min at 4°C. Cleared lysates were treated with 50 μg/mL RNase A (Qiagen) for 30 min at 4°C and then spun at 18,000g for 10 min at 4°C. Supernatants were pooled together and then incubated with 60 μL of IgG sepharose beads (50% slurry; GE Healthcare) for 1 h at 4°C with gentle mixing followed by an overnight digestion with 1 μg/μL PreScission protease at 4°C. The IgG beads were removed by centrifugation, and the supernatant was incubated with 70 μL of streptavidin beads (50% slurry; GE Healthcare) for 2 h at 4°C. The beads were washed seven times with lysis buffer, and the proteins were eluted with 50 μL of NuPage LDS sample buffer (Invitrogen). Samples were analyzed by mass spectrometry.
Coimmunoprecipitation, pull-down assays, and Western blotting
For pull-down and coimmunoprecipitation assays, HEK293T cells were grown in 10-cm plates and transfected using the calcium phosphate method, except for the experiments shown in Figure 2A, which used Lipofectamine 2000 according to the manufacturer's protocol. Cells were transfected with 24 μg of total plasmid DNA. Two days after transfection, cells were lysed for 15 min on ice in NET lysis buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1% Triton-X-100, 10% glycerol, 2.5 mM MgCl2, 1 mM DTT) supplemented with protease inhibitor (Roche) and 200 μg/mL RNase A (Qiagen). Cell lysates were spun at 16,000g for 15 min at 4°C. For SBP-tagged proteins, the cleared lysate was rotated for 30 min at 4°C with 25 μL of streptavidin sepharose (GE Healthcare). The beads were washed three times with NET buffer. Bound proteins were eluted with 100 μL of protein sample buffer and analyzed by Western blotting. For the PNRC2 and UPF1 pull-downs, cells were incubated with 50 nM okadaic acid for 4 h before harvest, and NET buffer supplemented with PhosSTOP phosphatase inhibitor (Roche) was used for lysis and washing. Coimmunoprecipitations using anti-GFP antibodies and antibodies used in this study are described in the Supplemental Material and Supplemental Table S3, respectively. All Western blots were developed with the ECL Western blotting detection system (GE Healthcare) as recommended by the manufacturer.
Protein expression, purification, and pull-down assays
BL21 star cells (Invitrogen) harboring plasmids encoding GST, GST-POP2, or MBP-SMG7 732–1091 were grown at 37°C in LB medium until reaching OD600 = 0.4. Protein expression was induced with IPTG, and all proteins were expressed overnight at 20°C. After harvest, cells were resuspended in lysis buffer (50 mM HEPES at pH 7.5, 200 mM NaCl, 1 mM DTT) supplemented with EDTA-free protease inhibitor (Roche), 1 mg/mL lysozyme, and 5 μg/mL DNase I and then lysed by sonication. The cleared lysates were incubated for 1 h with 5 mL of pre-equilibrated amylose resin (New England Biolabs) or glutathione agarose beads (Macherey-Nagel). The beads were washed with lysis buffer, and the proteins were eluted after a 15-min incubation with lysis buffer containing 25 mM maltose or L-glutathione. The proteins were subsequently purified over a gel filtration column (Superdex 200, GE Healthcare) and stored at −80°C after flash-freezing in liquid nitrogen. Pull-down assays were performed as described in the Supplemental Material.
Tethering and complementation assays
For tethering assays, HEK293T cells were cultured in six-well plates and transiently transfected with a mixture of three plasmids: 0.5 μg of the control plasmid (β-globin-GAP), 0.5 μg of the plasmid encoding the β-globin-6xMS2bs, and various amounts of the pCN-MS2-HA plasmid for the expression of MS2-HA fusion proteins. The amount of total DNA was adjusted to 3 μg with the pcDNA3 plasmid. When indicated, the transfection mixtures also contained plasmids encoding GFP-DCP2 (wild type or mutant), HA-MBP, or catalytically inactive HA-POP2 mutant. Complementation assays were performed as previously described (Jonas et al. 2013). Transfection mixtures contained 0.4 μg of plasmid pSUPERpuro expressing shRNAs. Total RNA was isolated using TriFast (Peqlab) and analyzed as described previously (Huntzinger et al. 2008). β-Tubulin was used as normalization control.
Acknowledgments
We are grateful to Dr. Oliver Mühlemann for the kind gift of HeLa cell lines expressing the β-globin NMD reporters and plasmids for the expression of MS2-SMG5, Dr. Jens Lykke-Andersen for providing the MS2 reporters, and Dr. Shigeo Ohno for antibodies to SMG6. We thank Dr. Boris Macek at the Proteome Center of the University of Tübingen for performing the mass spectrometry analysis, and Catrin Weiler for technical assistance. This study was supported by the Max Planck Society and grants from the Deutsche Forschungsgemeinschaft (DFG; FOR855 and the Gottfried Wilhelm Leibniz Program awarded to E.I.) and the European Union Seventh Framework Programme through a Marie Curie Fellowship to S.J. (FP7, n°275343). S.J. is the recipient of an EMBO long-term fellowship.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226951.113.
Freely available online through the Genes & Development Open Access option.
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Anders KR, Grimson A, Anderson P 2003. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J 22: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AT, Mangus DA, He F, Jacobson A 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol 21: 7366–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 23: 4805–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H 2007. Structural and functional insights into the human Upf1 helicase core. EMBO J 26: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE 2003. Characterization of human Smg5/7a, a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK 2013. SMG5–PNRC2 is functionally dominant compared with SMG5–SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res 41: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S 2009. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Grange T 2004. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Mauxion F, Seraphin B 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Fiorini F, Boudvillain M, Le Hir H 2013. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res 41: 2404–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 18: 537–547 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E 2003. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E 2006. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol 21: 1515–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- Hogg JR, Goff SP 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoa. RNA 14: 2609–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Weichenrieder O, Izaurralde E 2013. An unusual arrangement of two 14-3-3-like domains in the SMG5–SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 27: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A 2012. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE 2013. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci 110: 3357–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Cho H, Liu Z, Bowler MW, Piao S, Parker R, Kim YK, Song H 2012. Structural basis of the PNRC2-mediated link between mRNA surveillance and decapping. Structure 20: 2025–2037 [DOI] [PubMed] [Google Scholar]
- Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT 2009. Human Ccr4–Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687 [DOI] [PubMed] [Google Scholar]
- Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J 2007. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res 35: 7688–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Metze S, Herzog VA, Ruepp M-D, Mühlemann O 2013. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 19: 1432–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′ degradation. Mol Cell 11: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Muhlemann O, Lykke-Andersen J 2010. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 7: 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581 [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE 1998. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199 [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L 2008. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher KD, Parker R 2011. Interactions between Upf1 and the decapping factors Edc3 and Pat1 in Saccharomyces cerevisiae. PLoS ONE 6: e26547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Araki Y, Sakuno T, Katada T 2003. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J 22: 3951–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Yamashita A 2013. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 18: 161–175 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Zünd D, Gruber AR, Zavolan M, Mühlemann O 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943 [DOI] [PubMed] [Google Scholar]